Abstract
With over one million copies, Alu elements are the most abundant repetitive elements in the human genome. When transcribed, interaction between two Alus that are in opposite orientation gives rise to double-stranded RNA (dsRNA). Although the presence of dsRNA in the cell was previously thought to only occur during viral infection, it is now known that cells express many endogenous small dsRNAs, such as short interfering RNA (siRNAs) and microRNA (miRNAs), which regulate gene expression. It is possible that long dsRNA structures formed from Alu elements influence gene expression. Here, we report that human mRNAs containing inverted Alu elements are present in the mammalian cytoplasm. The presence of these long intramolecular dsRNA structures within 3′-UTRs decreases translational efficiency, and although the structures undergo extensive editing in vivo, the effects on translation are independent of the presence of inosine. As inverted Alus are predicted to reside in >5% of human protein-coding genes, these intramolecular dsRNA structures are important regulators of gene expression.
INTRODUCTION
It has been known for >40 years that multiple copies of DNA sequences termed repetitive elements are abundant in eukaryotic genomes (1). The bulk of human repetitive elements are derived from transposable elements (2,3). These elements have commonly been referred to as ‘junk’ or ‘selfish’ DNA, useful only for determining phylogenetic relationships between organisms (4,5). However, since the time of their discovery, the fact that repetitive sequences are interspersed with coding sequences in the genome, has lead to speculation that these elements may play a regulatory function in gene expression (6,7). In fact, multiple recent studies have identified functional roles of repetitive elements of the Alu family in regulating gene expression (8–10).
Alu elements are ∼300 nt sequences that are expressed either as autonomous RNAs or embedded within an mRNA (11). Autonomous Alu RNAs are expressed at low levels in normal cells, but increase in expression in response to numerous stress conditions, such as heat shock and cycloheximide treatment (8,12). During heat shock Alu RNAs bind to RNA polymerase II, preventing binding to promoters and thus repressing transcription (13). In addition, Alu RNAs act as translational activators, as overexpression stimulates translation of co-expressed mRNAs both in vitro and in vivo (14,15). In contrast to the low levels of ‘free’ Alus, embedded Alu elements are present in a large number of cellular mRNAs, with enrichment in UTRs (16,17). As UTRs are molecular ‘hotspots’ for elements that regulate gene expression, including mRNA stability, localization and translation, embedded Alus are likely to affect many of these processes (18–20). In fact, embedded Alus are known to be important mediators of alternative splicing (21). Inclusion of alternatively spliced Alu elements in 5′-UTRs results in upstream open reading frames (uORFs) that decrease translational efficiency (22). Thus, Alu elements are important regulators of gene expression both as autonomous RNAs and when present in UTRs.
In addition to their individual function, as Alu elements are very similar in sequence and randomly orientated within the human genome, interaction between two Alu elements that are in opposite orientation gives rise to double-stranded RNA (dsRNA) (23). Recently, intermolecular base-pairing of Alu elements present in the 3′-UTR of cytoplasmic human mRNAs and oppositely orientated Alus in long non-coding RNAs have been shown to target the mRNAs for Staufen-mediated degradation (24). While these intermolecular duplexes are predicted to affect ∼0.2% of human protein-coding genes, intramolecular dsRNA structures are far more common in mammalian cells (23). An abundance of these structures in the human transcriptome was uncovered by bioinformatic searches for mRNAs that undergo adenosine to inosine RNA editing (25–28). Adenosine deaminases that act on RNA (ADARs) are a family of enzymes that catalyze the hydrolytic deamination of adenosine to inosine in dsRNA (29,30). The presence of neighboring Alu elements in inverted orientation gives rise to an intramolecular dsRNA structure that is an ideal ADAR substrate (25). As editing sites are enriched in 3′-UTRs, these inverted Alu structures, and the inosines within them, are poised to regulate gene expression (31). However, the presence of long synthetic and viral dsRNA molecules in the mammalian cytoplasm are well known to activate an immune response (32,33). Therefore, to avoid detrimental effects on the cell, it has been proposed that endogenous mRNAs with these extensive dsRNA structures are sequestered from the cytoplasm (23,34,35). Consistent with this, extensive editing within inverted repeats of mouse CTN-RNA is reported to promote retention in nuclear paraspeckles (36). Furthermore, in human cell lines, reporter mRNAs with edited, inverted Alu 3′-UTRs were detected in paraspeckles (37). However, our previous studies demonstrated that endogenous Caenorhabditis elegans and human mRNAs with edited 3′-UTRs are present on cytoplasmic ribosomes (38). Thus, it is unclear if nuclear retention is indeed a general mechanism for modulating expression of genes bearing hyper-edited 3′-UTRs and/or required to prevent improper activation of the immune response (30,39).
Here, we report that endogenous human mRNAs with inverted Alus are exported to the cytoplasm to an equivalent level as mRNAs lacking these structures. In addition, we demonstrate that the presence of these intramolecular dsRNA structures within 3′-UTRs affects translation. Furthermore, although the structures undergo extensive editing, effects on translation are independent of the presence of inosine.
MATERIALS AND METHODS
Cells and viral infection
HeLa and HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 µg/ml penicillin and 100 units/ml streptomycin (MediaTech). For the ADAR1 knockdown, human ADAR1 sequences (Supplementary Table S5) were cloned into pLKO.1, a kind gift from Bob Weinberg (Addgene No 8453). The scrambled short hairpin RNA (shRNA) construct was a kind gift from Sheila Stewart (Addgene No 17920). shRNA plasmids were co-transfected with GAG/POL, REV and G -VSV plasmids (Addgene No 12251, 12253 and 12259, kind gifts of Didier Trono) into HEK-293 T cells using Trans-IT LT1 (Mirus) to generate lentiviruses. The lentivirus containing media and 8 µg/ml polybrene (Sigma) were incubated with HeLa cells. After 2 h incubation, 8 ml DMEM media with 10% FBS was added and cells were incubated for 24 h at 37°C. Fresh media was added and cells were allowed to grow without selection for 24 h. To select for infected cells, puromycin (7.5 µg/ml) containing media was used over the course of several days. After selection, HeLa cells with the stable knockdown ADAR1 (ADAR1kd) or the scrambled shRNA (HeLaSCR) were maintained in DMEM media with 10% FBS and 5 µg/ml puromycin.
Luciferase reporters and luciferase assay
PSMB2 (1478 nt), BPNT1 (1362 nt) and MPST (4712 nt) 3′-UTRs were amplified from HeLa genomic DNA. The MPST 3′-UTR and its controls were cloned downstream of Renilla luciferase in the psi-CHECK-1 vector (Invitrogen). The PSMB2 and BPNT1 3′-UTRs were cloned downstream of Firefly luciferase (from the pGL4.17 vector) in the pcDNA3 vector (Invitrogen). For all control Alu plasmids, the reverse complement of the first or second Alu for each 3′-UTR was polymerase chain reaction (PCR) amplified and inserted in place of the wild-type Alu sequence. Reporters were transfected using FuGENE HD transfection reagent, and cells were assayed using the Dual Luciferase Assay System (Promega).
Western analysis
Cells were resuspended in 4% sodium dodecyl sulfate (SDS), 100 mM Tris–HCl pH 6.8, 20% glycerol, sonicated. Protein concentrations were determined by Bradford (Sigma), and equivalent amounts of lysate were subjected to SDS–PAGE and western blotting with antibodies to Human ADAR1 (kind gift of Brenda Bass) and tubulin (Sigma).
RNA isolation, northern analysis, qRT-PCR and editing assays
RNA was isolated from cell pellets using Trizol (Invitrogen) or TRIreagent (Sigma). RNA was further purified by treating with TURBO DNase (Ambion) followed by RNeasy chromatography (Qiagen). Northern analyses used standard protocols for 1.5% formaldehyde agarose gel electrophoresis and nylon membrane blotting. Northern probes for GAPDH, PSMB2, firefly and Renilla luciferase were synthesized using in vitro transcription with radioactive ATP and a PCR template. Probes for tRNAlys and U3 small nuclear RNA (U3 snRNA) were created by addition of a 5′ radioactive phosphate to DNA oligos (Supplementary Table S5) using T4 polynucleotide kinase (New England Biolabs).
To synthesize complementary DNA (cDNA) for qRT-PCR and editing assays, DNase treated total RNA were reverse transcribed with SuperScript II reverse transcriptase (Invitrogen) and gene-specific primers (Supplementary Table S5). Samples were treated with RNaseH (New England Bioloabs). cDNA levels were measured in an Eppendorf Realplex instrument using KAPA SYBR Fast Universal Master Mix. qRT-PCR primers spanned at least one exon boundary and produced products of ∼150 bp. Quality of the qRT-PCR products was assessed using melting curve analysis and gel electrophoresis. For editing assays, cDNA, minus reverse transcriptase controls and genomic DNA were amplified by PCR with gene specific primers and sequenced using Ambion Big Dye reagents (Indiana Molecular Biology Institute, Indiana University).
RESULTS
mRNAs with edited double-stranded structures are present in the mammalian cytoplasm
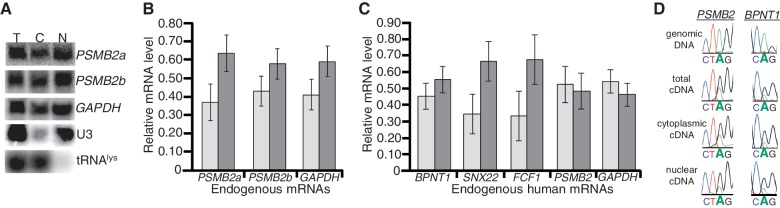
The extensive dsRNA formed from inverted Alu elements, and their editing state, has been proposed to alter mRNA export from the nucleus (36,37). However, we previously reported that two human mRNAs (PSMB2 and NDUFC2) and multiple C. elegans mRNAs with edited 3′-UTRs were present on cytoplasmic ribosomes (38). Furthermore, we have observed three additional endogenous human mRNAs with inverted Alu-containing 3′-UTRs, SNX22, FCF1 and BPNT1, on ribosomes (Supplementary Figure S1). These data suggest that nuclear retention is not a general mechanism used to regulate endogenous hyper-edited mRNAs. However, we considered the possibility that most edited mRNAs are retained, and the ribosome-associated mRNAs represented a small portion that escaped the nucleus. To address this possibility, HeLa cells were subjected to subcellular fractionation and the quantity of endogenous mRNA localized to the nucleus and cytoplasm was measured by northern blot or quantitative RT-PCR (qRT-PCR). Nuclear and cytoplasmic fractions were prepared from the same cells and identical cell equivalents were analyzed. The vast majority of U3 snRNA and tRNAlys were in nuclear and cytoplasmic fractions, respectively, consistent with clean fractionation (Figure 1A). PSMB2, an mRNA with a double-stranded 3′-UTR and the housekeeping mRNA, GAPDH, which does not contain Alu elements, had similar localization with ∼60% of total mRNA present in the cytoplasm (Figure 1A and B). In addition, two isoforms of endogenous PSMB2, one containing (PSMB2a) and one lacking (PSMB2b) 3′-UTR inverted Alu elements, localize to the cytoplasm to a similar extent (Figure 1A). Furthermore, qRT-PCR indicated that the majority of endogenous SNX22, FCF1 and BPNT1 mRNAs, which contain inverted Alu 3′-UTR structures, were cytoplasmic (Figure 1C).
Figure 1.
Cytoplasmic localization of endogenous mammalian mRNAs with inverted Alu 3′-UTR structures. (A) Northern blots were performed on total (T), cytoplasmic (C) and nuclear (N) HeLa RNA and hybridized with probes specific to the indicated RNAs. (B and C) Quantification of endogenous mRNA levels present in nuclear (light gray) and cytoplasmic (dark gray) fractions as determined by northern blot (B) or qRT-PCR (C) for three independent fractionations. Error bars represent SEM. (D) Total, cytoplasmic and nuclear mRNA from fractionation experiments shown in (A) was reversed transcribed, PCR amplified and sequenced. In addition, HeLa genomic DNA was PCR amplified and sequenced to serve as a negative control. One editing event for the PSMB2 and BPNT1 3′-UTRs is shown, but editing was measured across the 3′-UTR (Supplementary Tables S1 and S2).
The similar nuclear/cytoplasmic distribution of mRNAs with and without dsRNA structures suggested the presence of inosine in these regions did not affect export. However, it was possible that the cytoplasmic mRNA represents a hypo-edited fraction that escaped nuclear retention. To test this, cDNA was prepared from nuclear and cytoplasmic RNA. As reverse transcriptase reads inosine as guanosine, editing sites within an mRNA appear as adenosine to guanosine changes in cDNA. Representative sequence traces for PSMB2 and BPNT1 from total nuclear and cytoplasmic cDNA illustrate that editing of individual sites occurs to a similar extent in both fractions (see G peaks within A peaks, Figure 1D; Supplementary Tables S1 and S2). These data indicate that endogenous human mRNAs with inosine-containing inverted Alus are localized to the cytoplasm similar to mRNAs lacking these structures and inosines. Therefore, we conclude that while a small subset of edited mRNAs may be enriched in mammalian nuclei, in general, mRNA export is not inhibited by double-stranded structures or inosines.
Inverted Alu structures affect gene expression independent of RNA editing
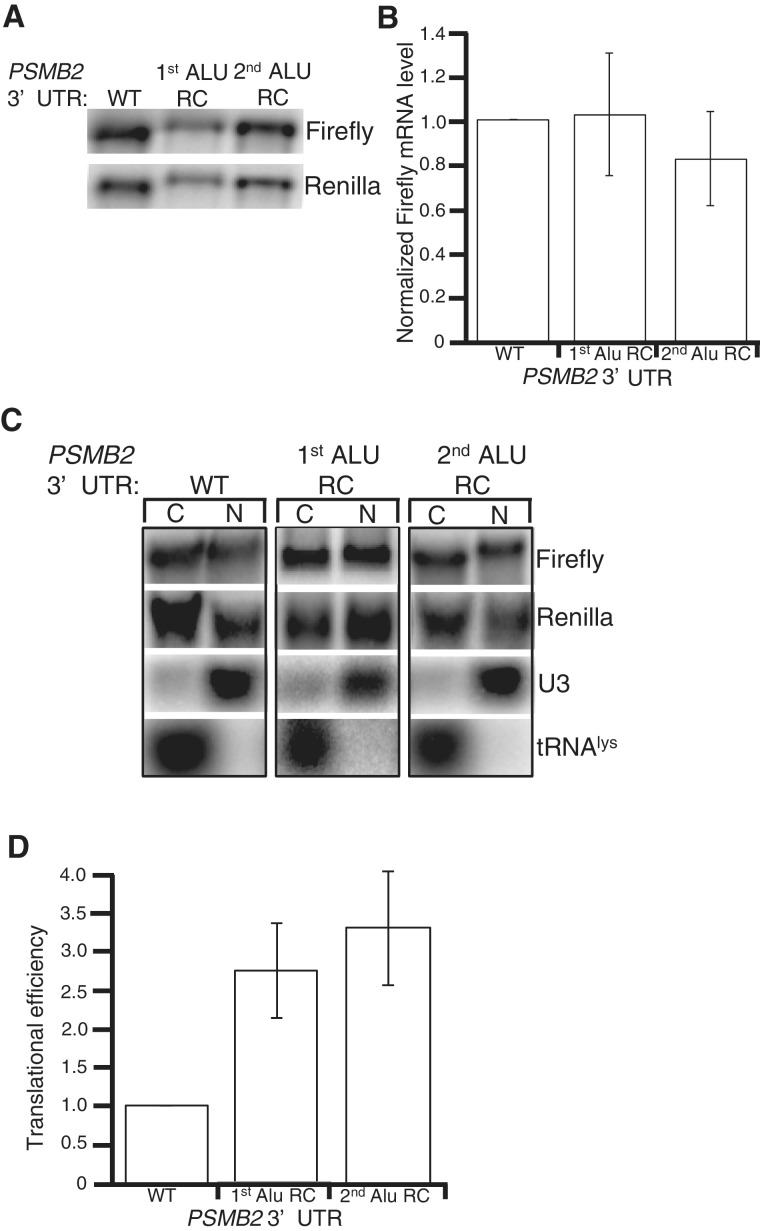
As 3′-UTR elements often direct post-transcriptional gene expression, and inverted Alus are abundant in the 3′-UTRs of human mRNAs, we tested whether these dsRNA structures regulate gene expression in human cell lines. As the PSMB2 locus produces isoforms with identical coding potential, but differing 3′-UTRs (Figure 1A), it is difficult to assess effects of the inverted Alus on endogenous PSMB2 expression. Therefore, the sequence of the entire PSMB2 3′-UTR present in HeLa cells was determined by rapid amplification of cDNA ends (RACE) (Supplementary Figure S2) and cloned immediately downstream of a firefly luciferase reporter (Figure 2A). When transfected into HeLa cells, the reporter was edited similar to endogenous PSMB2 (data not shown), suggesting the reporter 3′-UTR adopts the native dsRNA structure. To monitor effects of the PSMB2 3′-UTR on gene expression, we compared it to reporters with 3′-UTRs of the same length but containing the reverse complement (RC) sequence of either the first or second Alu. The controls do not form dsRNA as the two Alu elements are in the same orientation (Figure 2A) and consistent with this, we did not detect editing (data not shown). A Renilla luciferase plasmid was co-transfected to normalize for extract concentrations and transfection efficiency. HeLa cells transfected with the wild-type PSMB2 3′-UTR reporter had ∼2.5-fold less firefly luciferase activity compared to cells transfected with the RC controls (Figure 2B). These data raised the possibility that human double-stranded 3′-UTRs and/or the inosines within them inhibit gene expression in cis.
Figure 2.
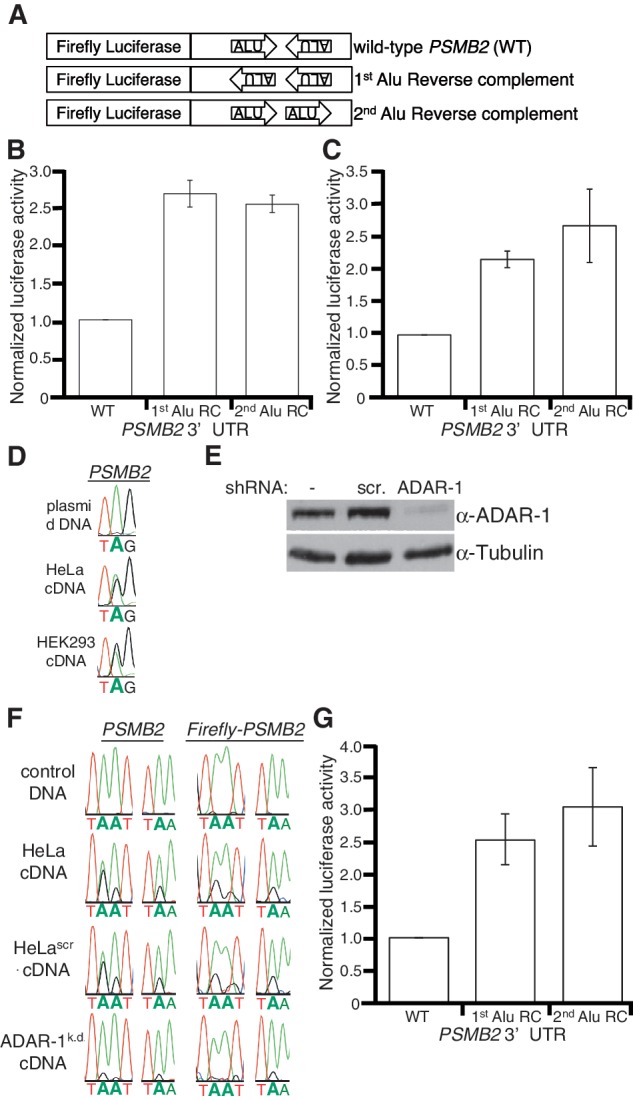
Inverted Alus affect gene expression independent of RNA editing. (A) Schematic of PSMB2 3′-UTR reporters. The wild-type (WT) PSMB2 3′-UTR was cloned immediately downstream of firefly luciferase. Arrows indicate location and orientation of Alu sequences. Control firefly reporters have individual Alus replaced with their corresponding reverse complement (RC) sequence. Thus, the Alus in the controls are in the same orientation. (B, C and G) Bar height represents luciferase activity of the indicated firefly PSMB2 3′-UTR reporters relative to a co-transfected Renilla luciferase plasmid, normalized to WT. Error bars show SEM for at least three independent biological replicates. (B) In HeLa cells, the mean of ten independent biological replicates indicates significant differences in luciferase activity between WT and 1st (P < 0.0001) and 2nd (P < 0.0001) Alu RC controls. (C) In HEK293 cells, significant differences between WT and 1st (P = 0.0009) and 2nd (P = 0.04) Alu RC controls were observed. (D) Reporter plasmid DNA and reporter cDNA was amplified and sequenced. One editing event is shown, but editing was observed at multiple sites. (E) HeLa cells stably transfected with no (−), scrambled (scr.) or an ADAR1 shRNA were subjected to SDS–PAGE and western blotting for ADAR1 and Tubulin. (F) Control DNA and cDNAs from endogenous and reporter PSMB2 were amplified from the indicated cells. Three editing events are shown, but editing was measured across the 3′-UTR (Supplementary Tables S3 and S4). (G) In ADAR1k.d. cells, significant differences in luciferase activity of WT PSMB2 3′-UTR and 1st (P = 0.014) and 2nd (P = 0.019) Alu RC controls were observed.
To test whether editing of the 3′UTR or the dsRNA structure per se was required for the effects on gene expression, we sought to analyze reporters in HEK293 cells, which were previously reported to exhibit very little editing (40). Similar to HeLa cells, transfection of the reporters into HEK293 cells resulted in ∼2-fold lower luciferase activity for the PSMB2 3′UTR compared to the controls (Figure 2C). Surprisingly, the reporter bearing the PSMB2 3′-UTR was edited to a similar extent in HEK293 and HeLa cells (Figure 2D). This was not unique to the reporter as we observed extensive editing within the 3′-UTRs of multiple endogenous mRNAs (PSMB2, BPNT1 and NDUFC2) in HEK293 cells (data not shown). Thus, in contrast to previous studies that characterized codon editing, our data indicate that editing of non-coding regions in HEK293 cells is similar to other cell lines and suggests that there may be cellular factors important for regulation of these two types of ADAR editing. Importantly, the similar gene regulation by the PSMB2 dsRNA structure in multiple cell lines indicates a conserved function.
To test whether editing was required for the regulatory function of the PSMB2 3′-UTR, a HeLa cell line in which ADAR1 expression is stably knocked down by RNA interference was created. The ADAR1 knockdown cells have <10% of ADAR1 levels of the parental HeLa cells or the scrambled shRNA control HeLa cell line (Figure 2E). Compared to both control cells, overall editing of both endogenous PSMB2 and the firefly reporter expressing the PSMB2 3′-UTR was decreased in the ADAR1k.d. cells (Figure 2F; Supplementary Tables 3 and 4). Although editing in the ADAR1k.d. cells was decreased, the reporter bearing the double-stranded PSMB2 3′-UTR had a similar translational output as in wild-type cells (Figure 2G). Therefore, the extent of editing is not critical for repression of gene expression by intramolecular Alu dsRNA.
Inverted Alu elements affect mRNA translational efficiency
Our data suggest that intramolecular Alu base-pairing does not affect nuclear export of endogenous mRNAs, but decreases reporter protein expression. As intermolecular Alu base-pairing was recently shown to target some mRNAs for degradation (24), we first tested whether the intramolecular duplex affected luciferase activity by altering mRNA stability. Northern blots were performed on RNA isolated from HeLa cells transfected with the control Renilla luciferase plasmid and firefly PSMB2 reporters (Figure 3A). The firefly luciferase activity of the reporter bearing the double-stranded PSMB2 3′-UTR is nearly 3-fold lower than RC controls in HeLa cells (Figure 2B), but the mRNA levels are similar (Figure 3A). In fact, quantitation of Northern blots from five independent transfections did not reveal a significant difference in mRNA levels of the PSMB2 reporters (Figure 3B). Consistent with this, we did not observe differences in mRNA levels of the PSMB2 reporters in either HEK293 or ADAR-1 k.d. cells (Supplementary Figure S3). Furthermore, similar to endogenous mRNAs, all of the reporter mRNAs were efficiently exported to the cytoplasm (Figure 3C). Therefore, the decrease in protein expression without a concomitant change in either mRNA stability or localization, indicates that the PSMB2 intramolecular dsRNA structure decreases translation efficiency (Figure 3D).
Figure 3.
Double-stranded 3′-UTRs do not affect mRNA export or stability. (A) Northern blots of total RNA from HeLa cells transfected with the indicated firefly PSMB2 3′-UTR reporters and a control Renilla luciferase reporter, hybridized with indicated probes. (B) Quantification of five independent biological replicates of experiment in (A). Height of the bar represents levels of firefly mRNA relative to Renilla mRNA, normalized to WT. (C) Northern blots of equal cell equivalents of cytoplasmic (C) and nuclear (N) RNA from HeLa cells transfected with the indicated firefly PSMB2 reporters and Renilla luciferase, hybridized with specific probes. (D) Translational efficiency of a firefly luciferase reporter with the indicated PSMB2 3′-UTR in HeLa cells. Each bar represents the mean and SEM of five biologically independent experiments. In each experiment, protein and RNA were isolated at the same time and levels measured by luciferase assay and northern blot, respectively. Efficiency is reported as the ratio of the normalized luciferase activity to the normalized mRNA level and is reported relative to the efficiency of the WT PSMB2 3′-UTR. Both luciferase activity and RNA levels are normalized to the co-transfected Renilla control reporter. Significant differences in translational efficiency between the PSMB2 3′-UTR (WT) and 1st (P = 0.02 and 2nd (P = 0.01) Alu RC controls were observed.
Effects of dsRNA structures on translation depend upon location within 3′-UTR
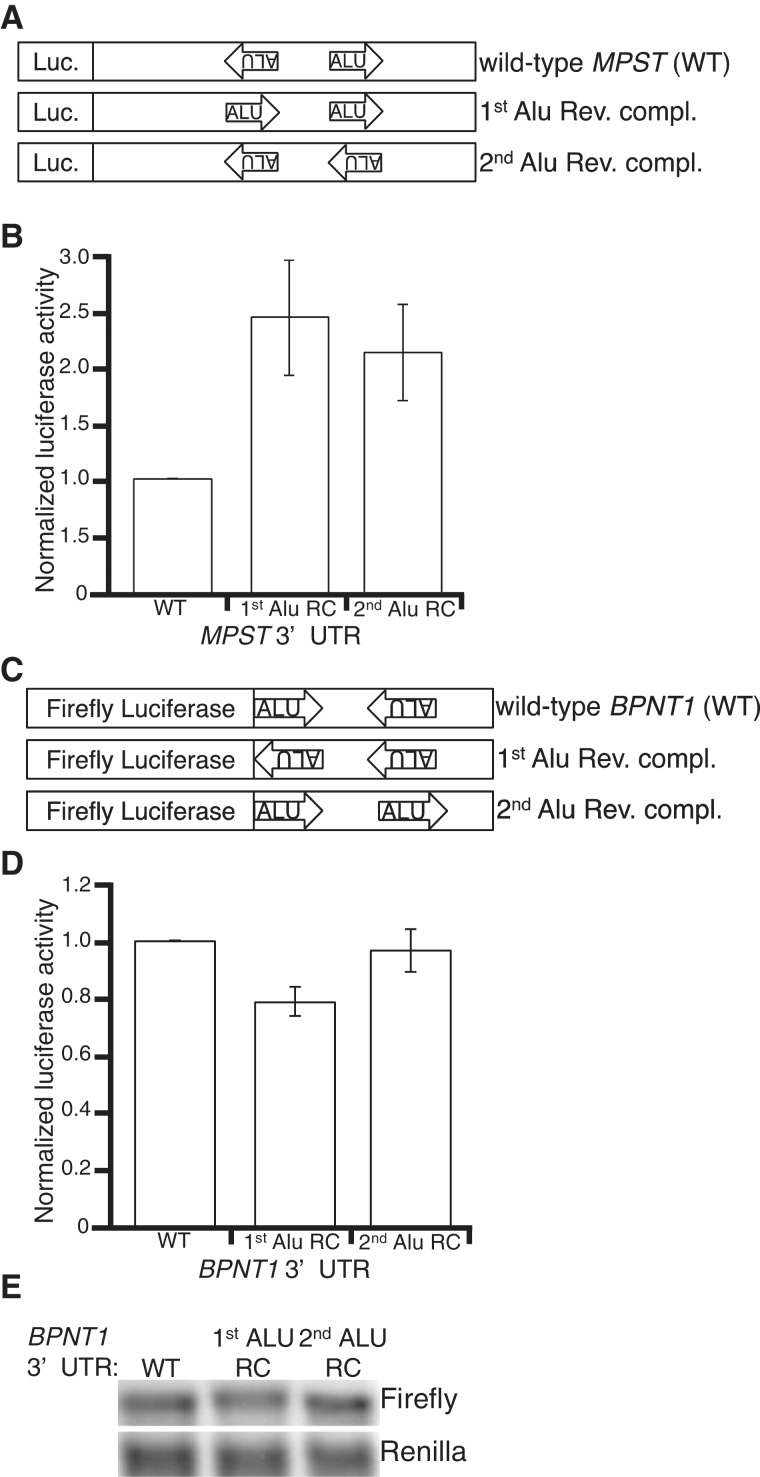
As inverted Alu elements are present in the 3′-UTRs of a large number of human protein-coding genes, we sought to determine whether the effects of the PSMB2 3′-UTR on translational efficiency extend to other inverted Alu elements. First, reporter constructs similar to those described above were created for the human genes MPST (Figure 4A and Supplementary Figure S4) and BPNT1 (Figure 4C and Supplementary Figure S5). Upon transfection into HeLa cells, the reporter bearing the MPST double-stranded 3′-UTR resulted in ∼2-fold lower luciferase activity compared to its control reporters (Figure 4B). In contrast, the reporter bearing the BPNT1 double-stranded 3′-UTR resulted in similar luciferase activity as Alu RC controls in both HeLa and HEK293 cells (Figure 4D and Supplementary Figure S6). It is possible that the BPNT1 3′-UTR affects translation, but these effects are masked by unequal expression of the wild-type reporter compared to its controls. However, northern blot analyses of the reporters in both cell lines suggest that the wild-type BPNT1 reporter is expressed similar to the Alu RC controls (Figure 4E and Supplementary Figure S6). In addition, we confirmed that the lack of regulatory function was not due to an absence of the dsRNA structure, as a similar editing pattern was observed for endogenous and reporter BPNT1 (data not shown).
Figure 4.
Some, but not all intramolecular Alu dsRNA structures affect gene expression. (A) Schematic of MPST 3′-UTR reporters. (B) Bar height represents luciferase activity of the indicated Renilla reporters relative to a co-transfected firefly luciferase, normalized to WT. Significant differences in luciferase activity between the MPST 3′-UTR (WT) and 1st (P = 0.04) and 2nd (P = 0.04) Alu RC controls were observed. (C) Schematic of BPNT1 3′-UTR reporters. (D) HeLa cells transfected with BPNT1 reporters described in (C). Bar height represents luciferase activity of the indicated firefly reporters relative to co-transfected Renilla luciferase, normalized to WT. (E) Northern blots of total RNA from the same HeLa cells assayed for luciferase activity in (D), hybridized with indicated probes.
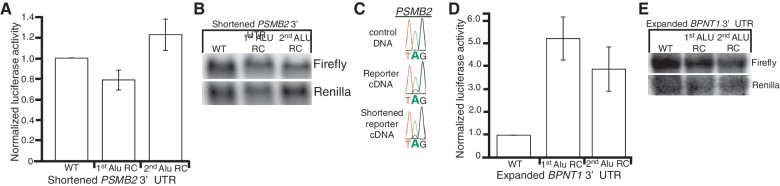
To understand the regulatory function of the inverted Alus in the PSMB2 and MPST 3′-UTRs compared with the lack of function of those in the BPNT1 3′-UTR, we took a closer examination of the Alu elements. One striking difference between the 3′-UTRs was the location of the inverted Alus; the PSMB2 and MPST structures located significantly downstream of the stop codon (472 nt and 1842 nt, respectively), whereas the BPNT1 structure is only 15 nt from the stop. The reporters were created similar to the endogenous context with the PSMB2, MPST and BPNT1 3′-UTRs located 481, 1855 and 21 nt downstream of the luciferase stop codon, respectively. To test whether this difference was critical for the effects on translation, we prepared reporters lacking the first 415 nt of the PSMB2 3′-UTR, which places the first Alu of the dsRNA or the controls, 65 nt from the stop. This shortened PSMB2 3′-UTR eliminated the effect of the inverted Alus on luciferase expression (Figure 5A). Importantly, as we observe similar mRNA levels of the shortened PSMB2 reporter bearing the inverted Alus compared with the shortened Alu RC controls (Figure 5B), the effects on luciferase activity are not due to an increase in reporter mRNA levels. In addition, the loss of translational control was not due to altered base-pairing, as the shortened PSMB2 reporter was edited similar to wild-type PSMB2 (Figure 5C). Finding that shortening the PSMB2 3′-UTR alters translational regulation, but not editing, further indicates that editing is neither sufficient, nor required, for this mechanism of gene regulation.
Figure 5.
Distance of inverted Alus from stop codon affects translation efficiency. (A, B) HeLa cells were transfected with firefly reporters bearing shortened PSMB2 3′-UTRs. Protein and RNA were isolated from the same cells and measured by luciferase assay (A) and Northern blot (B), respectively. (A) Bar height represents luciferase activity of the indicated firefly reporters relative to co-transfected Renilla luciferase, normalized to WT. Error bars show SEM for three independent biological replicates. (B) Northern blots of total RNA from HeLa cells transfected with the indicated shortened PSMB2 3′-UTR reporters and Renilla luciferase, hybridized with indicated probes. (C) Plasmid DNA and the indicated reporter cDNAs were amplified and sequenced. One editing event is shown, but editing was observed at multiple sites. (D and E) HeLa cells were transfected with firefly reporters bearing expanded BPNT1 3′-UTRs. Protein and RNA were isolated from the same cells and measured by luciferase assay (D) and Northern blot (E), respectively. (D) Bar height represents luciferase activity of the indicated firefly reporters relative to co-transfected Renilla luciferase, normalized to WT. Error bars show SEM for five independent biological replicates. In HeLa cells, significant differences in luciferase activity between the expanded BPNT1 3′-UTR (WT) and expanded 1st (P = 0.002) and 2nd (P = 0.0018) Alu RC controls were observed. (E) Northern blots of total RNA from HeLa cells transfected with the indicated expanded BPNT1 3′-UTR reporters and Renilla luciferase, hybridized with indicated probes.
Next, we tested whether moving the BPNT1 structure away from the stop codon would enable the inverted Alus to regulate translation. To avoid introducing unknown sequence elements, the endogenous PSMB2 sequence upstream of the first Alu was placed upstream of the BPNT1 3′-UTR and RC control reporters. The expanded BPNT1 3′-UTR resulted in 3- to 5-fold less luciferase activity (Figure 5D), but similar mRNA levels as the controls (Figure 5E). Thus, the ability of inverted Alus to regulate translation is not dependent upon sequence, but likely depends on the distance of the duplex from the stop codon. From these experiments, we cannot exclude the possibility that both inverted Alus and the first 415 nt of the PSMB2 3′-UTR are required for translational control. However, as the inverted Alus in the MPST 3′-UTR also decrease luciferase activity and are located significantly downstream of the stop codon, we favor the hypothesis that location of the intramolecular dsRNA structures within the 3′-UTR are likely important for translational control.
DISCUSSION
Our study of the fate of endogenous human mRNAs with inosine-containing 3′-UTRs support three main conclusions that advance our understanding of the in vivo effects of long dsRNA molecules. First, a surprising number of mRNAs containing long intramolecular duplexes are present in the mammalian cytoplasm. Second, neither editing nor the presence of the dsRNA structure affected mRNA export or stability. Third, these 3′-UTR intramolecular duplexes, but not the inosines within them, decrease mRNA translation in human cell lines.
Mammalian cells tolerate long dsRNA molecules in endogenous mRNAs
It has been known for >50 years that the presence of foreign dsRNA in the mammalian cytoplasm induces expression of interferon and activates the immune response (33,41). However, it is now known that cells produce a large amount of endogenous dsRNA. The common view is that short endogenous dsRNA molecules, such as siRNAs and miRNAs, are tolerated in the mammalian cytoplasm, whereas long endogenous dsRNA molecules undergo editing by ADARs, are retained in the nucleus to avoid activating an improper immune response (23,42). However, we observed a similar cytoplasmic localization of endogenous human mRNAs with and without dsRNA structures suggesting that the presence of inosine in these regions does not affect nuclear export. Consistent with this, the editing levels of both the cytoplasmic and nuclear localized mRNAs were similar, indicating that the extent of editing is not critical for nuclear export. Thus, nuclear retention is not a general mechanism used to regulate hyper-edited mRNAs. Furthermore, a large number of cellular mRNAs expressing these extensive dsRNA structures are present in the mammalian cytoplasm, and overexpression of these structures in reporter mRNAs did not result any detrimental effects on cell growth. Consistent with this, a recent study determined that overexpression of a reporter bearing a long hairpin 3′-UTR structure in mice did not cause any developmental defects or activate the interferon response (43). These data indicate that long dsRNA structures within human mRNAs are well tolerated in the mammalian cytoplasm. The characteristics of these dsRNA structures that allow the immune system to distinguish these ‘self’ molecules from foreign dsRNA are unclear. Our data suggest that length, subcellular localization and ADAR modification of endogenous dsRNA are not critical for ‘self’ recognition. It is possible that 5′-end modifications, binding of cellular factors or other modifications are important (44,45). Future work to elucidate the characteristics of these endogenous dsRNA structures that allow them to evade cytoplasmic dsRNA sensors will provide key insights into regulation of the innate immune response.
Intramolecular dsRNA structures function as translation control elements across species
Although repetitive elements have long been thought of as genomic junk, recent studies have suggested that Alu elements can influence gene expression in a variety of different manners (8,10,12). We observed that two Alu elements in inverted orientation within the 3′-UTR of human mRNAs give rise to dsRNA. Although the structures are extensively edited in vivo, the presence of inosine did not affect mRNA export, stability or translatability. However, the dsRNA structure itself decreased translational efficiency. Furthermore, as we detected translational regulation by inverted Alu structures from multiple human genes, and we previously observed that double-stranded 3′-UTR structures formed from inverted repeats decrease translational efficiency of endogenous C. elegans mRNAs (38), we propose that intramolecular dsRNA structures can function as translational control elements across species. However, it is important to note that we also observed double-stranded 3′-UTR structures that do not alter translation. Furthermore, Alu elements in 3′-UTRs have also been shown to impact gene expression by regulating mRNA production, localization and stability (8,10), and inverted Alus may have a similar diversity.
It is interesting to draw parallels between the long intramolecular Alu dsRNA structures and small intermolecular dsRNA structures that form between miRNAs and 3′-UTRs. miRNAs are well known to affect translation, however, the exact mechanisms that miRNAs use to regulate translation are controversial (46–48). miRNAs are best known for regulating translation initiation (49,50). However, the presence miRNA-repressed mRNAs on translating ribosomes indicates that other mechanisms after initiation also occur (51–53). As we detect endogenous mRNAs with double-stranded 3′-UTRs on translating ribosomes (Supplementary Figure S1), we also predict that these long intramolecular dsRNA structures regulate translation after initiation. However, in general, specific mechanisms to regulate translation post-initiation are not well understood. Two recent reports have suggested that interactions of RNA binding proteins with both eukaryotic elongation factor 1A (eEF1A) and small 3′-UTR stem-loop structures attenuates translation elongation (54,55). Future work to identify factors that interact with inverted Alus will be critical to understanding how these dsRNA structures regulate translation.
In addition to mechanism, it is important to determine whether there are specific cellular conditions that exploit these structures to regulate gene expression. It is interesting to note that, in the case of PSMB2, we have identified two isoforms that have alternative 3′-UTRs, one containing and one lacking the inverted Alus. Alternative 3′-UTRs have been suggested to be important in cancer, presumably because shortened UTRs lack key regulatory elements (56,57). Interestingly, the majority of mRNAs with inverted Alu-containing 3′-UTRs have been reported to contain alternative polyadenylation signals (37). It is possible that production of two different isoforms allow for the fine-tuning of PSMB2 expression and that in certain cell types or conditions the ratios of the isoforms vary. Identification of conditions in which expression of these isoforms vary will be a critical step in understanding the biological function of these intramolecular dsRNA structures and the inosines within them.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–6 and Supplementary Methods.
FUNDING
Start-up funds and a Research Enhancement Grant, Indiana University School of Medicine (to H.A.H.). Funding for open access charge: Indiana University Start-up Funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brenda Bass and Peter Hollenhorst for critical reading of the manuscript. We also thank Cheng Kao, Amy Pasquinelli and Mark Parnell for helpful comments during the preparation of the manuscript. Author Contributions: C.R.C., K.L.D. and H.A.H. performed experiments. H.A.H. designed the experiments and wrote the manuscript.
REFERENCES
- 1.Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 4.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 6.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 7.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 8.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr. Opin. Genet. Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 10.Gong C, Maquat LE. “Alu”strious long ncRNAs and their role in shortening mRNA half-lives. Cell Cycle. 2011;10:1882–1883. doi: 10.4161/cc.10.12.15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 12.Berger A, Strub K. Multiple roles of Alu-related noncoding RNAs. Prog. Mol. Subcell. Biol. 2011;51:119–146. doi: 10.1007/978-3-642-16502-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Hasler J, Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006;34:2374–2385. doi: 10.1093/nar/gkl246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin CM, Kimura RH, Schmid CW. Selective stimulation of translational expression by Alu RNA. Nucleic Acids Res. 2002;30:3253–3261. doi: 10.1093/nar/gkf419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 17.Hasler J, Samuelsson T, Strub K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell. Mol. Life Sci. 2007;64:1793–1800. doi: 10.1007/s00018-007-7084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 19.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat. Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 21.Lin L, Shen S, Tye A, Cai JJ, Jiang P, Davidson BL, Xing Y. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008;4:e1000225. doi: 10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y. Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl Acad. Sc.i USA. 2011;108:2837–2842. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LL, Carmichael GG. Nuclear editing of mRNA 3′-UTRs. Curr. Top. Microbiol. Immunol. 2012;353:111–121. doi: 10.1007/82_2011_149. [DOI] [PubMed] [Google Scholar]
- 24.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 28.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogg M, Paro S, Keegan LP, O'Connell MA. RNA editing by mammalian ADARs. Adv. Genet. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 30.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010;20:4–22. doi: 10.1002/rmv.633. [DOI] [PubMed] [Google Scholar]
- 33.Gantier MP, Williams BR. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 37.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlstedt H, Ohman M. Site-selective versus promiscuous A-to-I editing. Wiley Interdiscip. Rev. RNA. 2011;2:761–771. doi: 10.1002/wrna.89. [DOI] [PubMed] [Google Scholar]
- 40.Maas S, Melcher T, Herb A, Seeburg PH, Keller W, Krause S, Higuchi M, O'Connell MA. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 41.Lampson GP, Tytell AA, Field AK, Nemes MM, Hilleman MR. Inducers of interferon and host resistance. I. Double-stranded RNA from extracts of Penicillium funiculosum. Proc. Natl Acad. Sci. USA. 1967;58:782–789. doi: 10.1073/pnas.58.2.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeWitte-Orr SJ, Mossman KL. dsRNA and the innate antiviral immune response. Future Virol. 2010;5:325–341. [Google Scholar]
- 43.Nejepinska J, Malik R, Filkowski J, Flemr M, Filipowicz W, Svoboda P. dsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Res. 2012;40:399–413. doi: 10.1093/nar/gkr702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5′-end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5:140–144. doi: 10.4161/rna.5.3.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito T, Gale M., Jr Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J. Exp. Med. 2008;205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 47.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 50.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 52.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 53.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 54.Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012;19:176–183. doi: 10.1038/nsmb.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.