Abstract
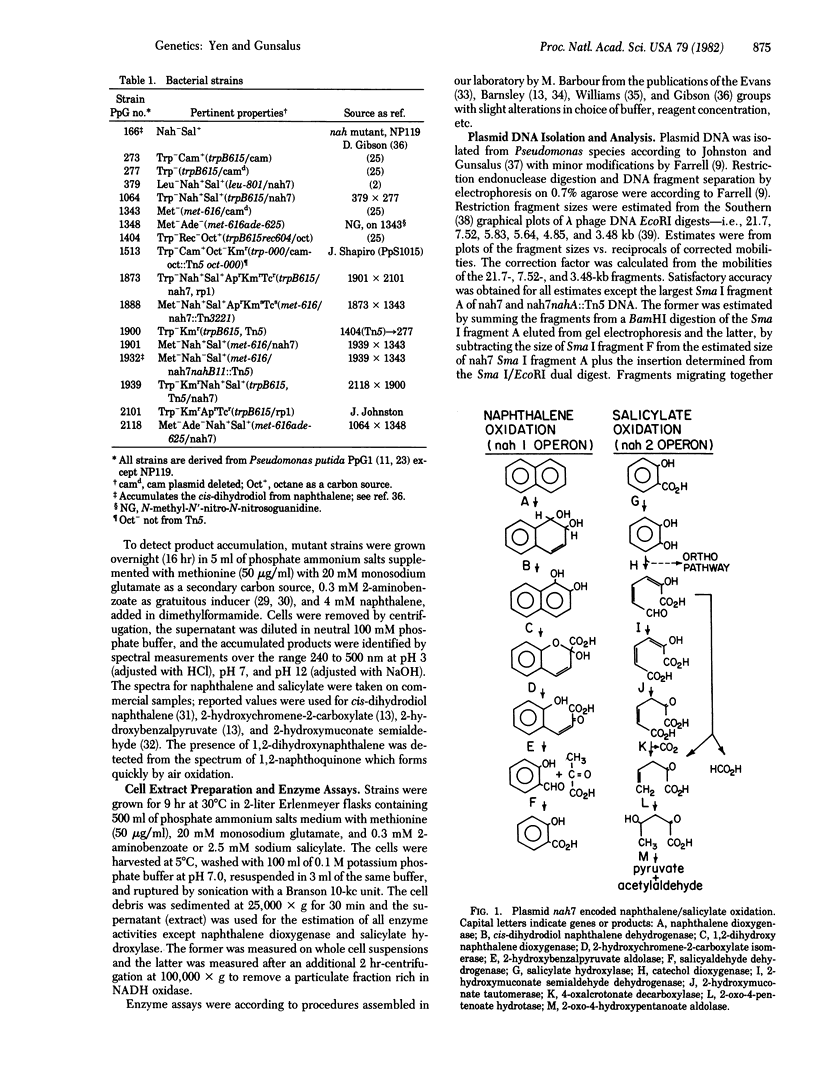
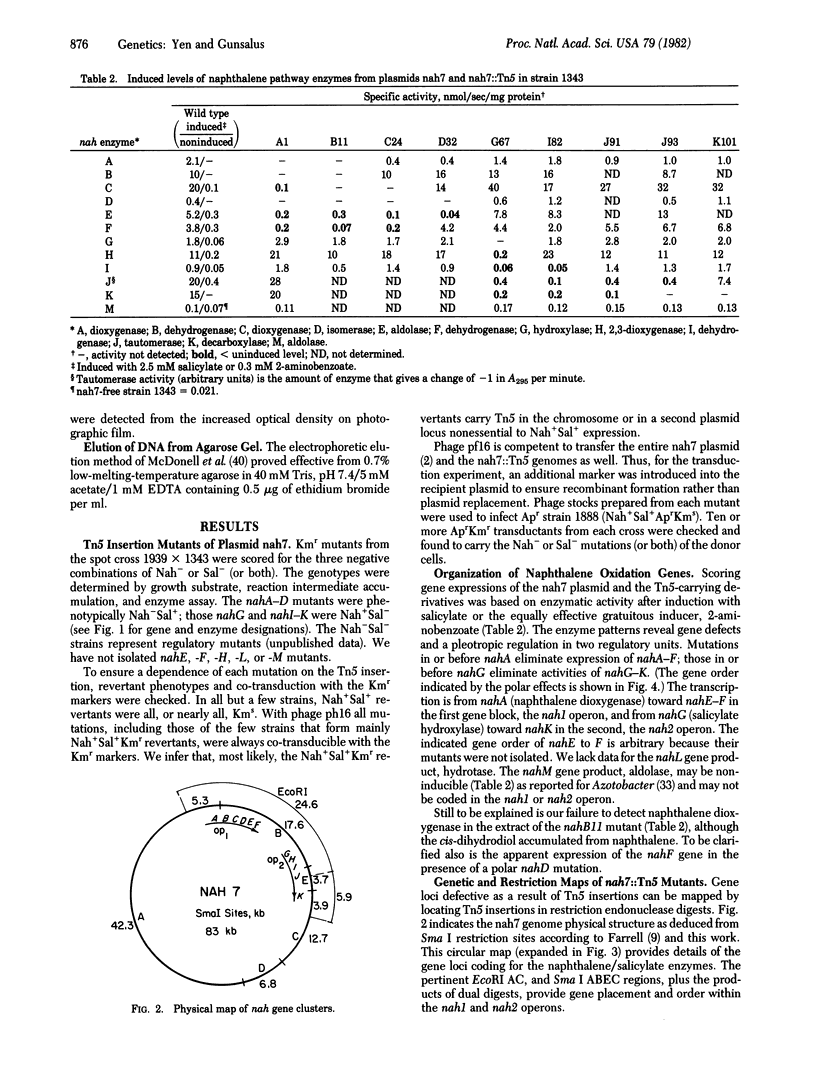
Genes for naphthalene metabolism are localized on nah7, an 83-kilobase (kb) plasmid, in two gene clusters under salicylate control. Polar mutations formed by insertion of the transposon Tn5 permit detection of the transcription direction and the gene organization within two approximately 10-kb DNA segments separated by a approximately 7-kb regulatory gene region. The gene cluster specifying conversion of naphthalene to salicylate lies near the left initiation of a 25-kb DNA fragment A released by EcoRI; that for the salicylate pathway via catechol meta-fission lies near the right terminus with extension into the adjoining 5.9-kb fragment C. The genetic organization and regulation resemble the tol plasmid-encoded "upper" and "lower" pathways of toluene/xylene oxidation in Pseudomonas putida mt2.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnsley E. A. Naphthalene metabolism by pseudomonads: the oxidation of 1,2-dihydroxynaphthalene to 2-hydroxychromene-2-carboxylic acid and the formation of 2'-hydroxybenzalpyruvate. Biochem Biophys Res Commun. 1976 Oct 4;72(3):1116–1121. doi: 10.1016/s0006-291x(76)80247-1. [DOI] [PubMed] [Google Scholar]
- Barnsley E. A. Role and regulation of the ortho and meta pathways of catechol metabolism in pseudomonads metabolizing naphthalene and salicylate. J Bacteriol. 1976 Feb;125(2):404–408. doi: 10.1128/jb.125.2.404-408.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnsley E. A. The induction of the enzymes of naphthalene metabolism in pseudomonads by salicylate and 2-aminobenzoate. J Gen Microbiol. 1975 May;88(1):193–196. doi: 10.1099/00221287-88-1-193. [DOI] [PubMed] [Google Scholar]
- Bayly R. C., Dagley S., Gibson D. T. The metabolism of cresols by species of Pseudomonas. Biochem J. 1966 Nov;101(2):293–301. doi: 10.1042/bj1010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Weiss A., Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980 May;142(2):439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A., Berg D. E., Botstein D., Lederberg E. M., Novick R. P., Starlinger P., Szybalski W. Nomenclature of transposable elements in prokaryotes. Gene. 1979 Mar;5(3):197–206. doi: 10.1016/0378-1119(79)90078-7. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M., Chou G., Gunsalus I. C. Genetic regulation of octane dissimilation plasmid in Pseudomonas. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1137–1140. doi: 10.1073/pnas.70.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol. 1972 Nov;112(2):815–823. doi: 10.1128/jb.112.2.815-823.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M., Gunsalus C. F., Gunsalus I. C. Transduction and the clustering of genes in fluorescent Pseudomonads. Proc Natl Acad Sci U S A. 1968 May;60(1):168–175. doi: 10.1073/pnas.60.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Plasmids in Pseudomonas. Annu Rev Genet. 1976;10:7–30. doi: 10.1146/annurev.ge.10.120176.000255. [DOI] [PubMed] [Google Scholar]
- Chou G. I., Katz D., Gunsalus I. C. Fusion and compatibility of camphor and octane plasmids in Pseudomonas. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2675–2678. doi: 10.1073/pnas.71.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. I., Evans W. C. Oxidative metabolism of naphthalene by soil pseudomonads. The ring-fission mechanism. Biochem J. 1964 May;91(2):251–261. doi: 10.1042/bj0910251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973 Jun;114(3):974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell R., Gunsalus I. C., Crawford I. P., Johnston J. B., Ito J. Restriction endonuclease sites and aromatic metabolic plasmid structure. Biochem Biophys Res Commun. 1978 May 30;82(2):411–416. doi: 10.1016/0006-291x(78)90891-4. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Williams P. A. Construction of a partial diploid for the degradative pathway encoded by the TOL plasmid (pWWO) from Pseudomonas putida mt-2: evidence for the positive nature of the regulation by the xyIR gene. Mol Gen Genet. 1980 Jan;177(2):321–328. doi: 10.1007/BF00267445. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Heinaru A. L., Duggleby C. J., Broda P. Molecular relationships of degradative plasmids determined by in situ hybridisation of their endonuclease-generated fragments. Mol Gen Genet. 1978 Apr 17;160(3):347–351. doi: 10.1007/BF00332979. [DOI] [PubMed] [Google Scholar]
- Hermann M., Garg G. K., Gunsalus I. C. Fertility factors in Pseudomonas putida: selection and properties of high-frequency transfer and chromosome donors. J Bacteriol. 1979 Jan;137(1):28–34. doi: 10.1128/jb.137.1.28-34.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey A. M., Yeh H. J., Jerina D. M., Patel T. R., Davey J. F., Gibson D. T. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry. 1975 Feb 11;14(3):575–584. doi: 10.1021/bi00674a018. [DOI] [PubMed] [Google Scholar]
- Johnston J. B., Gunsalus I. C. Isolation of metabolic plasmid DNA from Pseudomonas putida. Biochem Biophys Res Commun. 1977 Mar 7;75(1):13–19. doi: 10.1016/0006-291x(77)91282-7. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Patel T. R., Gibson D. T. Purification and propeties of (plus)-cis-naphthalene dihydrodiol dehydrogenase of Pseudomonas putida. J Bacteriol. 1974 Sep;119(3):879–888. doi: 10.1128/jb.119.3.879-888.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Kramer R. A., Davis R. W. Cloning of the yeast ribosomal DNA repeat unit in SstI and HindIII lambda vectors using genetic and physical size selections. J Mol Biol. 1978 Aug 15;123(3):371–386. doi: 10.1016/0022-2836(78)90085-2. [DOI] [PubMed] [Google Scholar]
- Rheinwald J. G., Chakrabarty A. M., Gunsalus I. C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci U S A. 1973 Mar;70(3):885–889. doi: 10.1073/pnas.70.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Reznikoff W. S. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191–199. doi: 10.1016/0092-8674(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Murray K., Williams P. A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972 Jul 24;28(3):347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Shamsuzzaman K. M., Barnsley E. A. The regulation of naphthalene metabolism in pseudomonads. Biochem Biophys Res Commun. 1974 Sep 23;60(2):582–589. doi: 10.1016/0006-291x(74)90280-0. [DOI] [PubMed] [Google Scholar]
- Shamsuzzaman K. M., Barnsley E. A. The regulation of naphthalene oxygenase in pseudomonads. J Gen Microbiol. 1974 Jul;83(0):165–170. doi: 10.1099/00221287-83-1-165. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Characterization of a spontaneously occurring mutant of the TOL20 plasmid in Pseudomonas putida MT20: possible regulatory implications. J Bacteriol. 1977 Jun;130(3):1149–1158. doi: 10.1128/jb.130.3.1149-1158.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga M. C., Durham D. R., Welch R. A. Plasmid- and chromosome-mediated dissimilation of naphthalene and salicylate in Pseudomonas putida PMD-1. J Bacteriol. 1981 Sep;147(3):836–843. doi: 10.1128/jb.147.3.836-843.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]