Abstract
The conserved heterodimeric endonuclease Mus81–Eme1/Mms4 plays an important role in the maintenance of genomic integrity in eukaryotic cells. Here, we show that budding yeast Mus81–Mms4 is strictly regulated during the mitotic cell cycle by Cdc28 (CDK)- and Cdc5 (Polo-like kinase)-dependent phosphorylation of the non-catalytic subunit Mms4. The phosphorylation of this protein occurs only after bulk DNA synthesis and before chromosome segregation, and is absolutely necessary for the function of the Mus81–Mms4 complex. Consistently, a phosphorylation-defective mms4 mutant shows highly reduced nuclease activity and increases the sensitivity of cells lacking the RecQ-helicase Sgs1 to various agents that cause DNA damage or replicative stress. The mode of regulation of Mus81–Mms4 restricts its activity to a short period of the cell cycle, thus preventing its function during chromosome replication and the negative consequences for genome stability derived from its nucleolytic action. Yet, the controlled Mus81–Mms4 activity provides a safeguard mechanism to resolve DNA intermediates that may remain after replication and require processing before mitosis.
INTRODUCTION
The maintenance of genome stability during chromosome replication and the fidelity of DNA synthesis are essential for cell survival and to prevent pathological cellular conditions that could lead to cancer and other diseases (1,2). Checkpoints and DNA repair pathways are key elements of the cellular response to replication perturbations that allows the preservation of genome integrity (3–8). Different specific endonucleases also contribute to genomic stability by cleaving DNA secondary structures that arise during replication-associated repair processes or during replication restart after fork blocks, thereby helping with successful chromosome replication (9,10). One of these nucleases is the heterodimeric Mus81–Eme1/Mms4 complex, which is widely conserved among eukaryotes and related to the XPF family of proteins (11,12). Mus81–Eme1/Mms4 is a structure-specific endonuclease composed of a catalytic subunit, Mus81, and a non-catalytic subunit, Eme1/Mms4 (Eme1 in mammals, Schizosaccharomyces pombe and plants; Mms4 in Saccharomyces cerevisiae and Drosophila), both of which are required for the activity of the complex (13).
Numerous studies have shown that Mus81–Eme1/Mms4 cleaves branched DNA substrates in vitro with different affinity, such as model replication forks, 3′-flaps, D-loops, Y- and X-shaped structures (13–24). All of these substrates are potential targets of this endonuclease in vivo, and, in fact, in mitotic cells Mus81–Eme1/Mms4 contributes to the processing of DNA intermediates produced during chromosome replication in the presence of DNA lesions, as mus81 or mms4 yeast mutants are sensitive to different agents that damage the DNA impairing the progression of replication forks (14,18,25–28). Moreover, this endonuclease is required during the repair of broken replication forks (29). It is also known that, in budding yeast, mus81 and mms4 mutants show synthetic lethality with mutations of the complex formed by the RecQ helicase Sgs1, Top3 and Rmi1 (BLM–TOPIIIα–RMI1–RMI2 in human cells, or BTR complex), which is suppressed by mutations that prevent the early steps of homologous recombination (13,14,30,31). The same requirement of Mus81 for cell viability in the absence of a RecQ helicase was also found in S. pombe, Drosophila and Arabidopsis (18,26,32,33). These data suggested that the RecQ and Mus81 complexes constitute alternative but overlapping ways to process joint molecules (JM) during recombination-mediated DNA repair (11). In addition, Mus81–Mms4 has also overlapping functions with the Holliday junction resolvase Yen1 during DNA repair in proliferating cells (34–37). Importantly, Mus81–Eme1/Mms4 is required even in the absence of exogenous DNA damaging agents, as the lack of this endonuclease causes high levels of chromosomal rearrangements in budding yeast (38,39), as well as various chromosome aberrations in mammalian cells (40–44). All of these data indicate that Mus81–Eme1/Mms4 substrates are produced as a consequence of induced DNA damage and also in normal cycling cells, and that their processing is necessary for preserving genome stability (11).
Despite the requirement of Mus81–Eme1/Mms4 for the maintenance of genome integrity, its nucleolytic activity during S-phase could, paradoxically, be a source of genomic instability. Thus, the uncontrolled cleavage of its potential targets, including replication forks and different DNA intermediates, would be detrimental for the completion of chromosomal replication and could induce the formation of chromosomal rearrangements. Likewise, as Mus81–Eme1/Mms4 is able to promote the formation of mitotic crossovers (35), the resolution of JM by the unregulated nucleolytic action of this complex during replication-associated DNA repair could lead to elevated levels of sister chromatid exchanges and loss of heterozygosity. Therefore, the activity of Mus81–Eme1/Mms4 should be under strict control during chromosomal replication; however, the mechanism by which cells regulate this function was largely unknown. Recently, it has been proposed that Mus81 is negatively regulated in human cells by Wee1 (45), and another recent work has also shown a regulated function of this endonuclease in the resolution of Holliday junctions during meiosis and mitosis (46). In this work, we have expanded these studies and have analysed further the regulation and function of budding yeast Mus81–Mms4 during the mitotic cell cycle. We show that its nuclease activity is inactive during S-phase, which avoids the potential cleavage of different DNA intermediates by Mus81–Mms4, and later, when the cells complete bulk DNA synthesis, the Cdc28- and Cdc5-dependent phosphorylation of the Mms4 subunit allows the activation and normal function of the Mus81–Mms4 endonuclease complex.
MATERIALS AND METHODS
Strains, media and cell cycle experiments
The yeast strains used in this work were constructed by standard techniques and are listed in Supplementary Table S1. The pML (47) and pYN (48) plasmid series were used as templates for PCR. The mms4-np mutant was obtained by transformation of an mms4Δ strain with a vector containing the PADH1-3HA-mms4-np construction (GeneArt), which was inserted into the MMS4 genomic locus. The yeast cells were grown routinely at 30°C in YP medium with 2% glucose. For expression using the GAL1–10 promoter, the YP medium was supplemented with 2% raffinose or 2% galactose. To synchronize cells in G1-phase, the α-factor mating pheromone was added to a final concentration of 5–10 µg/ml. To block cells in S-phase, hydroxyurea (HU) was used at 0.2 M. To synchronize cells in G2/M, nocodozale was used at 5 µg/ml. The percentage of binucleated cells was estimated by fluorescence microscopy, and cell visualization was as described (49). Samples for flow cytometry were processed as described (50) and analysed using a FACSCalibur flow cytometer (BD Biosciences).
Immunoblotting
Protein extracts for immunoblotting were prepared and analysed as described (49). HA-, Myc- and Tandem Affinity Purification (TAP)-tagged proteins were detected with the antibodies 12CA5 (CBMSO), 9E10 (Cancer Research UK) and PAP (Sigma-Aldrich), respectively. The anti-Cdc5 antibody (sc-6733) was from Santa Cruz Biotechnology. The HRP-coupled anti-mouse secondary antibody (Vector Labs) was used with 12CA5 and 9E10. The Horseradish Peroxidase (HRP)-coupled anti-goat secondary antibody (Santa Cruz Biotechnology) was used with anti-Cdc5. The immunoreactive bands were detected using enhanced chemiluminescence (ECL prime, GE Healthcare) according to the manufacturers’ instructions.
Phosphatase assays
Phosphorylated HA-Mms4 was immunoaffinity purified from 108 cells. The cells were disrupted using glass beads in 700 µl of lysis buffer: 40 mM Tris–HCl (pH 7.5), 100 mM NaCl, 4% glycerol, 0.1% NP40, Complete inhibitors cocktail (Roche), 8 mM EDTA, 8 mM EGTA, 2 mM benzamidine, 2 µg/ml pepstatin A, 5 mM NaF and 5 mM Na4P2O7. The extract was clarified by centrifugation after brief sonication. The supernatant was incubated with 12CA5 antibody for 180 min at 4°C, followed by 60-min incubation with 15 µl of protein G Sepharose 4 fast flow (GE Healthcare). The Sepharose-bound proteins were centrifuged, washed with PMP buffer (New England Biolabs) and used for the phosphatase assays. The reactions were carried out at 30°C for 30 min using 8 U of λ-phosphatase (New England Biolabs) in PMP buffer plus 1 mM MnCl2, 2 mM Na3VO4, 20 mM NaF and 2 mM sodium glycerophosphate, and stopped with Laemmli buffer. To inhibit λ-phosphatase, 50 mM Na3VO4, 250 mM NaF and 10 mM sodium glycerophosphate were added.
Synthetic DNA structures and nuclease activity assays
The oligonucleotides used to make the DNA substrates are listed in Supplementary Table S2. For the formation of the synthetic structures used as the substrates in the nuclease assays, the oligonucleotides 3′ FL-1, RF-1 and XO-1 were 5′-32P-labelled using [γ-32P]ATP (Perkin Elmer) and T4 Polynucleotide kinase (New England Biolabs) and then annealed with an excess of their complementary oligonucleotides. The annealing was performed by heating the DNA molecules for 10 min at 80°C in 200 mM NaCl plus 60 mM Tris–HCl (pH 7.5) buffer, followed by slow cooling to room temperature (RT).
Tagged-Mms4 was immunoaffinity purified from 7.5 × 108 cells, which were disrupted using glass beads in 800 µl of binding buffer (51). For Mms4-TAP, the supernatant was incubated for 90 min at 4°C with 15 µl of IgG Sepharose 6 fast flow (GE Healthcare). For HA-Mms4, the supernatant was incubated for 180 min at 4°C with 12CA5 antibody, followed by 60-min incubation with 15 µl of protein G Sepharose 4 fast flow (GE Healthcare). The Sepharose-bound proteins were centrifuged, washed extensively and used directly for the reactions.
Nuclease activity assays were based on a previously described method (51). The reaction mixtures (12.5 µl) contained 20 fmol of labelled DNA substrate in 100 mM NaCl (200 mM NaCl for nicked Holliday junctions, nHJs), 50 mM Tris–HCl (pH 7.5), 3 mM MgCl2 (5 mM for nHJs), 250–500 ng poly[dI-dC] plus the immunoaffinity purified Mms4 protein. The reactions were incubated for 1 h at 30°C. For RFs and 3′-FLs, the reactions were stopped with denaturing stop buffer (19% formamide, 4 mM EDTA, 0.01% xylene–cyanol and 0.01% bromophenol). For the nHJs, the reactions were stopped with 50 mM EDTA, 0.8% SDS, 2 mg/ml proteinase K, followed by incubation for 30 min at 30°C, and loading buffer (10% glycerol, 4 mM EDTA, 0.01% xylene–cyanol and 0.01% bromophenol) was added. The 32P-labelled products were analysed by 10% neutral TBE–PAGE (for nHJs), or by electrophoresis through 10% (for RFs) or 20% (for 3′-FLs) denaturing gels containing 7 M urea.
Drugs sensitivity assays
Logarithmic cultures growing in YPD medium at 30°C were normalized to 1 × 107 cells/ml, and 10-fold serial dilutions were spotted onto YPD plates with different concentrations of methyl methanesulphonate (MMS), HU, 4-nitroquinoline 1-oxide (4NQO) or cisplatin. All of the drugs were purchased from Sigma-Aldrich. The plates were incubated at 30°C for 48–72 h.
RESULTS
Mms4 is phosphorylated during the cell cycle
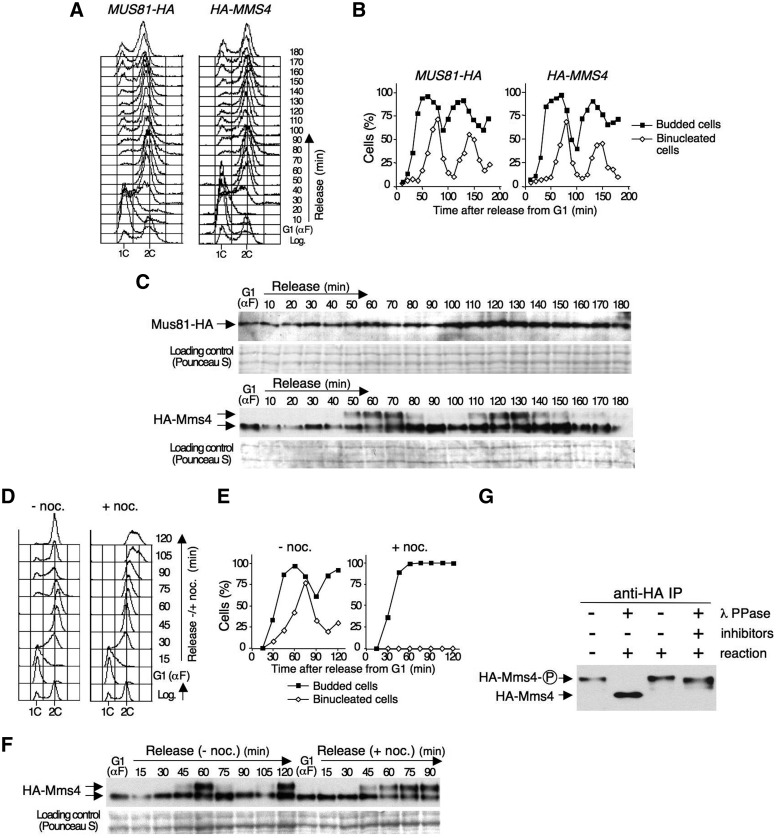
To begin to study the regulation of the Mus81–Mms4 endonuclease, we analysed the levels and possible modifications of Mus81 and Mms4 during the cell cycle, using strains in which these subunits were tagged with the HA-epitope. In each case, cycling cells were synchronized in G1 using α-factor and then released from this block in fresh medium. The cultures were allowed to progress through approximately two cell cycles, as judged by flow cytometry (Figure 1A) and the assessment of the percentage of budded and binucleated cells (Figure 1B). Proteins were prepared from cells harvested at regular intervals and analysed by immunoblot (Figure 1C). This analysis did not show any significant variations in the protein levels of the catalytic subunit, Mus81, during the experiment or changes in its electrophoretic migration (Figure 1C, upper panel). In contrast, the non-catalytic subunit, Mms4, suffered marked changes during the course of the experiment, clearly showing slower-migrating band(s) that appeared in a cell cycle-dependent manner (Figure 1C, lower panel). Thus, Mms4 underwent a shift from 50 to 70 min after release from G1, and the slow-migrating form of the protein decreased gradually afterwards to undetectable levels. The modified form of Mms4 was clearly detectable again in the second cell cycle, at 110–140 min after the release from the G1 block (Figure 1C, lower panel). These data, together with those in Figures 1A and B, indicate that Mms4 is modified at the end of S-phase or when cells enter mitosis.
Figure 1.
Mms4 undergoes cell cycle-dependent phosphorylation. (A) MUS81-HA and HA-MMS4 cells were blocked in G1 using α factor and then released from the block and monitored for approximately two cell cycles. The cells were collected at the indicated time points and the DNA content was determined by flow cytometry. (B) Percentage of budded and binucleated cells throughout the experiment. (C) Immunoblot analysis of the Mus81-HA and HA-Mms4 proteins throughout the experiment. A Pounceau S stained membrane coincident with Mus81/Mms4 migration was used as a loading control. (D) HA-MMS4 cells were blocked in G1 using α factor and then released into S-phase either in the absence or the presence of nocodazole in the medium. The cells were collected at the indicated time points, and the DNA content was determined by flow cytometry. (E) Percentage of budded and binucleated cells at each time point. (F) Immunoblot analysis of HA-Mms4 during the course of the experiment. (G) Phosphatase assay. HA-Mms4 was immunoprecipitated from extracts obtained from nocodazole-arrested cells. The protein was then incubated with or without λ-phosphatase and with λ-phosphatase plus phosphatase inhibitors prior to immunoblot analysis. Phosphorylated Mms4 is indicated as ‘Mms4-P’.
To determine more precisely the timing of the Mms4 changes, we analysed whether the modification of this protein requires the progression through mitosis or is an earlier event. To address this question, HA-MMS4 cells were synchronized in G1 using α factor and then released in a medium with or without nocodazole, a drug that blocks cells in G2/M. Flow cytometry (Figure 1D) and the estimation of the budding index and the percentage of binucleated cells (Figure 1E) indicated that the cells in both cultures entered and progressed normally through S-phase. In the absence of nocodazole, the cells continued to progress through the cell cycle; in the medium containing the drug, the cells were blocked with a 2C DNA content and remained large-budded, with single undivided nuclei, indicating a G2/M arrest. Immunoblot analysis (Figure 1F) showed that, in both cases, Mms4 was modified at 45–60 min after release from G1, when the cells reached the 2C DNA content based on flow cytometry, both in the presence and in the absence of nocodazole in the medium. In cells treated with nocodazole, the modification of Mms4 was held as a consequence of the G2/M block, unlike in untreated cells, in which the pattern was similar to that shown in Figure 1C. These results indicate that the cell cycle-dependent modification of Mms4 occurs after bulk DNA synthesis but before chromosome segregation and, therefore, is not a consequence of cells progressing to anaphase. The results also show that the modification of Mms4 is reversed during mitosis.
As the purification of Mms4 yielded a phosphorylated protein (17), it was possible that the modification of Mms4 we observed was due to phosphorylation. To test this hypothesis, Mms4 was immunoprecipitated from nocodazole-arrested cells and treated with λ-phosphatase. This treatment resulted in a shift of the modified Mms4 to the faster migrating form of the protein (Figure 1G), which was prevented by the inclusion of phosphatase inhibitors in the reaction, demonstrating that the cell-cycle dependent modification of the Mms4 subunit is due to phosphorylation.
Mms4 phosphorylation is Cdc28 (CDK)- and Cdc5 (Polo-like kinase)-dependent
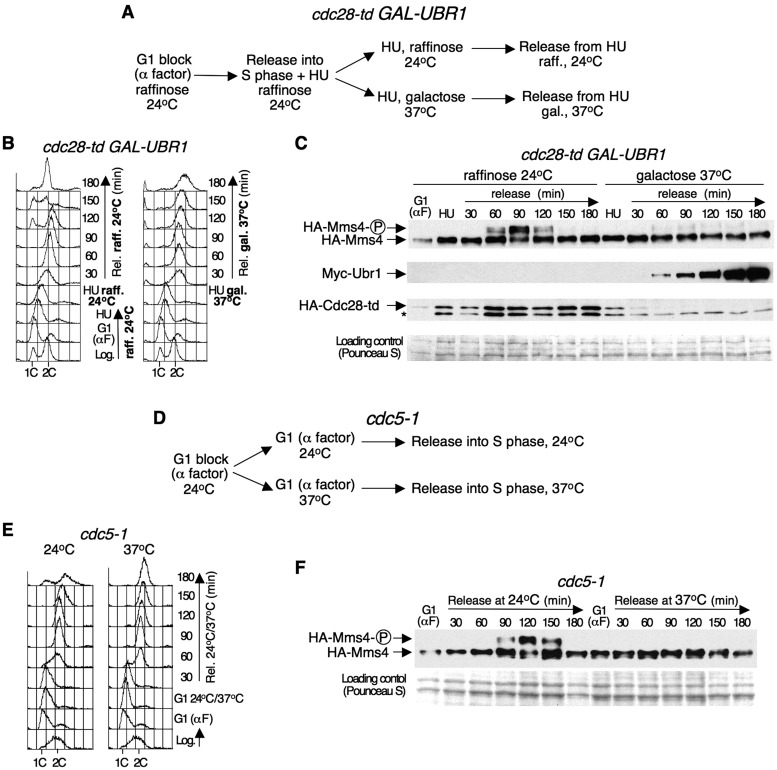
The amino acid sequence of Mms4 contains three full (T/S-P-X-K/R) and three minimal (T/S-P) Cyclin-dependent kinase (CDK) phosphorylation consensus sites (Supplementary Figure S1). In addition, Mms4 was identified as one of the possible targets of Cdk1 (budding yeast Cdc28) (52). It was, therefore, possible that Cdc28 was required for the cell cycle-dependent phosphorylation of Mms4 that we identified. To investigate this possibility, we used the heat-inducible degron strain cdc28-td PGAL1–10-UBR1. In this mutant, an NH2-terminal extension (the degron) is added to Cdc28, reducing the half-life of the fusion-protein at 37°C by proteasomal degradation (53). Moreover, UBR1 expression is controlled under the GAL1–10 promoter, allowing high levels of Ubr1 to enhance the degradation of the temperature-sensitive fusion (54). cdc28-td PGAL1–10-UBR1 HA-MMS4 cells were grown at 24°C in medium with raffinose as the carbon source, synchronized in G1 with α factor and released in medium with HU, which causes depletion of the dNTP levels. When cells are released from G1 in the presence of HU, DNA replication initiates from early firing origins and replication forks stall, causing a block in early S-phase; after this point, new replication initiation is dispensable for the completion of S-phase (55). The culture was split in two: one half was incubated under permissive conditions (medium with raffinose—GAL1–10 promoter OFF-, 24°C); in the other half, UBR1 expression was induced upon the addition of galactose after shifting the cells to 37°C, which allowed Cdc28 degradation. In both cases, the cells were released from the HU arrest (Figure 2A). Flow cytometry analysis (Figure 2B) indicated that, after the release from the HU block, the cells completed DNA replication under permissive conditions within 90 min and entered a new cell cycle. Under restrictive conditions, they also progressed and completed S-phase but, as a consequence of Cdc28 inactivation and its requirement for progression through mitosis, remained with a 2C DNA content as large-budded cells with undivided nuclei.
Figure 2.
Mms4 phosphorylation through the cell cycle is Cdc28 and Cdc5 dependent. (A) Mms4 phosphorylation depends on Cdc28. Scheme of the experiment as explained in the text. (B) cdc28-td PGAL1–10-UBR1 HA-MMS4 cells were collected at the indicated time points, and the DNA content was determined by flow cytometry. (C) Immunoblot analysis of HA-Mms4, Myc-Ubr1 and HA-Cdc28-td during the course of the experiment. The phosphorylated form of Mms4 is indicated as ‘Mms4-P’. The asterisk indicates an unrelated band that cross-reacted with the 12CA5 antibody. A Pounceau S-stained membrane was used as a loading control. (D) Mms4 phosphorylation requires Cdc5 activity. Scheme of the experiment as explained in the text. (E) The cell cycle progression of cdc5-1 HA-MMS4 cells was monitored by flow cytometry. (F) Immunoblot analysis of HA-Mms4 during the course of the experiment.
Immunoblot analysis (Figure 2C) showed that, under permissive conditions, Mms4 underwent cell cycle-dependent phosphorylation as described above (Figure 1): the modified band appeared when the cells reached the 2C DNA content, as judged by flow cytometry. Consistent with the permissive conditions, Ubr1 was not detectable, and Cdc28 was not degraded. However, at the restrictive conditions, Cdc28 was degraded by 60 min after release from the HU block, coincident with Ubr1 detection, and the slow migrating form of Mms4 was nearly undetectable (Figure 2C). These results were not due to UBR1 overexpression or incubation at 37°C, as a CDC28+ PGAL1–10-UBR1 HA-MMS4 strain used as a control showed that Mms4 was modified normally under the same experimental conditions (Supplementary Figure S2). These results indicate that the phosphorylation of Mms4 during the cell cycle depends upon the cyclin-dependent kinase Cdc28.
The window of the cell cycle in which Mms4 is phosphorylated coincides approximately with the time when the levels and activity of the Polo-like kinase Cdc5 peak in budding yeast (56,57). Indeed, in the experiment carried out with the cdc28-td mutant described above, the peak of expression of Cdc5 coincides with the timing of Mms4 phosphorylation at 24°C and occurs at the same time at 37°C (Supplementary Figure S3). Furthermore, the amino acid sequence of Mms4 contains three potential conditional docking sites for Polo-like kinases: S-pS/pT-P/X, where pS and pT are phospho-Ser and phospho-Thr, respectively (58,59) (Supplementary Figure S1). The Polo-box domain (PBD) of Polo-like kinases binds to these sites, which overlap with three of the CDK consensus sites of Mms4. Therefore, it was possible that Cdc5 was also involved in the modification of Mms4 that we observed. To study this possibility, the phosphorylation of Mms4 was analysed in the temperature-sensitive mutant cdc5-1. cdc5-1 HA-MMS4 cells growing exponentially at 24°C were synchronized in G1 with α factor, and the culture was then divided into two: one half was held in G1 at 24°C for 1 h, and the other half was maintained in G1 for 1 h at the 37°C non-permissive temperature. Afterwards, each culture was released from the G1-block at 24 or 37°C, respectively (Figure 2D). Flow cytometry analysis showed that the cells completed DNA replication between 90 and 120 min after release from G1 at both 24 and 37°C (Figure 2E). At the permissive temperature, the cells continued to progress through the cell cycle, whereas at 37°C, due to inactivation of Cdc5 and the requirement of this protein for mitosis, were arrested with a 2C DNA content and undivided chromatin. Mms4 was then analysed by immunoblot (Figure 2F). At the permissive temperature (24°C), the phosphorylated form of Mms4 was clearly detectable between 90 and 150 min after G1 release, when the cells had a 2C DNA content. However, when Cdc5 was inactivated (at the restrictive temperature), the modified band was not detected. These data indicate that, in addition to Cdc28, the Polo-like kinase Cdc5 is also required for the cell cycle-dependent phosphorylation of Mms4.
Mms4 phosphorylation correlates with the high nuclease activity of Mus81–Mms4
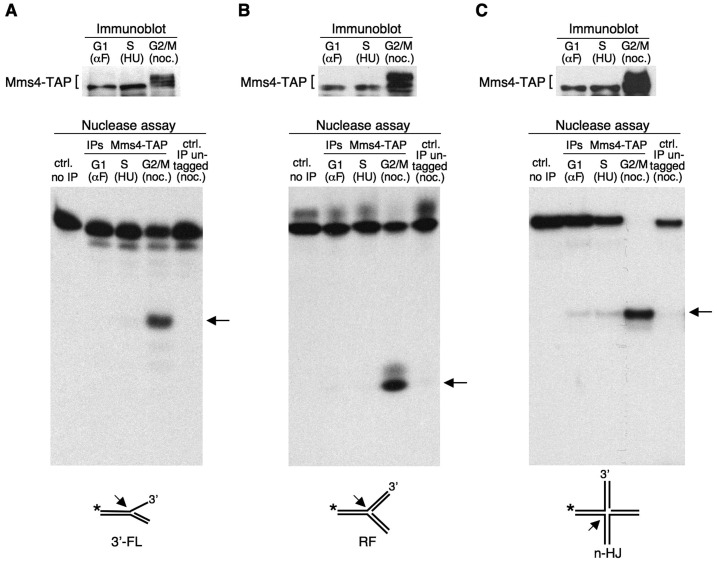
To investigate the biological significance of the cell cycle-dependent phosphorylation of Mms4, we analysed the nuclease activity of Mus81–Mms4 throughout the cell cycle. We used cells expressing TAP-tagged Mms4 (MMS4-TAP), which behaved as HA-MMS4 cells (Supplementary Figure S4). The MMS4-TAP cells were synchronized in G1 using α factor, in early S-phase using HU or in G2/M using nocodazole. As shown in the immunoblots (Figure 3, upper panels), Mms4-TAP was hyperphosphorylated in the cells blocked in G2/M, whereas the slow-migrating form of the protein was not detected in G1- or S-phase arrested cells. In each case, Mms4-TAP was immunoprecipitated from the cell extracts and the nuclease activity was tested (Figure 3, lower panels). As substrates, we used different 32P-labelled synthetic structures that can be generated under different situations and are potential targets during S-phase: a 3′-flap (Figure 3A), a model replication fork (Figure 3B) and a nHJ (Figure 3C). Although Mms4 does not have catalytic activity, it has been shown that the immunoprecipitation of Mms4-TAP from extracts prepared from asynchronous cultures can yield nuclease activity (60), indicating that the Mus81–Mms4 complex immunoprecipitates as a whole and is functional under these conditions.
Figure 3.
Mms4 phosphorylation correlates with the acquisition of nuclease activity by Mus81–Mms4. The extracts were prepared from MMS4-TAP cells synchronized in G1 with α factor, in S-phase with HU and in G2/M with nocodazole. Mms4-TAP was analysed by immunoblot (A–C, upper panel). In each case, Mms4-TAP was immunoaffinity purified from the extracts, and the nuclease activity (lower panel) was assayed by the resolution of three different 32P-labelled substrates: a 3′-flap (3′-FL) (A), a replication fork (RF) (B) and a nHJ (C). An arrow indicates the labelled-product resulting from the nucleolytic cleavage of each substrate. The controls were nuclease assays using the immunoprecipitated extracts from untagged cells blocked in G2/M or a reaction in the absence of extract.
Figure 3 (lower panels) shows that the nuclease activity of Mus81–Mms4 was markedly different at the distinct stages of the cell cycle. Thus, with the three assayed substrates, very low nuclease activity was found when Mms4 was immunoprecipitated from the extracts obtained from cells blocked in the G1- or S-phases. However, this activity was high when Mms4 was affinity-purified from the extracts obtained from cells arrested in G2/M, as determined by the appearance of clear nicked labelled DNA fragments in all cases. These results indicate a tight correlation between the phosphorylation of the non-catalytic subunit, Mms4, and the acquisition of nuclease activity by Mus81–Mms4, which allowed the cleavage of all of the assayed substrates. The data show a strict cell cycle regulation of the Mus81–Mms4 complex, which becomes active only when cells finish S-phase.
The nuclease activity of Mus81–Mms4 is reduced in a phosphorylation-defective mms4 mutant
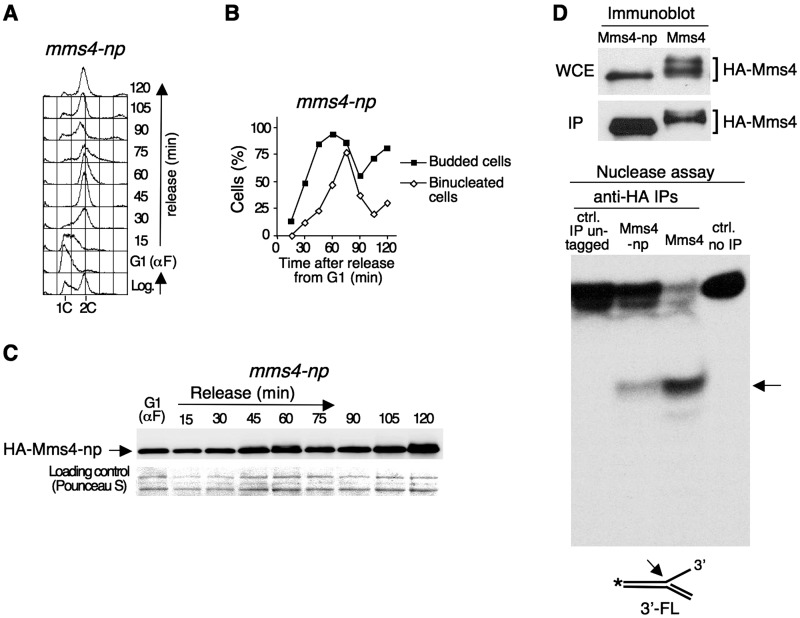
The results shown above indicate that the Cdc28/Cdc5-dependent phosphorylation of Mms4 is necessary for the regulation of Mus81/Mms4 activity. To analyse the consequences of Mms4 phosphorylation further, we constructed an mms4 mutant, mms4-np, in which all the serines and threonines at the CDK consensus sites of Mms4 and the three adjacent serines that could be part of the potential docking sites for Cdc5 were mutated to alanines (Supplementary Figure S1). To begin to characterize this mutant, mms4-np cells were synchronized in G1 with α factor and released afterwards in fresh medium. Flow cytometry analysis (Figure 4A) and the estimation of the percentage of budded and binucleated cells (Figure 4B) indicated that the mms4-np mutant behaved as the wild-type strain with regard to cell cycle progression (Figure 1). Protein samples were obtained from cells harvested throughout the experiment and analysed by SDS–PAGE and immunoblotting (Figure 4C). The immunoblot showed that, unlike wild-type Mms4, the Mms4-np mutant protein was not hyperphosphorylated during the cell cycle (Figure 1). This result reinforced the idea that Mms4 was a direct target for Cdc28/Cdc5 and provided a tool to analyse the functional significance of its phosphorylation directly.
Figure 4.
Reduced nuclease activity in a phosphorylation-defective mms4 mutant. (A) HA-mms4-np cells were blocked in G1 using α factor and released from the block in fresh medium. Cells were collected at the indicated time points and the DNA content throughout the cell cycle was monitored using flow cytometry. (B) Percentage of budded and binucleated cells during the experiment. (C) Immunoblot analysis of HA-Mms4-np during the course of the experiment. (D) Nuclease activity assay. The extracts were prepared from HA-mms4-np and HA-MMS4 cells blocked in G2/M with nocodazole. The phosphorylation of wild-type Mms4 and mutant Mms4-np in the whole cell extract (WCE), as well as the yield of the immunoprecipitation of each protein (IP) were monitored by immunoblot (upper panels). About 2% of the total amount of the immunoprecipitated protein used for the nuclease assays was loaded in each case. The nuclease activity was assayed using a 32P-labelled 3′-flap as a substrate (lower panel). An arrow indicates the labelled-product resulting from the nucleolytic cleavage. The controls were as in Figure 3.
Next, we performed a nuclease activity assay using the phosphorylation-defective mutant mms4-np (Figure 4D and Supplementary Figure S5). For this, HA-mms4-np cells were synchronized in G2/M using nocodazole, when wild-type cells have a robust Mus81–Mms4 nuclease activity (Figure 3). As shown in the immunoblot, unlike Mms4, Mms4-np was not hyperphosphorylated in G2/M, (Figure 4D, upper panel). Mms4-np was immunoprecipitated from these G2/M-arrested cells and the nuclease activity was assayed using a 32P-labelled 3′-flap as a substrate and compared to that of the wild-type MMS4 cells under the same experimental conditions. As shown in Figure 4D (lower panel), the nuclease activity of the mms4-np mutant was strongly reduced in comparison to the wild-type, as determined by the appearance of considerably less nicked-labelled product in the former. This result shows that the phosphorylation of the Mms4 subunit is required for the normal function of the Mus81–Mms4 nuclease complex.
Inability to phosphorylate Mms4 increases the sensitivity of cells lacking the RecQ-helicase Sgs1 to agents that cause DNA damage or replicative stress
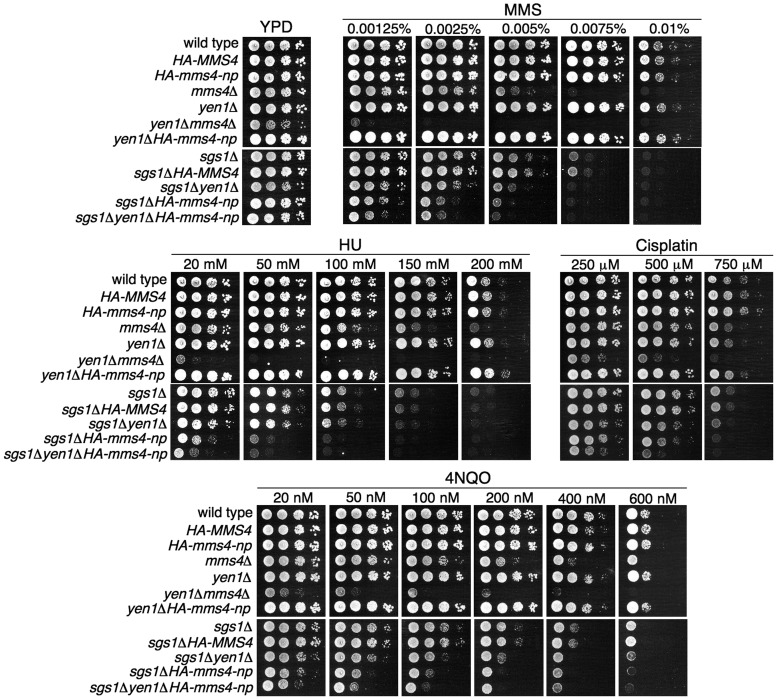
To gain further insight into the biological relevance of Mms4 phosphorylation, we studied the importance of the modification of this protein in the cellular response to different agents that perturb DNA replication. For this purpose, we analysed the sensitivity of mms4-np cells to several drugs that cause DNA damage or replicative stress and can interfere with the progression of replication forks: MMS, 4NQO, HU and cisplatin (Figure 5). Like mus81Δ, mms4Δ cells are sensitive to all of these agents, but mms4-np did not show a significant sensitivity to any of the treatments (Figure 5); unlike yen1Δmms4Δ cells, the yen1Δmms4-np mutant cells were not synthetically sick and did not show hypersensitivity to the agents assayed. These results suggest that, although the nuclease activity of Mus81–Mms4 is clearly reduced when Mms4 is not phosphorylated (Figure 4D), the remaining activity is sufficient for the resistance to the agents tested, even in the absence of the Yen1 resolvase, which cooperates with Mus81–Mms4 for DNA-damage repair (34–37).
Figure 5.
Cells lacking the RecQ-helicase Sgs1 increase the sensitivity to agents that cause DNA damage or replicative stress in the absence of Mms4 phosphorylation. Serial dilutions (10-fold) of normalized log-phase cultures of the different strains were spotted onto YPD plates containing the indicated amounts of MMS, 4NQO, HU or cisplatin and incubated at 30°C for 48–72 h. The HA-MMS4 and sgs1ΔHA-MMS4 strains, which behave like wild-type and sgs1Δ, respectively, were used as controls to show that the phenotypes of sgs1Δ-HA-mms4-np cells are not due to the tag on Mms4.
We next studied the sensitivity of the mms4-np cells to the same agents in the absence of the RecQ helicase Sgs1, with which Mus81–Mms4 functionally overlaps (13,61,62). Sgs1 is involved in the non-nucleolytic processing of some DNA intermediates that arise during replication in the presence of DNA damage or replicative stress, and is required for genome stability (63,64). Although the mus81Δ or mms4Δ mutants are synthetically lethal with sgs1Δ (30), the sgs1Δmms4-np cells were viable, suggesting that the residual activity conferred by Mms4-np is sufficient to keep them alive. This allowed a useful way to analyse the consequences of the defective function of Mus81–Mms4 for DNA repair in an sgs1Δ background. As shown in Figure 5, the sensitivity of the sgs1Δ cells to MMS, 4NQO and HU increased considerably when combined with the mms4-np mutant and moderately after cisplatin treatment. Consistent with the sensitivity of the yen1Δmms4-np and sgs1Δyen1Δ cells to these agents, the triple mutant sgs1Δyen1Δmms4-np did not show important differences with sgs1Δmms4-np. These data indicate that, although the low nuclease activity of the mms4-np mutant is sufficient to respond to DNA damage or replicative stress when Sgs1 is present, in the absence of this helicase the cells require full Mus81–Mms4 nuclease activity, for which Mms4 phosphorylation is necessary.
DISCUSSION
The structure-specific endonuclease Mus81–Eme1/Mms4 has an important role in DNA repair and the maintenance of genome integrity (10–12). However, similar to a ‘double-edged sword’, if the activity of this enzyme was uncontrolled during S-phase, it would lead to the undesired cleavage of some DNA structures during chromosome replication and, consequently, to genomic instability. Therefore, it is essential for cells to possess efficient tools to maintain the nucleolytic function of this endonuclease under strict control. In this work, we have established a regulatory mechanism for Mus81–Eme1/Mms4 that helps to understand how it functions in vivo. Using budding yeast, we have shown that Mus81–Mms4 is tightly regulated during the mitotic cell cycle by Cdc28 (CDK)- and Cdc5 (Polo-like kinase)-dependent phosphorylation of the Mms4 subunit, and that this phosphorylation is required for the nuclease activity of the complex.
We have found that, whereas Mus81, the catalytic subunit of budding yeast Mus81–Mms4, does not present significant changes throughout the mitotic cell cycle, the non-catalytic subunit, Mms4, undergoes cell cycle-dependent phosphorylation. Mms4 had been previously identified as one of the multiple possible targets of Cdc28 (52). Here, using a tight, conditional cdc28-td degron mutant, we have shown that the cell cycle-regulated modification of Mms4 we observed depends on Cdc28. Moreover, our results have also indicated that Mms4 lacking the CDK phosphorylation consensus sites cannot be phosphorylated. These data are consistent with Mms4 being a direct substrate of the cyclin-dependent kinase Cdc28. In addition, Mms4 contains potential conditional docking sites for Polo-like kinases (58), which overlap with three of the CDK phosphorylation consensus sites of this protein. Our results using a conditional cdc5 mutant have indicated that the Polo-like kinase Cdc5 is also necessary for the phosphorylation of Mms4 during the cell cycle. The involvement of Cdc5 in the modification of this protein is not merely an indirect consequence of the requirement of this kinase for the progression through mitosis, as we have shown that Mms4 is already phosphorylated in cells arrested in G2/M. Taken together, our data strongly suggest that Cdc28 phosphorylates the Mms4 subunit of the Mus81–Mms4 complex, priming this protein for the subsequent phosphorylation by Cdc5. Additionally, it is also possible that Cdc28 phosphorylates and activates Cdc5 (52,65), thereby enabling the phosphorylation of Mms4 by this kinase.
The nuclease activity assays carried out in this work have clearly shown that the cell cycle-dependent phosphorylation of Mms4 is required for the full activity of the Mus81–Mms4 complex. The results obtained indicate that Mms4 phosphorylation enables this endonuclease to cleave a variety of branched DNA structures that can be generated under different situations during chromosome replication, including replication forks. Nevertheless, our data have revealed that Mms4 phosphorylation and the subsequent Mus81–Mms4 activation only occur during a narrow window of the cell cycle, once the cells have finished bulk DNA synthesis but before they progress through mitosis. This mode of regulation prevents the nuclease activity of Mus81–Mms4 during S-phase, thus avoiding the potential problems for chromosome replication and genome stability derived from the cleavage of DNA substrates. Moreover, the regulation of this endonuclease provides an efficient safeguard mechanism to resolve, before mitosis, different DNA intermediates that cannot be processed or may escape resolution by other pathways and remain at the end of S-phase. This mechanism contributes to ensure the correct completion of chromosome duplication and the later chromosome segregation, both of which are essential for genome integrity and normal progression through the cell cycle.
The data obtained in the nuclease activity assays with the phosphorylation-defective mms4-np mutant have provided evidence of the requirement of Mms4 phosphorylation for the normal function of Mus81–Mms4. This mutant has also provided a system to analyse the consequences of defective Mus81–Mms4 activity in cells lacking the RecQ-helicase Sgs1, as the sgs1Δmms4-np double mutant is viable, whereas the mus81 or mms4 null mutants are synthetically lethal with sgs1Δ (30). The nuclease assays have shown that the mms4-np cells exhibit reduced endonuclease activity, but this residual function allows cell survival in an sgs1Δ background. Moreover, unlike mms4Δ cells, the mms4-np mutant did not show a significant sensitivity to MMS, HU, 4NQO or cisplatin in SGS1+ cells. This result suggests that the reduced nuclease activity of the mms4-np cells is sufficient for the cellular response to these agents when Sgs1 is present. However, cells lacking Sgs1 significantly increase their sensitivity to MMS, HU, 4NQO and cisplatin when Mms4 cannot be phosphorylated, indicating that, in the absence of the RecQ helicase, the low nuclease function provided by Mms4-np is not sufficient to cope with the problems originated by these drugs, which results in a hypersensitivity to them. These data indicate that a lack of RecQ-helicase activity makes the function of Mus81–Mms4 crucial, and show that Mms4 phosphorylation is required to confer full activity to the Mus81–Mms4 complex in vivo. These results also suggest that the non-nucleolytic resolution pathway mediated by the Sgs1/Top3/Rmi1 complex would be the primary choice for cells to resolve intermediates that originate during replication-associated DNA repair or fork stalling, and that Mus81–Mms4, the mode of regulation of which restricts its period of action, would operate later to cleave the unresolved DNA structures.
Our results agree with recent findings on the regulation of Mus81–Mms4 (46) and expand these studies, providing new insights into the control of Mus81–Mms4 activity during the mitotic cell cycle and its relevance for genome integrity, both under normal conditions and in response to exogenous agents that perturb DNA replication. The data presented here indicate that precise Mus81–Mms4 regulation through the cell cycle plays an essential role in preventing genomic instability, a hallmark of cancer (66). As Mus81–Mms4 is evolutionarily conserved, it would be interesting to study whether some tumour cells show uncontrolled Mus81 nuclease activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
Spanish Ministry of Science and Innovation [Grants BFU2010-16989 and Consolider Ingenio CSD2007-00015]; Fundación Ramón Areces (Institutional Grant to the Centro de Biología Molecular Severo Ochoa); Universidad Autónoma de Madrid (predoctoral fellowship to M.G.-F.); Spanish Ministry of Science and Innovation (predoctoral fellowships to M.A.O.-B. and M.V.V.). Funding for open access charge: Spanish Ministry of Science and Innovation [Grants BFU2010-16989 and Consolider Ingenio CSD2007-00015].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Benito Baños, Miguel de Vega and Luis Blanco for advice on the formation and resolution of DNA synthetic structures, John Diffley for providing yeast strains and Pedro San Segundo for the antibody against Cdc5. We also thank Crisanto Gutiérrez and Sergio Moreno for useful comments on the article.
REFERENCES
- 1.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell. Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 3.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell. Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 4.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair. 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biol. Cell. 2009;101:617–627. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- 7.Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair. 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair. 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Marti TM, Fleck O. DNA repair nucleases. Cell. Mol. Life Sci. 2004;61:336–354. doi: 10.1007/s00018-003-3223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 13.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 16.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 19.Taylor ER, McGowan CH. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc. Natl Acad. Sci. USA. 2008;105:3757–3762. doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciccia A, Constantinou A, West SC. Identification and characterization of the human Mus81-Eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 21.Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 22.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human MUS81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 24.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, III, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 25.Xiao W, Chow BL, Milo CN. Mms4, a putative transcriptional (co)activator, protects Saccharomyces cerevisiae cells from endogenous and environmental DNA damage. Mol. Gen. Genet. 1998;257:614–623. doi: 10.1007/s004380050689. [DOI] [PubMed] [Google Scholar]
- 26.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer HD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Interthal H, Heyer HD. MUS81 encodes a novel Helix-hairpin-Helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 28.Kai M, Boddy MN, Russell P, Wang TS. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 2005;19:919–932. doi: 10.1101/gad.1304305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartung F, Suer S, Bergmann T, Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRECQ4A. Nucleic Acids Res. 2006;34:4438–4448. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trowbridge K, McKim K, Brill SJ, Sekelsky J. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics. 2007;176:1993–2001. doi: 10.1534/genetics.106.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco MG, Matos J, Rass U, Ip SC, West SC. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair. 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tay YD, Wu L. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J. Biol. Chem. 2010;285:11427–11432. doi: 10.1074/jbc.M110.108399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agmon N, Yovel M, Harari Y, Liefshitz B, Kupiec M. The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage. Nucleic Acids Res. 2011;39:7009–7019. doi: 10.1093/nar/gkr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair. 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 42.Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell. Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiyama T, Katsura M, Yoshihara T, Ishida M, Kinomura A, Tonda T, Asahara T, Miyagawa K. Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells. Nucleic Acids Res. 2006;34:880–892. doi: 10.1093/nar/gkj495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domínguez-Kelly R, Martín Y, Koundrioukoff S, Tanenbaum ME, Smits VAJ, Medema RH, Debatisse M, Freire R. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 2011;194:567–579. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory Control of the Resolution of DNA Recombination Intermediates during Meiosis and Mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 48.Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez MV, Rojas V, Tercero JA. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair. 2008;7:1693–1704. doi: 10.1016/j.dnarep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Labib K, Diffley JFX, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell. Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 51.Rass U, West SC. Synthetic junctions as tools to identify and characterize Holliday junction resolvases. Methods Enzymol. 2006;408:485–501. doi: 10.1016/S0076-6879(06)08030-X. [DOI] [PubMed] [Google Scholar]
- 52.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 53.Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- 54.Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 55.Bousset K, Diffley JFX. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng L, Hunke L, Hardy CFJ. Cell cycle regulation of the Saccharomyces cerevisiae Polo-like kinase Cdc5p. Mol. Cell. Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 58.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 59.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 60.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 61.Hickson ID, Mankouri HW. Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell Cycle. 2011;10:3078–3085. doi: 10.4161/cc.10.18.16919. [DOI] [PubMed] [Google Scholar]
- 62.Ashton TM, Mankouri HW, Heidenblut A, McHugh PJ, Hickson ID. Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 2011;31:1921–1933. doi: 10.1128/MCB.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cobb JA, Bjergbaek L. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 2006;34:4106–4144. doi: 10.1093/nar/gkl557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortensen EM, Haas W, Gygi M, Gygi SP, Kellogg DR. Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr. Biol. 2005;15:2033–2037. doi: 10.1016/j.cub.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 66.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell. Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.