Abstract
The conserved bacterial anticodon nuclease (ACNase) RloC and its phage-excluding homolog PrrC comprise respective ABC-adenosine triphosphatase (ATPase) and ACNase N- and C-domains but differ in three key attributes. First, prrC is always linked to an ACNase silencing, DNA restriction–modification (R–M) locus while rloC rarely features such linkage. Second, RloC excises its substrate’s wobble nucleotide, a lesion expected to impede damage reversal by phage transfer RNA (tRNA) repair enzymes that counteract the nick inflicted by PrrC. Third, a distinct coiled-coil/zinc-hook (CC/ZH) insert likens RloC’s N-region to the universal DNA damage checkpoint/repair protein Rad50. Previous work revealed that ZH mutations activate RloC’s ACNase. Data shown here suggest that RloC has an internal ACNase silencing/activating switch comprising its ZH and DNA-break-responsive ATPase. The existence of this control may explain the lateral transfer of rloC without an external silencer and supports the proposed role of RloC as an antiviral contingency acting when DNA restriction is alleviated under genotoxic stress. We also discuss RloC’s possible evolution from a PrrC-like ancestor.
INTRODUCTION
Bacteria disable translation in response to various stress situations including phage infection (1). A case in point is imparted by the anticodon nuclease (ACNase) PrrC, a potential phage-excluding device counteracted by phage-induced transfer RNA (tRNA) repair (2,3). RloC is a novel ACNase of unknown function whose similarity to PrrC and salient features portray it as a stronger antiviral device responsive to an added stress cue (4). Evaluating RloC’s distinctive traits and purported role necessitates prior description of its better documented homolog PrrC, particularly the prototype encoded by a rare Escherichia coli strain (EcoPrrC) (5–7).
EcoPrrC’s ACNase is silenced in the uninfected host by the physically associated Type Ic DNA restriction–modification (R–M) protein EcoprrI (6,8). During phage T4 infection, it is activated by the co-opted T4 anti-DNA restriction peptide Stp (9). The activated ACNase nicks tRNALys 5′ to the wobble base yielding 3′-cyclic P and 5′-OH termini. This damage could disable T4 late translation and contain the infection (2,10). However, T4’s tRNA repair proteins 3′-phosphatase/5′-polynucleotide kinase (Pnk) and RNA ligase 1 (Rnl1) offset it. Specifically, Pnk converts the cleavage termini into a 3′-OH and 5′-P pair that Rnl1 joins (2). The above restriction/anti-restriction cascade may be shared by other PrrC-encoding bacteria, judged from prrC’s consistent linkage to a Type Ic R–M locus, the ACNase activities of PrrC orthologs looked at (3,4,11) and a case of coincident inactivation of the ACNase and linked R–M system (12). The idea that Pnk and Rnl1 evolved as ACNase antidotes (13,14) is reinforced by their ubiquity among T4-like phage of PrrC/RloC-encoding bacteria but absence from T4-like cyanophage not expected to encounter these ACNases (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi and http://phage.ggc.edu/).
In vitro activation of the latent EcoPrrC ACNase (EcoPrrC–EcoprrI complex) requires besides Stp the hydrolysis of GTP in the presence of dTTP. ATP inhibits the activation but whether it exerts this effect through EcoPrrC or EcoprrI’s adenosine triphosphatase (ATPase)/DNA translocase is not known (15–17). Isolated EcoPrrC has overt ACNase activity refractory to Stp and GTP but unstable without dTTP or a non-hydrolyzable dTTP analogue. The significance of this protection is indicated by the importance of the T4-induced accumulation of dTTP for the manifestation of EcoPrrC’s ACNase activity (3,15,18–20). These observations underlie a scheme where GTP hydrolysis drives the activation of the ACNase and the accumulated dTTP stabilizes the activated form. GTP and dTTP likely exercise their distinct functions through EcoPrrC’s ABC-ATPase N-domains, to which these nucleotides bind with vastly differing affinities without displacing each other (3,15 and our unpublished data). EcoPrrC’s remaining C-proximal third harbours residues implicated in tRNALys recognition (21–24) and a putative catalytic ACNase triad (3) that is conserved by RloC (4). EcoPrrC may act as a tetramer (3) whose N-domains dimerize head-to-tail like typical ABC-ATPases and the C-domains in parallel (24). Another view is that EcoPrrC is a dimer whose ACNase domains do not interact (11).
RloC shares PrrC’s organization into ABC-ATPase and ACNase domains but differs in three key features. First, RloC rarely interacts with an R–M system in cis although teaming of RloC with an R–M system in trans is not excluded (4). Second, RloC excises its substrate’s wobble nucleotide, a lesion expected to frustrate damage reversal by phage tRNA repair enzymes (4). Third, a coiled-coil/zinc-hook (CC/ZH) insert in RloC’s ATPase domain likens its N-region to the universal DNA damage checkpoint/repair protein Rad50 (25–27). Rad50’s CC/ZH folds back into an anti-parallel CC protruding from the ATPase head domain with the ZH motif Cys–X–X–Cys at its apex. Two ZH apices join by coordinating Zn++ to the four Cys of their dimerization interface. Such ZH joints form between two flexible CC/ZH protrusions of the same Rad50 dimer or between two different DNA-borne Rad50 dimers (26,27). The latter mode bridges distant DNA molecules and is essential for Rad50-mediated DNA transactions (28,29). Other DNA bridging structure maintenance of chromosomes (SMC) proteins join their CC protrusions through hydrophobic apices (30).
RloC is so far the only protein other than Rad50 known to harbour a CC/ZH insert in its ABC-ATPase domain. A regulatory role of this extension is suggested by ZH mutations that activate RloC’s ACNase and exacerbate its toxicity (4). This fact underlies a model where RloC responds to DNA insults by disabling translation, benefiting its host as an antiviral contingency as DNA restriction is alleviated under genotoxic stress (4,31,32).
In this work, we investigated the role of RloC’s Rad50-like N-region in regulating its C-proximal ACNase, specifically the anticipated involvement of the ZH motif and ABC-ATPase head domain in this function. To accomplish this feat, we used the RloC ortholog from the thermophile Geobacillus kaustophilus (GkaRloC) (4) and model ACNase substrates that also served to further characterize the RloC excision reaction. The data suggest that RloC’s ACNase is regulated by its coupled ZH and DNA-break-responsive ATPase, in keeping with the above model. The existence of such internal control may also account for the lateral transfer of rloC without a linked ACNase silencer.
MATERIALS AND METHODS
GkaRloC mutagenesis
Mutations in GkaRloC’s ATPase motifs were introduced by Quick Change (33) and verified by DNA sequencing.
GkaRloC expression and purification
Escherichia coli Rosetta™ (DE3) pLysS (Novagen) encoding inducible T7 RNA polymerase and rare tRNA species served as host cell for expressing plasmid pGkaRloC-L-His6 or the indicated mutant derivatives (4). Cells transformed by them were grown to 1.5 OD600 at 37°C (wt, K44N, D572N) or 25°C (C291G) in modified TY medium (3.2% trypton, 2.4% yeast extract, 35 mM NaCl, 89 mM potassium phosphate buffer, pH 7.5 and 0.4% glycerol) containing 100 µg/ml ampicillin and 34 µg/ml chloramphenicol. Protein expression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside. The culture was shifted then to 16°C and shaken for 16 h. The cells were harvested and washed twice with buffer I (10 mM Tris–HCl, pH 7.5, 15 mM MgCl2, 1 M KCl and 10% glycerol) and once in buffer II (10 mM Tris–HCl, pH 7.5, 15 mM MgCl2, 50 mM KCl and 10% glycerol). The protein expression level and in vivo ACNase activity were assessed as previously described (4). Protein purification steps were performed at 0–4°C. The bacterial pellet was suspended in 1.5 vol of buffer III (10 mM Na-HEPES, pH 7.5, 10 mM MgCl2, 5 mM β-mercaptoethanol and 10% glycerol) containing ethylenediaminetetraacetic acid-free protease inhibitor cocktail (Roche). Following passage through an Aminco pressure cell at 18 000 psi, the lysate was centrifuged 30 min at 30 000 g. The supernatant was made up to 5 mM imidazole and loaded onto an immobilized cobalt affinity resin (TALON, Clontech). The column was washed with 10 vol of buffer III containing 5 mM imidazole and 250 mM NaCl followed by 10 vol of buffer III plus 5 mM imidazole. GkaRloC was eluted in buffer III plus 0.5 M imidazole. Peak fractions were concentrated in Vivaspin 20 concentrator 30000 MWCO (Sartorius), loaded on Q-trap anion exchange column (GE-Healthcare) and eluted in a linear 0–1 M NaCl gradient in buffer III. Peak fractions emerging at ∼140 mM NaCl were stored in small aliquots at −80°C.
ACNase substrates
Internally labelled Saccharomyces cerevisiae tRNAGlu(UUC) was prepared by treating the total cellular tRNA fraction with the Kluyveromyces lactis tRNase γ-zymocin (34) in 20 mM Tris–HCl, pH 7.5 and 1 mM MgCl2. The resultant tRNAGlu(UUC) fragments were ligated back within the total tRNA fraction by T4 Pnk and Rnl1 (NEB) in the presence of 1 µM [γ-32P]ATP (3000 Ci/mmol, NEN) essentially as described (35). The ligated back tRNAGlu(UUC) was gel purified along with similar-sized non-labelled ‘carrier’ tRNA species. The presence of these carrier species did not affect the GkaRloC cleavage pattern of tRNAGlu(UUC). This was indicated by an identical pattern obtained with the ligated back gel-purified tRNAGlu(UUC) fragments. Derivatives of the labelled sc-tRNAGlu(UUC) partially incised 3′ or 5′ to the wobble position were prepared by incubating this substrate with γ-zymocin (34) or EcoPrrC-D222E (3).
GkaRloC ACNase assays
The standard in vitro ACNase assay mixtures (10 µl) contained 100–500 ng GkaRloC protein, 70 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 10 mM dithiothreitol, 0.5 mM ATP, 10 ng/µl double-stranded DNA fragments (BstEII-digested λ DNA, NEB) and 0.05–0.1 pmol of [5′-32P]Lys3-ASL or 0.01–0.02 pmol of the internally labelled sc-tRNAGlu(UUC). After 5–10 min pre-incubation at 25°C, the reaction was started by adding the substrate. Where indicated, other DNA species substituted the standard λ DNA fragments. They included DNA duplexes or hairpins of defined size (Supplementary Table S1) and pUC19 DNA (Fermentas) used as such, or after relaxation with E. coli DNA topoisomerase I or linearized with Sma I endonuclease, both provided by NEB. The reaction was stopped with 2 vol of 10 M urea, 0.01% each of xylene cyanol and bromphenol blue. The products were separated by denaturing polyacrylamide gel electrophoresis, monitored by autoradiography and quantified by Scion Image software (NIH). In vivo ACNase activity was monitored by in vitro 5′-end labelling tRNA fragments generated in cells expressing the indicated GkaRloC allele, as described (4)
RESULTS
GkaRloC excises the wobble nucleotide by successive cleavages in the 3′ to 5′ order
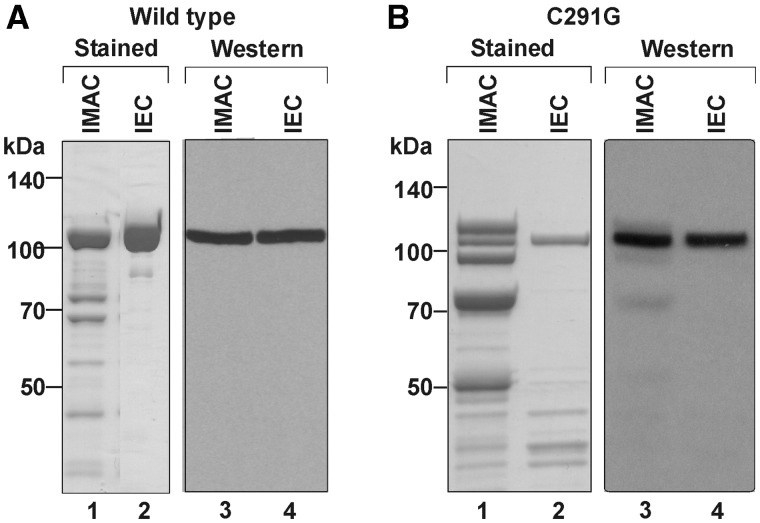
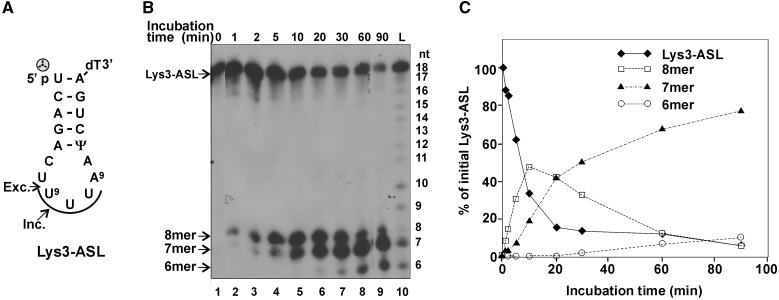
The regulation of GkaRloC’s ACNase by its Rad50-like N-domain was studied using a His6-tagged form expressed in E. coli and purified by immobilized-metal affinity chromatography (IMAC) followed by ion-exchange chromatography (IEC), respectively (Figure 1A). Its ACNase activity was assayed with two convenient model substrates that were cleaved in a manner similar to that seen with E. coli tRNAs GkaRloC targets in vitro and in vivo. A minimal model ACNase substrate was a [5′-32P] anticodon stem-loop of sequence and base modifications of mammalian tRNALys3 (Lys3–ASL) (23,36) (Figure 2A). Lys3–ASL was preferred over a less reactive ASL with E. coli tRNALys modifications or hypomodified counterparts cleaved differently from the full-sized tRNA substrates (not shown). In contrast, Lys3–ASL was cleaved by GkaRloC first 3′ and then 5′ to the wobble base, yielding in respective order labelled 8- and 7mers. Prolonged incubation yielded also a labelled 6mer (Figure 2B and C). Such further trimming has not been observed with full-sized tRNAs (4). Therefore, it was ascribed to greater flexibility of the cleaved minimal substrate.
Figure 1.
Isolation of GkaRloC alleles. Aliquots of the indicated fractions of wild-type GkaRloC (A) or its ZH mutant C291G (B) were separated by SDS–PAGE and monitored by staining or immunoblotting using an anti-His tag monoclonal antibody (4).
Figure 2.
GkaRloC-mediated digestion of a 5′-end labelled ASL substrate. (A) Lys3–ASL sequence (the 3′-dT extension facilitated the chemical synthesis of this ASL (36). The arrows indicate incision (Inc.) and excision (Exc.) sites. U9 is the modified wobble base mcm5s2U. The arc highlights the anticodon triplet.  denotes the 5′-label. (B) GkaRloC ACNase activity was assayed using its IMAC fraction and the [5′-32P]Lys3–ASL substrate as detailed in ‘Materials and Methods’ section. The reaction products were separated by denaturing PAGE. L-size ladder of partially hydrolyzed Lys3–ASL. (C) Time course of Lys3–ASL decay and formation of the labelled products.
denotes the 5′-label. (B) GkaRloC ACNase activity was assayed using its IMAC fraction and the [5′-32P]Lys3–ASL substrate as detailed in ‘Materials and Methods’ section. The reaction products were separated by denaturing PAGE. L-size ladder of partially hydrolyzed Lys3–ASL. (C) Time course of Lys3–ASL decay and formation of the labelled products.
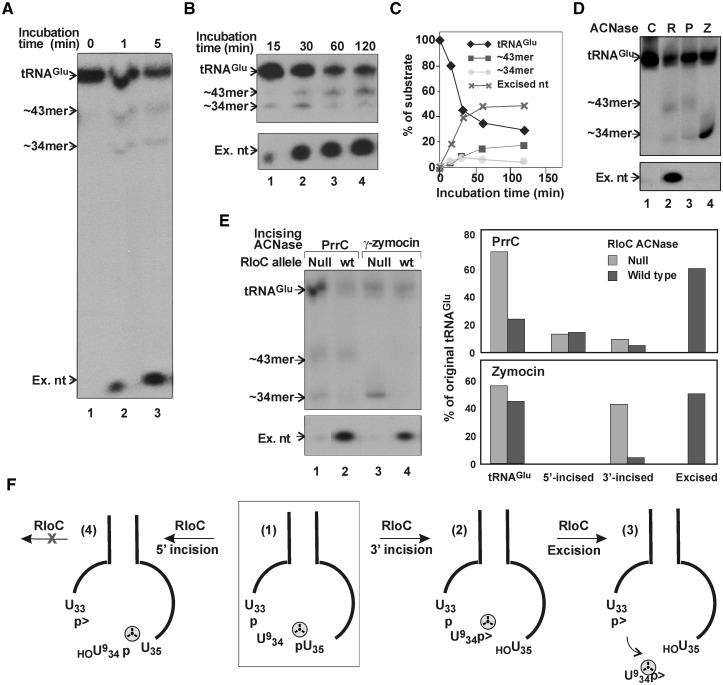
A full-sized model substrate used was the major S. cerevisiae tRNAGlu [sc-tRNAGlu(UUC)]. It resembles the E. coli counterpart preferentially cleaved by GkaRloC (4). Importantly, sc-tRNAGlu(UUC) can be readily tagged 3′ to the wobble base, allowing direct visualization of the excised nucleotide. To introduce this tag, we exploited the specificity of γ-zymocin, the poisonous subunit of the toxin secreted by killer strains of the dairy yeast K. lactis. Within target S. cerevisiae cells γ-zymocin incises tRNAGlu(UUC) 3′ to the 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) wobble base (34). Such a reaction performed in vitro and followed by T4 Pnk and Rnl1-mediated repair in the presence of [γ-32P]ATP yielded the desired internally labelled sc-tRNAGlu(UUC). GkaRloC converted this substrate into three labelled products: ∼34 and ∼43 nt fragments formed in relatively low amounts and the excised nucleotide that accumulated (Figure 3A and B). The ∼34mer migrated with the labelled fragment released by γ-zymocin (Figure 3D, lanes 2 and 4), indicating that GkaRloC incised sc-tRNAGlu(UUC) similarly. The ∼43mer migrated with the major labelled fragment formed by EcoPrrC (Figure 3D, lanes 2 and 3), which normally incises its substrate 5′ to the wobble base. Thus, GkaRloC incised sc-tRNAGlu(UUC) also at this site. Note that EcoPrrC nicked sc-tRNAGlu(UUC) also 3′ to the wobble base, albeit, to a lesser extent than at the 5′ site (Figure 3D, lane 3). Such a shift in cleavage specificity has been observed with some EcoPrrC mutants and substrate analogues (21,22). The excised wobble nucleotide was seen with GkaRloC but not EcoPrrC (compare lanes 2 and 3). This discrepancy confirmed that GkaRloC rather than a contaminating E. coli activity catalyzed the excision because both ACNases were expressed and purified similarly.
Figure 3.
GkaRloC excises the wobble nucleotide from sc-tRNAGlu(UUC). (A and B) The sc-tRNAGlu(UUC) substrate radiolabeled 3′ to the wobble base was incubated with the IMAC fraction of GkaRloC for the indicated times and the products separated by denaturing PAGE. ∼34- and ∼43mer are the respective labelled fragments resulting from 3′ and 5′ incisions; Ex.nt is the excised nucleotide. (C) Time course of substrate decay and product formation in (B). (D) Cleavage products of the sc-tRNAGlu(UUC) substrate (C) formed by GkaRloC (R), PrrC (P) or γ-zymocin (Z). (E) GkaRloC cleaves the pre-formed 3′ but not 5′ incision product of sc-tRNAGlu(UUC). Sc-tRNAGlu(UUC) was incubated with PrrC (lanes 1 and 2) or γ-zymocin (lanes 3 and 4) followed by incubation with the ACNase-null mutant E696A (4) (lanes 1 and 3) or wild-type GkaRloC (wt) (lanes 2 and 4). The chart on the right shows the proportions of intact sc-tRNAGlu(UUC), the derived 5′- or 3′-incision products formed respectively by PrrC or by PrrC and γ-zymocin as well as the excised wobble nucleotide formed in the subsequent incubation with wild-type GkaRloC. (F) The GkaRloC-mediated incising and excising cleavages of sc-tRNAGlu(UUC). The internally labelled substrate highlighted by an open square (1) is incised by GkaRloC 3′ to the wobble base to yield compound (2) followed by an excising cleavage upstream that yields the tRNA fragments 1-33 and 35-76 and excised wobble nucleotide (3). Inadvertent incision 5′ to the wobble base yields a dead-end product (4). The tRNA substrates and products are schematically depicted by their anticodon stem loop region. Indicated in it are the wobble and two flanking bases.  denotes phosphate label.
denotes phosphate label.
The ability of GkaRloC to incise sc-tRNAGlu(UUC) on either side of the wobble base could be taken to indicate that the excision occurs by successive cleavages not only in the 3′ → 5′ direction, as previously assumed (4) but also in the opposite. Yet, more likely seemed that the 5′ incision yielded a dead-end product since the ∼34mer declined with the overall reaction, as would a bona fide intermediate, while the ∼43mer accumulated (Figure 3B and C). Moreover, GkaRloC effectively removed the wobble nucleotide from a preformed 3′ incision product generated by γ-zymocin or as EcoPrrC’s minor product but not from EcoPrrC’s major, 5′ incision product (Figure 3E). These data indicated that GkaRloC excised the wobble nucleotide by successive cleavages in the 3′ → 5′ order only (Figure 3F). It is also noteworthy that the 3′ pre-incised tRNA was preferentially cleaved in the presence of the intact (Figure 3E, lanes 3 and 4). This result hinted that the incision is rate-limiting and the overall reaction processive.
GkaRloC’s ATPase activates its ACNase
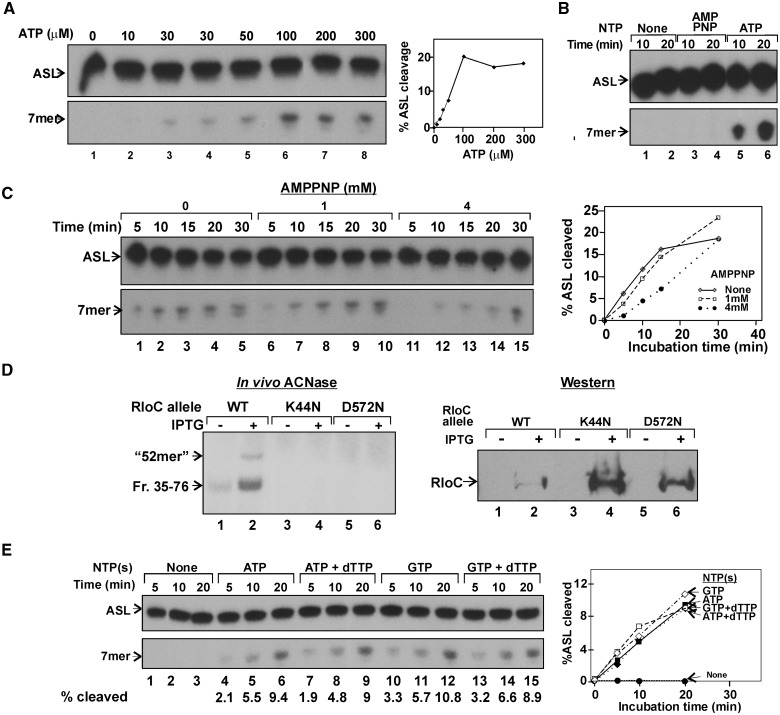
Unlike the overt ACNase activity of isolated EcoPrrC (3) purified GkaRloC was virtually devoid of ACNase activity in the absence of a hydrolyzable nucleotide. As shown, adding ATP to GkaRloC’s IMAC fraction dramatically enhanced the cleavage of Lys3–ASL in a sigmoid dose dependence (Figure 4A). In contrast, adenosine 5′-(β,γ-imido) triphosphate (AMPPNP) did not elicit such activation (Figure 4B), suggesting that nucleotide hydrolysis is required. However, when ATP was also present AMPPNP not only delayed the activation of the ACNase in a dose-dependent manner but also stabilized the ACNase once activated (Figure 4C). In this regard GkaRloC superficially resembled EcoPrrC whose latent form is activated by GTP hydrolysis and the activated stabilized by dTTP binding (3,15,20).
Figure 4.
GkaRloC’s ATPase activates its ACNase. GkaRloC’s ACNase of the IMAC fraction was assayed in vitro in panels (A)–(C) and (E) essentially as described in Materials and Methods but in the absence of added DNA. (A) Dependence of GkaRloC’s ACNase activity on ATP’s level. (B) GkaRloC’s ACNase activity was assayed in the presence of 500 µM of the indicated nucleotides. (C) Time courses of GkaRloC’s ACNase activity in the presence of 0.5 mM ATP and indicated amounts of AMPPNP. (D) In vivo ACNase activity of the indicated GkaRloC alleles. Left panel—RNA extracted from cells expressing these alleles was 5′-end labelled using T4 Pnk and separated by denaturing PAGE. Right panel—the expression of the indicated GkaRloC alleles were monitored by Western using an anti-His tag monoclonal antibody (4). (E) Nucleotide specificity of GkaRloC’s ACNase activation. The activation reaction was performed in the presence of the indicated nucleotides (GTP and ATP at 0.5 mM each, dTTP at 5 µM).
To determine whether GkaRloC harbours the ACNase-activating ATPase, we singly mutated its respective Walker A and B residues Lys44 and Asp572 as corresponding lesions inactivate other ABC-ATPases (37–39). When cells expressing wild-type GkaRloC or either ATPase mutant were assayed for in vivo ACNase activity (Materials and Methods) only the former yielded the expected labelled ∼42mers emanating from mature tRNA substrates and a minor ∼52mer possibly derived from tRNA precursor(s) carrying a 3′-tail (4) (Figure 4D, left panel). The ATPase mutants were also expressed at a higher level than wild type, in keeping with their ACNase-null phenotypes (right panel). These data indicated that GkaRloC harbours the ACNase-activating ATPase.
GTP (but not ATP) hydrolysis activates in vitro the latent EcoPrrC–EcoprrI ACNase holoenzyme but does not augment the overt ACNase of free EcoPrrC. Moreover, dTTP stabilizes both the activated and overt PrrC ACNases (15). In contrast, ATP and GTP activated GkaRloC’s ACNase similarly and regardless of dTTP’s presence (Figure 4E).
ACNase activation by GkaRloC’s ATPase requires DNA
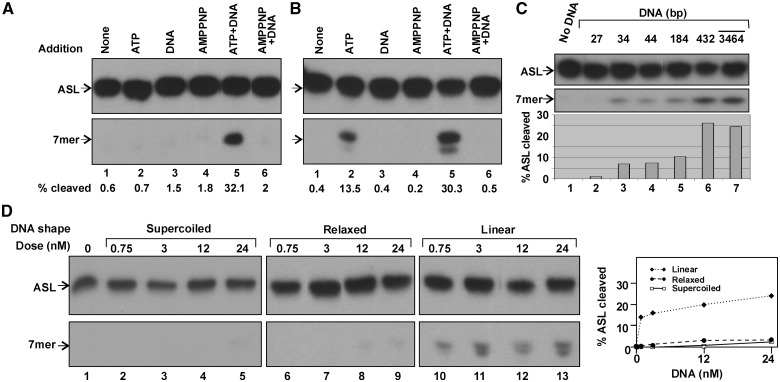
When GkaRloC’s purer IEC fraction (Figure 1A) was assayed, we noticed that adding ATP did not activate its ACNase (Figure 5A). This suggested that an essential activating factor was fractionated away. DNA seemed the culprit since ACNase-enhancing ZH mutations (4) were expected to modulate GkaRloC’s interaction with DNA, as with Rad50 (26). Moreover, other SMC proteins harbour a DNA-dependent ATPase (30,39,40). Indeed, GkaRloC’s ACNase was activated in the IEC fraction only when both ATP and DNA were added (Figure 5A, lanes 2, 3 and 5). AMPPNP failed to activate the ACNase regardless of the presence of DNA (lanes 4 and 6). The partial activation seen when only ATP was added to the cruder IMAC fraction (Figure 5B; lanes 1 and 2) was attributed to co-purifying DNA, since prior DNase I treatment abolished it (Supplementary Figure S1, lane 6). Moreover, adding the standard DNA dose to the treated fraction restored the activation (lane 8). This indicated that the DNase I treatment did not inactivate the ACNase and the minimal DNase I dose needed to degrade the endogenous DNA was overwhelmed by the added DNA.
Figure 5.
GkaRloC’s ACNase-activating ATPase depends on DNA. (A and B) Effect of the indicated additions on ACNase activity in IEC (A) or IMAC (B) fractions of GkaRloC. (C and D) Effect of size (C) or shape (D) of the DNA added to the IEC fraction on the activation of GkaRloC’s ACNase.
Defined duplexes of 5–432 bp (Supplementary Table S1) were compared in ACNase activation with the routinely used mixture of large DNA fragments averaging ∼3.5 kb. The former were used at 100 nM, the latter at an optimal ∼4 nM level and GkaRloC at ∼50 nM. Duplexes of 27 bp and less did not detectably activate the ACNase (Figure 5C, lanes 1 and 2 and data not shown). Duplexes of 34–184 bp activated it modestly (lanes 3–5), ∼3-fold less than the larger duplexes (lanes 6 and 7). Presumably, the non-activating were smaller than GkaRloC’s minimal DNA target, which may be close to the ∼23 bp DNA binding channel visualized in a Rad50/AMPPNP co-crystal structure (41). We assume that the biphasic size dependence of the activating duplexes reflects a minimal length and/or bendability needed for the coalescence of GkaRloC molecules in cis. The effect of DNA termini on the ACNase activating potential was investigated by comparing supercoiled and relaxed circular forms of the 2686 bp plasmid pUC19 DNA with a linear counterpart (Figure 5D). As shown, the linear form was by far more reactive, suggesting that DNA termini figure in the activation.
GkaRloC’s anticipated DNA-dependent ATPase activity was not detected over the K44N or D572N ATPase mutant backgrounds when IEC fractions were assayed (not shown). We assume that traces of co-purifying DNA-dependent E. coli ATPase(s) masked that of GkaRloC. This assumption concurs with that inferred from AMPPNP’s dual effect (Figure 4C); i.e. once activated by its ATPase, the ACNase is stabilized by nucleotide binding. Noteworthy in this regard is also the weak DNA-dependent ABC-ATPase (0.2 min−1) reported for RecF, which is related to Rad50 (42) and, by implication, to RloC.
Mutating GkaRloC’s ZH short circuits the ACNase switch
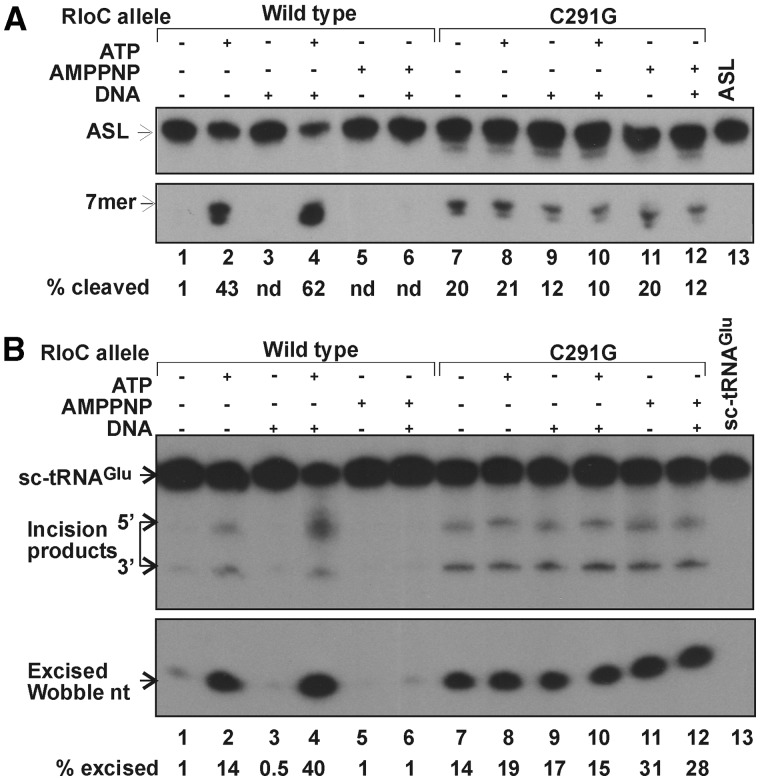
It seemed conceivable that ACNase-activating ZH mutations (4) uncouple the ACNase from its activating ATPase. This premise was tested by comparing wild-type GkaRloC with the ZH mutant C291G in their responses to ATP or AMPPNP, without or with DNA. GkaRloCC291G was expressed and isolated similar to wild type, albeit, with poorer yield and purity (Figure 1B). Unlike the wild-type protein, it featured overt ACNase activity that was refractory to the DNA-dependent ATPase, both when Lys3–ASL (Figure 6A) or sc-tRNAGlu(UUC) (Figure 6B) were used as a substrate, and whether the IMAC (Figure 6) or IEC fraction was used (Supplementary Figure S2).
Figure 6.
The ZH mutation C291G uncouples GkaRloC’s ACNase from its DNA-dependent ATPase. The IMAC fractions of the indicated GkaRloC alleles were assayed for ACNase activity in the presence of the indicated additions using as a substrate Lys3–ASL (A) or sc-tRNAGlu(UUC) (B).
The latter result suggested that ZH joints formed by the wild-type protein silenced its ACNase. Unexpectedly, using IEC fractions we found that Zn++ abolished both the wild-type and ZH mutant ACNases as well as EcoPrrC’s (Supplementary Figure S3). Presumably, Zn++ mediated these inhibitions through another or an added site, shared perhaps by RloC and PrrC. Thus, it remained uncertain whether Zn++ coordination by the ZH of the wild-type ACNase accounted for its silencing. Finally, our previous report that Zn++ fails to inhibit the ZH mutant ACNase (4) must have been in error. Namely, it turned out that imidazole left in the IMAC fraction used in the former study titrates the zinc ions and thus prevents the inhibition.
DISCUSSION
GkaRloC’s excision mechanism
GkaRloC’s ability to cleave Lys3–ASL 3′ and then 5′ to the wobble base (Figure 2) and direct visualization of the wobble nucleotide GkaRloC excises from sc-tRNAGlu(UUC) (Figure 3) support the conclusion that GkaRloC is a wobble nucleotide-excising ACNase (4). Unexpectedly, GkaRloC incised sc-tRNAGlu(UUC) both 3′ and 5′ to the wobble base. However, the latter incision yielded a dead-end product that was not further cleaved (Figure 3). It is noteworthy that GkaRloC generates in vitro a 5′ incision product also from E. coli tRNALys. This result was obtained in an experiment intended to examine whether GkaRloC occludes its incision intermediate from the T4 tRNA repair enzymes. The resultant cleavage ligation junctions labelled from [γ-32P]ATP were mostly of defective products lacking the wobble nucleotide. The remaining junctions were of two comparable fractions of intact tRNA molecules formed by reversal of the 5′ or 3′ incision (Supplementary Figure S4). Nonetheless, an in vivo 5′ incision product derived from tRNALys or any other E. coli tRNA targeted by GkaRloC has not been detected (4). These facts and selective inhibition of the 5′-incision and excising cleavage (Supplementary Figure S5) indicate that under the in vitro conditions used GkaRloC could occasionally skip the initial 3′ cleavage site.
GkaRloC’s tRNA substrate specificity
GkaRloC disrupts a variety of bacterial and eukaryal tRNAs and their analogues (4; Figures 2 and 3 and data not shown). It differs in this regard from the fungal ACNase γ-zymocin, which is highly sensitive to changes in the modifying side chain of the sc-tRNAGlu(UUC) wobble base and poorly cleaves other tRNA species sharing this base (34). Nonetheless, it is conceivable that GkaRloC is less promiscuous in nature. This is suggested by the observation that EcoPrrC’s natural specificity for tRNALys is compromised when this ACNase is ectopically over-expressed or assayed in vitro (21,23). Moreover, inactivation of a single tRNA species could suffice to disable the synthesis of phage proteins while minimizing the potential hazard to the uninfected bacterial host. Testing this expectation requires a natural RloC encoding host and means to activate its ACNase. On the other hand, from a practical perspective, GkaRloC’s in vitro promiscuity and unique cleavage site specificity may facilitate artificial replacements of tRNA wobble bases and identification of novel ones.
RloC’s internal ACNase switch
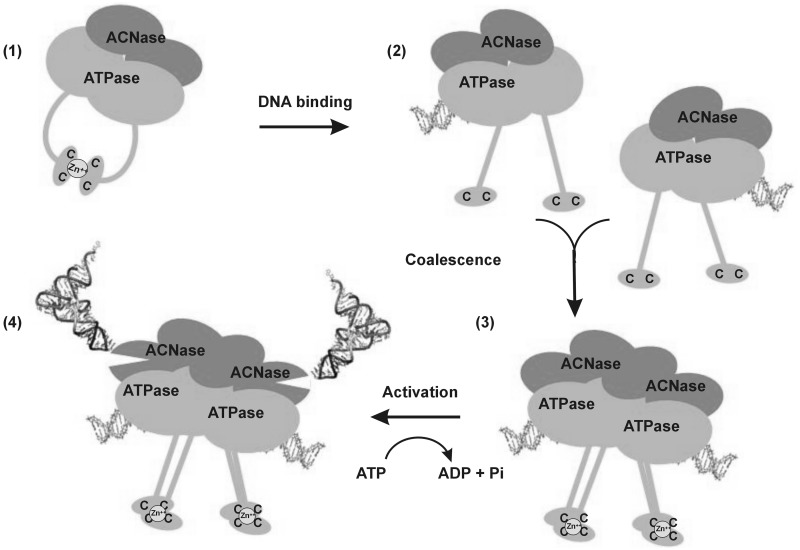
A unique trait of GkaRloC and, by implication, of RloC in general is an internal ACNase regulating device comprising the coupled ZH and DNA-dependent ATPase. The existence of this device is inferred from the activation of GkaRloC’s ACNase by ZH mutations (4) that also render this ACNase independent of the otherwise activating DNA-dependent ATPase (Figure 6). These facts suggest that intramolecular ZH joints accounted for the silencing of the wild-type ACNase in the absence of ATP and DNA (Figure 5). By analogy with the changes Rad50 undergoes upon DNA binding (27), it may be proposed that DNA binding straightens GkaRloC’s CC protrusions and thus disrupts the inhibitory intramolecular ZH joints. The affinity of GkaRloC’s ZH for zinc is not known. However, if similar to that of a zinc finger protein (43) it could suffice to capture zinc ions released from the intramolecular hook within an intermolecular. This possibility and the observed effects of DNA size and shape on GkaRloC’s ACNase activation (Figure 5C and D) underlie a proposed scheme where association of RloC dimers bound at proximal DNA termini triggers ATP hydrolysis and consequent activation of the ACNase (Figure 7). It could be argued that a double-stranded DNA break (DSB) satisfies such a need for proximal DNA termini in vivo. However, DSBs did not elicit ACNase activity in the natural RloC encoding Acinetobacter sp. ADP1 although over-expressing the AciRloC in E. coli showed it to be potentially active (our unpublished data). Therefore, DSBs may be only one of the physiological triggers needed to activate RloC’s ACNase.
Figure 7.
Model of GkaRloC’s ACNase activation by its DNA-dependent ATPase. By analogy with Rad50 (26,27), it is proposed that a free GkaRloC dimer forms intramolecular ZHs that silence its ACNase (1). Upon DNA binding to the ATPase head domains the CC protrusions straighten liberating the coordinated zinc ion (2) such that only intermolecular zinc hooks can form between dimers bound to different DNA ends (3). At this state, the ATPase is turned on and unleashes the ACNase (4). It is also assumed that zinc ions released from the intramolecular ZH could be trapped by the intermolecular.
RloC’s internal ACNase switch may confer advantages
The advent of an internal ACNase silencer could have provided RloC with important advantages over PrrC, added to the assumed frustration of phage-induced tRNA repair. First, an internal ACNase silencer could free RloC from dependence on PrrC’s external silencer; namely, the R–M system to which RloC is only rarely linked. It is noteworthy that rloC often appears as the single cargo of integrated phage, transposon or plasmid elements (4), suggesting it can be laterally inherited without a linked silencer. The advent of an internal silencer could have compensated for this deficiency, enabling such transfer without damage to the recipient cell. Nonetheless, we cannot exclude that an R–M protein interacting in trans partakes in the activation of GkaRloC’s ACNase, e.g. relaying foreign DNA and/or genotoxic signals transduced by anti-DNA restriction (9) and/or DNA restriction alleviation (32,44) factors.
Second, RloC’s internal silencer could have superseded the dTTP gauging device of its assumed PrrC-like progenitor. PrrC’s sensitivity to dTTP’s level helps confine the toxicity of this ACNase to its viral target by co-opting the phage-induced dTTP accretion to stabilize its activated form and, possibly precluding potential cytotoxicity of any free PrrC molecules translated in excess over, or inadvertently released from the silencing R–M partner (20). In contrast, the activated form of free RloC is likely stabilized by binding a purine NTP rather than dTTP (Figure 4C and E). Moreover, since RloC lacks overt ACNase activity, it need not be inactivated in the uninfected cell. The indifference to dTTP’s level could expand the range of RloC’s viral targets to include phage that do not induce dTTP’s accretion.
RloC’s evolution
RloC’s evolution from a PrrC-like progenitor is favoured to the converse by the greater complexity of RloC and the above-mentioned selective advantages that could have enhanced its antiviral potential and distribution among bacteria. Moreover, comparing matching regions of Photorhabdus luminescens PrrC and E. coli APECO1 RloC (Figure 8A) suggests that a consensus sequence implicated with PrrC’s unusual nucleotide specificity (PrrC Box; 15 and our unpublished data) degenerated in RloC (Figure 8B). Another determinant associated with PrrC’s unusual nucleotide specificity, the 3′ terminal Arg of PrrC’s Walker A motif (marked by arrow) (45) is missing from RloC. An alternative scenario where RloC arose by fusion of an ACNase domain with the bacterial Rad50 homolog SbcC seems less likely since the two ACNases resemble each other in ATPase motifs more than SbcC or any other known ABC-ATPases.
Figure 8.
RloC’s putative degenerated PrrC Box. (A) Sequence alignment of Photorhabdus luminescence PrrC and parts of E. coli APEC01 RloC flanking the CC/ZH domain. (B) The aligned consensus PrrC Box and putative degenerated counterpart of RloC were derived from respective ortholog cohorts of bacteria listed in Supplementary Table S2.
RloC’s biological role
The idea that RloC responds to genotoxic stress by disabling translation (4) appears to contradict the requisite synthesis of DNA repair proteins (46). Yet, such response could benefit bacteria that alleviate Type I DNA restriction during recovery from DNA damage (31). This measure precludes degradation of fully unmodified cell DNA made during the recovery (44) but at the cost of increased susceptibility to phage infection (32). Activating RloC’s ACNase in this situation could prevent the spread of the infection to vulnerable sibling cells. This model, inferred from the harsh lesion inflicted by RloC, the ACNase-activating ZH mutations (4) and Rad50’s functions (26,27) was reinforced by observations indicating that GkaRloC’s ACNase is regulated by its coupled ZH and DNA-dependent ATPase, the importance of DNA termini but failure of DSBs alone to detectably activate an RloC ACNase in vivo. Testing this model and the expectation that RloC defies phage-induced tRNA repair call for experimental systems based on natural RloC expressing hosts and cognate phage endowed with tRNA repair enzymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
The Israel Science Foundation—Jerusalem (to G.K.); United States–Israel Binational Science Foundation (to G.K.); the Israeli Ministry of Science, Israel–Taiwan cooperation (to G.K.). Funding for open access charge: Israel Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Anders S. Byström for the GST-zymocin plasmid, Darrell R. Davis for Lys3–ASL and Ezra Yagil for comments.
REFERENCES
- 1.Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 2.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanga-Kanfi S, Amitsur M, Azem A, Kaufmann G. PrrC-anticodon nuclease: functional organization of a prototypical bacterial restriction RNase. Nucleic Acids Res. 2006;34:3209–3219. doi: 10.1093/nar/gkl415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidov E, Kaufmann G. RloC: a wobble nucleotide-excising and zinc-responsive bacterial tRNase. Mol. Microbiol. 2008;69:1560–1574. doi: 10.1111/j.1365-2958.2008.06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depew RE, Cozzarelli NR. Genetics and physiology of bacteriophage T4 3′-phosphatase: evidence for the involvement of the enzyme in T4 DNA metabolism. J. Virol. 1974;13:888–897. doi: 10.1128/jvi.13.4.888-897.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann G. Anticodon nucleases. Trends Biochem. Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]
- 8.Tyndall C, Meister J, Bickle TA. The Escherichia coli prr region encodes a functional type IC DNA restriction system closely integrated with an anticodon nuclease gene. J. Mol. Biol. 1994;237:266–274. doi: 10.1006/jmbi.1994.1230. [DOI] [PubMed] [Google Scholar]
- 9.Penner M, Morad I, Snyder L, Kaufmann G. Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 1995;249:857–868. doi: 10.1006/jmbi.1995.0343. [DOI] [PubMed] [Google Scholar]
- 10.Sirotkin K, Cooley W, Runnels J, Snyder L. A role in true-late gene expression for the T4 bacteriophage 5′-polynucleotide kinase 3′-phosphatase. J. Mol. Biol. 1978;123:221–233. doi: 10.1016/0022-2836(78)90322-4. [DOI] [PubMed] [Google Scholar]
- 11.Meineke B, Schwer B, Schaffrath R, Shuman S. Determinants of eukaryal cell killing by the bacterial ribotoxin PrrC. Nucleic Acids Res. 2011;39:687–700. doi: 10.1093/nar/gkq831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meineke B, Shuman S. Determinants of the cytotoxicity of PrrC anticodon nuclease and its amelioration by tRNA repair. RNA. 2012;18:145–154. doi: 10.1261/rna.030171.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galburt EA, Pelletier J, Wilson G, Stoddard BL. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure. 2002;10:1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 14.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SF, Stammers DK. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 15.Amitsur M, Benjamin S, Rosner R, Chapman-Shimshoni D, Meidler R, Blanga S, Kaufmann G. Bacteriophage T4-encoded Stp can be replaced as activator of anticodon nuclease by a normal host cell metabolite. Mol. Microbiol. 2003;50:129–143. doi: 10.1046/j.1365-2958.2003.03691.x. [DOI] [PubMed] [Google Scholar]
- 16.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Noort J, van der Heijden T, Dutta CF, Firman K, Dekker C. Initiation of translocation by Type I restriction-modification enzymes is associated with a short DNA extrusion. Nucleic Acids Res. 2004;32:6540–6547. doi: 10.1093/nar/gkh999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg GR, He P, Hilfinger J, Tseng MJ. Deoxyribonucleoside triphosphate synthesis and phage T4 DNA replication. In: Karam JD, Drake JW, Kreuzer KN, Mosig G, Hall DW, Eiserling FA, Black LW, Spicer EK, Kutter E, Carlson K, et al., editors. Molecular Biology of Bacteriophage T4. Washington DC: American Society for Microbiology; 1994. pp. 14–27. [Google Scholar]
- 19.Sargent RG, Mathews CK. Imbalanced deoxyribonucleoside triphosphate pools and spontaneous mutation rates determined during dCMP deaminase-defective bacteriophage T4 infections. J. Biol. Chem. 1987;262:5546–5553. [PubMed] [Google Scholar]
- 20.Klaiman D, Kaufmann G. Phage T4-induced dTTP accretion bolsters a tRNase-based host defense. Virology. 2011;414:97–101. doi: 10.1016/j.virol.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Meidler R, Morad I, Amitsur M, Inokuchi H, Kaufmann G. Detection of anticodon nuclease residues involved in tRNALys cleavage specificity. J. Mol. Biol. 1999;287:499–510. doi: 10.1006/jmbi.1999.2634. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Meidler R, Amitsur M, Kaufmann G. Specific interaction between anticodon nuclease and the tRNALys wobble Base. J. Mol. Biol. 2001;305:377–388. doi: 10.1006/jmbi.2000.4282. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y, Blanga S, Amitsur M, Meidler R, Krivosheyev E, Sundaram M, Bajji AC, Davis DR, Kaufmann G. Structural features of tRNALys favored by anticodon nuclease as inferred from reactivities of anticodon stem and loop substrate analogs. J. Biol. Chem. 2002;277:3836–3841. doi: 10.1074/jbc.M110072200. [DOI] [PubMed] [Google Scholar]
- 24.Klaiman D, Amitsur M, Blanga-Kanfi S, Chai M, Davis DR, Kaufmann G. Parallel dimerization of a PrrC-anticodon nuclease region implicated in tRNALys recognition. Nucleic Acids Res. 2007;35:4704–4714. doi: 10.1093/nar/gkm494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 27.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 28.Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 2010;9:1299–1306. doi: 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, Sildas S, Hammel M, Russell P, et al. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano T. SMC proteins and chromosome mechanics: from bacteria to humans. Philos. Trans. R. Soc. Lond B Biol. Sci. 2005;360:507–514. doi: 10.1098/rstb.2004.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoms B, Wackernagel W. Genetic control of damage-inducible restriction alleviation in Escherichia coli K12: an SOS function not repressed by lexA. Mol. Gen. Genet. 1984;197:297–303. doi: 10.1007/BF00330977. [DOI] [PubMed] [Google Scholar]
- 32.Blakely GW, Murray NE. Control of the endonuclease activity of type I restriction-modification systems is required to maintain chromosome integrity following homologous recombination. Mol. Microbiol. 2006;60:883–893. doi: 10.1111/j.1365-2958.2006.05144.x. [DOI] [PubMed] [Google Scholar]
- 33.Ansaldi M, Lepelletier M, Mejean V. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal. Biochem. 1996;234:110–111. doi: 10.1006/abio.1996.0060. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Huang B, Esberg A, Johansson MJ, Byström AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amitsur M, Morad I, Kaufmann G. In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J. 1989;8:2411–2415. doi: 10.1002/j.1460-2075.1989.tb08371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundaram M, Crain PF, Davis DR. Synthesis and characterization of the native anticodon domain of E. coli tRNALys: simultaneous incorporation of modified nucleosides mnm(5)s(2)U, t(6)A, and pseudouridine using phosphoramidite chemistry. J. Org. Chem. 2000;65:5609–5614. doi: 10.1021/jo000338b. [DOI] [PubMed] [Google Scholar]
- 37.Webb BL, Cox MM, Inman RB. ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J. Biol. Chem. 1999;274:15367–15374. doi: 10.1074/jbc.274.22.15367. [DOI] [PubMed] [Google Scholar]
- 38.Reyes ED, Patidar PL, Uranga LA, Bortoletto AS, Lusetti SL. RecN is a cohesin-like protein that stimulates intermolecular DNA interactions in vitro. J. Biol. Chem. 2010;285:16521–16529. doi: 10.1074/jbc.M110.119164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lammens A, Schele A, Hopfner KP. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 40.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 41.Möckel C, Lammens K, Schele A, Hopfner KP. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007;26:867–877. doi: 10.1038/sj.emboj.7601537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt JA, Fierke CA. Selection of carbonic anhydrase variants displayed on phage. Aromatic residues in zinc binding site enhance metal affinity and equilibration kinetics. J. Biol. Chem. 1997;272:20364–20372. doi: 10.1074/jbc.272.33.20364. [DOI] [PubMed] [Google Scholar]
- 44.Makovets S, Powell LM, Titheradge AJ, Blakely GW, Murray NE. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol. Microbiol. 2004;51:135–147. doi: 10.1046/j.1365-2958.2003.03801.x. [DOI] [PubMed] [Google Scholar]
- 45.Meineke B, Shuman S. Structure–function relations in the NTPase domain of the antiviral tRNA ribotoxin Escherichia coli PrrC. Virology. 2012;427:144–150. doi: 10.1016/j.virol.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedberg EC, Walker GC, Siede W, Wood RD, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]