Abstract
Double-strand breaks (DSBs) are repaired by two distinct pathways, non-homologous end joining (NHEJ) and homologous recombination (HR). The endonuclease Artemis and the PIK kinase Ataxia-Telangiectasia Mutated (ATM), mutated in prominent human radiosensitivity syndromes, are essential for repairing a subset of DSBs via NHEJ in G1 and HR in G2. Both proteins have been implicated in DNA end resection, a mandatory step preceding homology search and strand pairing in HR. Here, we show that during S-phase Artemis but not ATM is dispensable for HR of radiation-induced DSBs. In replicating AT cells, numerous Rad51 foci form gradually, indicating a Rad51 recruitment process that is independent of ATM-mediated end resection. Those DSBs decorated with Rad51 persisted through S- and G2-phase indicating incomplete HR resulting in unrepaired DSBs and a pronounced G2 arrest. We demonstrate that in AT cells loading of Rad51 depends on functional ATR/Chk1. The ATR-dependent checkpoint response is most likely activated when the replication fork encounters radiation-induced single-strand breaks leading to generation of long stretches of single-stranded DNA. Together, these results provide new insight into the role of ATM for initiation and completion of HR during S- and G2-phase. The DSB repair defect during S-phase significantly contributes to the radiosensitivity of AT cells.
INTRODUCTION
Exposure of cells to ionizing radiation (IR) induces a broad spectrum of DNA damage including double-strand breaks, which are potentially lethal but can also lead to genomic instability, thereby increasing the cancer risk for the whole organism. Thus, a complex DNA damage response evolved to coordinate DNA repair, cell cycle regulation and eventually cell death for preventing those consequences. The key player of the radiation-induced damage response is the PI3-kinase-like kinase Ataxia-Telangiectasia Mutated (ATM), which is mutated in individuals suffering from the human syndrome Ataxia-telangiectasia (AT) (1). AT is a most severe neurodegenerative disorder that is associated with immunodeficiency, early cancer proneness and limited life span (2). Importantly, ATM is involved in double-strand break (DSB) recognition and activation of the appropriate signalling cascade, via phosphorylation of the histone component H2AX, recruitment and stabilization of the adaptor molecules MDC1 and 53BP1. ATM-mediated signalling contributes to the activation of downstream repair and cell cycle checkpoint proteins such as NBS1, SMC1, Chk2, p53 and BRCA1. Accordingly, ATM deficiency leads to abrogation of the cell cycle checkpoints G1/S, G2/M and intra-S (3–6), resulting in the most characteristic phenotype of radioresistant DNA synthesis (RDS), the failure to reduce the rate of DNA synthesis in response to IR (7,8). RDS is caused when the Chk2-p53-p21 axis is not properly activated, and by a failure to delay origin firing or to interrupt ongoing replication after IR (the intra-S-phase checkpoint). Besides checkpoint activation, ATM is also implicated in DSB repair. Accordingly, loss of ATM precludes successful rejoining of a small fraction of DSBs, which is considered to be responsible for the greatly enhanced radiosensitivity of AT cells (9–12).
Recently, it was demonstrated that Artemis, the nuclease defective in RS-SCID (13), displays partly overlapping features with AT with respect to DSB repair. In contrast to ATM, Artemis appears not to be implicated in direct checkpoint activation (5). However, it was suggested that ATM/ATR-catalyzed phosphorylation of Artemis facilitates the recovery from G2-block through regulation of CyclinB/Cdk1 activation (14,15). Biochemically, the Artemis protein is an exo- and endonuclease critically involved in the resolution of hairpin DNA structures (16), which are regular intermediates of V(D)J recombination. In addition, Artemis is capable of removing modified DNA termini such as radiation-induced phosphoglycolates (17). Although ATM and Artemis display defined enzymatical differences, both ATM- and Artemis-deficient cells share similar hypersensitivity to IR. Furthermore, epistatic and kinetic analyses of DSB repair revealed that ATM and Artemis might act in the same pathway, which includes the Mre11/Nbs1/Rad50 complex (MRN) and 53BP1 (12,18). Recently, this pathway was linked to the repair process in heterochromatic regions of the genome (18–20). Defects in ATM and/or Artemis affect the slow component of DSB repair in G1/G0, which led to the conclusion that mainly non-homologous end joining (NHEJ) is concerned. However, solid evidence was provided that ATM is also involved in homologous recombination (HR) (21–25). Very recently, ATM, Artemis, BRCA2 and Rad51 were placed in the same HR pathway that is required for the repair of 10–15% of DSBs in the G2-phase (20). Relaxation of heterochromatic DNA, i.e. by Kap1 depletion, rendered ATM and Artemis dispensable for the repair, suggesting that ATM- and Artemis-dependent HR is responsible for DSB repair in heterochromatic regions in G2. Significant recombination activity also takes place during the S-phase to cope with directly induced DSBs as well as replication-associated DSBs. However, it is not known whether and how ATM and Artemis are involved in HR-directed repair during the S-phase.
Here, we investigated DSB repair, in particular HR, in the context of the cell cycle in AT and Artemis cells. In those cells, we found similar repair phenotypes in G2, confirming a common defect in the HR pathway. However, we observed strikingly different repair phenotypes during the S-phase. Artemis cells showed no HR defect during replication particularly with regards to Rad51 foci kinetics, indicating that the nuclease Artemis is not essential for HR in S-phase. AT cells, in contrast, displayed a pronounced HR defect with significant accumulation of Rad51 foci, which fail to decline later suggesting an initiated but incomplete recombination process during replication in the absence of ATM.
MATERIALS AND METHODS
Cells
The human fibroblast cell lines (kindly provided by P.A. Jeggo) 1BR.3 (wild-type), AT1BR (ATM deficient), FO2-385 and CJ179 (Artemis deficient) were grown in alpha-medium (Gibco-Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum (FCS). All of the utilized human fibroblast cell lines displayed a similar proliferation rate with population doubling times 26.5, 27.2, 24.4 and 27.8 h, respectively. The human cervical carcinoma cell lines HeLa and HeLa-pGC (containing the gene conversion substrate) were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco-Invitrogen) supplemented with 10% FCS. CV-1, an African green monkey kidney cell line, was grown in DMEM supplemented with 5% FCS.
Small interfering RNA and inhibitors
Rad51, Artemis, ATM and control (scrambled, CyclophilinB, GAPDH) small interfering RNA (siRNA) oligonucleotides were obtained from Dharmacon SMARTpool (Artemis: GUACGGAGCCAAAGUAUAA, GCACAACUAUGGAUAAAGU, UGAAUAAGCUAGACAUGUU, CACCAAAGCUUUUCAGUGA; ATM: GCAAAGCCCUAGUAACAUA, GGUGUGAUCUUCAGUAUAU, GAGAGGAGACAGCUUGUUA, GAUGGGAGGCCUAGGAUUU). ATR siRNA was obtained from MWG Biotech (ATR: AAGCCAAGACAAAUUCUGUGU). HiPerFect transfection reagent (Qiagen) was used for the knock down in HeLa. Cells were incubated with the siRNA complexes twice (first time with 25 nM, second time after 48 h with 50 nM). CV-1 cells were transfected using Roti Fect Plus (Roth) transfection reagent. ATM and Chk1 were chemically inhibited using 10 µM KU55933 (Biozol) and 0.1 µM UCN-01 (Sigma), respectively.
DSB-repair reporter assay for gene conversion
To induce DSBs, HeLa cells containing the stably integrated reporter construct for gene conversion pGC (26) were transfected with the I-SceI expression vector pCMV3xnls-I-SceI (2 µg) using LipoFectamin2000 (Invitrogen) as transfection reagent. Seventy-two hours after transfection the cells were assessed for green fluorescence by flow cytometry (FACScan, BD Bioscience). Treatment with siRNA (50 nM) for protein depletion had been started 48 h prior to DSB induction. A second boost of siRNA was transfected together with the I-SceI expression plasmid.
Western blotting
Western blot analyses were performed with whole-cell extracts using the freeze and thaw method for lysis (27). Proteins were resolved by gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (4–15%), transferred onto a polyvinyl difluoride (PVDF) membrane and probed with the following primary antibodies: anti-Artemis (Novus Biologicals), anti-ATM (Epitomics), anti-Rad51 (Abcam), anti-pChk2, anti-pCHk1, anti-Chk1 (Cell Signaling) and anti-β-actin (Sigma–Aldrich). Bound primary antibodies were visualized with horseradish peroxidase -conjugated anti-rabbit and anti-mouse antibodies (Amersham).
Immunofluorescent microscopy
Cells were grown on coverslips, fixed with 2% formaldehyde (Merck) in phosphate-buffered saline (PBS) for 10 min and washed with PBS three times. Fixed cells were permeabilized (5 min on ice) with 0.2% Triton X-100 (Serva) in 1% bovine serum albumin (BSA, PAA)-PBS and blocked with 3% BSA for 1 h. In addition to DNA staining with 4′,6-diamidin-2-phenylindol (DAPI), the respective proteins were visualized with anti-Rad51 (Abcam), anti-γ-H2AX (Cell Signaling), anti-Cenp-F (Lifespan Biosciences) and secondary anti-mouse AlexaFluor594 (Invitrogen) or anti-rabbit fluorescein (Amersham) antibodies. Slides were mounted in Vectashield mounting medium (Vector Laboratories). Fluorescent microscopy was performed using the Zeiss AxioObserver.Z1 microscope (objectives: EC PlnN 40×/0.75 DICII, resolution 0.44 µm; PlnN Apo 63×/1.4 Oil DICII, resolution 0.24 µm; EC PlnN 100×/1.3 Oil DICII, resolution 0.26 µm and filters: Zeiss 43, Zeiss 38, Zeiss 49). Semi-confocal images were obtained using the Zeiss Apotome, Zeiss AxioCam MRm and Zeiss AxioVision Software.
Cell cycle analysis
For flow cytometry, cells were harvested and fixed with 80% ethanol (−20°C). The fixed cells were washed with PBS and DNA was stained with PI solution containing RNase A. Cell cycle distribution was monitored by flow cytometry (FACScan, BD Bioscience) and analyzed by the ModFit software (Verity Software House).
To monitor cell cycle distribution by fluorescence microscopy, cells were incubated with 5-ethynyl-2′-deoxyuridine (EdU, 1:2000; Click-iT Assay Kit, Invitrogen) for 24 h and then fixed and stained for EdU (following the manufacturer’s protocol) and additionally for centromeric protein F (Cenp-F) and DNA (DAPI). Differentially stained sub-fractions of the entire cell population were enumerated by eye. DNA repair in irradiated S-phase cells was monitored by pulse-labeling the cells with EdU 30 min before irradiation and only EdU-positive cells were evaluated. Additional Cenp-F staining was performed to identify G2-phase cells. For evaluating cells in G2, progression of EdU-pulse labeled S-phase cells into G2 was blocked by 5 µM aphidicolin (Sigma–Aldrich). EdU-positive cells were now excluded from analysis, monitoring Cenp-F-positive and accordingly G2-phase cells only.
Colony formation assay
For colony formation, cells were seeded and allowed to adhere before drug treatment or irradiation. The specific ATM inhibitor (10 µM KU55933) was added 30 min prior to irradiation (200 keV, 15 mA, additional 0.5 mm Cu filter at a dose rate of 0.8 Gy/min) and kept in the medium for 24 h. To temporarily block replication, cells were exposed to 5 µM aphidicolin for 5–6 h, commencing 30 min before IR. Cells were then incubated in drug-free medium for colony formation and stained with crystal violet. Colonies of 50 cells or more were counted manually and survival curves were derived from triplicates of at least three independent experiments.
Graphs and statistics
If not stated otherwise, experiments were independently repeated at least three times. Data points represent the mean (±SEM) of all individual experiments.
Statistical analysis, data fitting and graphics were performed with the GraphPad Prism 5.0 program (GraphPad Software).
RESULTS
AT and Artemis cells share hyper-radiosensitivity and repair defects but display important differences in the cell cycle response
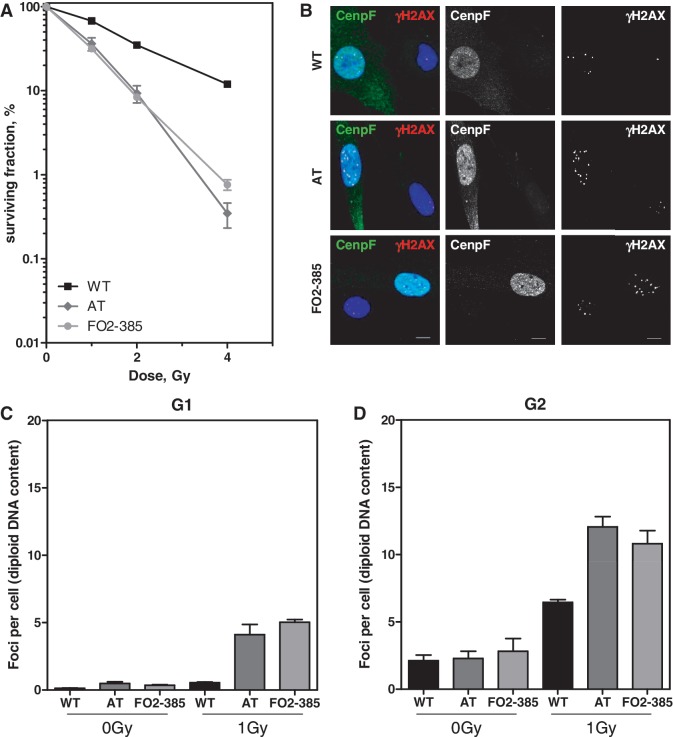
The colony formation assay was applied to confirm hypersensitivity of ATM and Artemis-deficient human fibroblasts. As expected, radiation-induced cell death of AT and Artemis cells was largely enhanced when compared with normal wild-type (WT) cells. The degree of radiosensitivity was surprisingly similar (Figure 1A), although ATM and Artemis proteins have widely divergent biochemical functions with respect to cell cycle regulation and DNA repair.
Figure 1.
Similarly enhanced radiosensitivity in AT and Artemis fibroblasts due to an enhanced number of residual DSBs. (A) Radiosensitivity of exponentially growing WT, AT and Artemis cells (FO2-385) was measured by colony formation after various X-ray doses. Error bars represent the SEM of three independent experiments. (B) Differential staining for G2-phase cells (Cenp-F-positive) 24 h after irradiation with 1 Gy in addition to detection of γH2AX foci. Bars, 10 µm. Quantification of residual γH2AX foci in (C) Cenp-F-negative G1-phase cells and (D) Cenp-F-positive G2-phase cells 24 h after IR (1 Gy). S-phase cells showing a strong pan-nuclear γH2AX signal were excluded from the analysis. The AT cell line AT1BR proved to be hyperploid (1.5-fold enhanced compared with WT and Artemis cells (data not shown)). Their foci number/nucleus was normalized to a diploid DNA content.
Cell cycle distribution 24 h after IR showed a significantly reduced fraction of S-phase cells in both WT and Artemis strains, indicating a sustained G1-arrest. In contrast, AT cells predominantly accumulated in the G2-phase (Supplementary Figure 1A) although ATM is known to be required for the G2-checkpoint activation and maintenance (4,5,28). Since different repair pathways may apply, depending on the cell cycle phase, we determined the number of non-resolved γH2AX foci (Figure 1B), which correspond to the number of unrepaired DSBs in either the G1- or the G2-phase of the cell cycle 24 h after IR. To distinguish between both phases, cells were additionally stained for the G2 marker Cenp-F. Separate analyses of G1- (only DAPI-positive, Figure 1B and C) and G2-phase cells (in addition Cenp-F-positive, Figure 1B and D) revealed for AT and Artemis cells in comparison to WT cells enhanced numbers of residual γH2AX foci in both cell cycle phases. Artemis cells showed 5.0 ± 0.2 foci/nucleus in the G1- and 10.8 ± 1.0 foci/nucleus in the G2-phase compatible with the 2-fold higher DNA content. In contrast, AT cells showed 4.1 ± 0.8 foci in the G1-phase and 12.1 ± 0.8 in the G2-phase, suggesting that additional foci are formed besides those expected from doubling the DNA content. Since AT cells but not Artemis cells continue DNA synthesis after IR, new DSBs may arise during the S-phase in AT cells.
To distinguish between cells that have passed replication from those that were immediately arrested in the respective cell cycle phases, cells were incubated with the thymidine analog EdU (S-phase) and additionally stained for Cenp-F (G2-phase) (Supplementary Figure 1B, (20,29,30)). During the 24-h observation period following irradiation with 1 Gy, 11% of WT and 40% of Artemis, cells failed to incorporate EdU and therefore did not pass through the S-phase, indicating a radiation-induced primary G1-arrest (Supplementary Figure 1B, Example (1) blue bars; at 2 Gy the respective numbers were 28 and 52%). In contrast, all AT cells were EdU positive, displaying their defect in the G1-checkpoint activation. The fraction of AT cells that were in addition Cenp-F-positive increased upon irradiation (Supplementary Figure 1B, Example (3), yellow bars), confirming the accumulation of AT cells in G2 (Supplementary Figure 1A). Together these results demonstrate that after irradiation AT cells pass through replication and arrest in the following G2-phase, without being blocked in either G1- or S-phase.
AT and Artemis cells show distinct HR defects
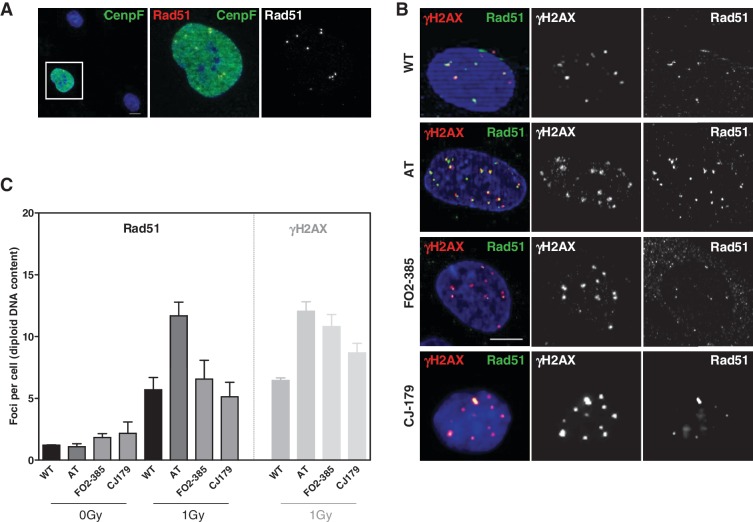
The similar AT and Artemis repair phenotype was hitherto linked to deficient repair of a common subset of DSBs (10–15%) in both the G1- and 2-phase of the cell cycle (5,12). We now observed a different amount of residual damage when the 24-h observation period included the S-phase, which may implicate HR of replication-associated damage to a yet unknown extent. This possibility was addressed by monitoring Rad51 focus formation as a marker of recombination activity 24 h after IR (Figure 2). Differential staining showed discrete nuclear Rad51 foci only in Cenp-F and/or EdU-positive cells (S- and G2-phase, respectively, see below) but never in G1 or confluent G0 cells (Figure 2A and Supplementary Figure 4) confirming that recombination processes are restricted to both the S- and G2-phase of the cell cycle. Enumeration after 1 Gy revealed in AT cells on average 11.7 ± 1.1 Rad51 foci but only 5.7 ± 0.5 and 6.6 ± 0.5 in WT and FO2-385 Artemis cells, respectively. Importantly, in WT and AT cells Rad51 and γH2AX foci were frequently co-localized giving thus identical numbers while in FO2-385 Artemis cells 39% of residual γH2AX foci were not decorated with Rad51 (Figure 2B and C). To confirm these results, we examined a second Artemis-deficient cell line, CJ179, which displayed essentially the same phenotype although on a slightly lower level (Figure 2B and C). γH2AX foci are generally considered the equivalent of unrepaired DSBs while Rad51 foci represent an early step in HR, which involves recruitment and loading of this major recombinase onto single-stranded DNA (ssDNA) (31). However, persistent Rad51 foci can also indicate non-completed recombination processes. Thus, the lower number of Rad51 foci in Artemis (compared with γH2AX foci) as well as the enhanced amount of Rad51 foci in AT cells could reflect HR defects, perhaps affecting different steps in the recombination process, respectively.
Figure 2.
The number of Rad51 foci in G2 is elevated in AT but not in Artemis cells. (A) Differential staining for G2-phase cells (Cenp-F-positive) in addition to detection of Rad51. Rad51 foci were visible only in Cenp-F-positive G2-phase cells (left panel). Magnification revealed distinct Rad51 foci (yellow as merged from green and red). The right panel depicts the red channel only as recorded in black/white. (B) Cells were co-stained for γH2AX and Rad51, which showed that in AT cells all Rad51 foci co-localized with γH2AX but not in both Artemis cell lines. Shown are cells in G2. G1 cells did not stain for Rad51 and S-phase cells with hyperintense γH2AX signals were not considered. Bars, 10 µm. (C) Quantification of Rad51 and γH2AX foci 24 h after IR revealed identical numbers for both damage markers in WT and AT but not in both Artemis cell lines.
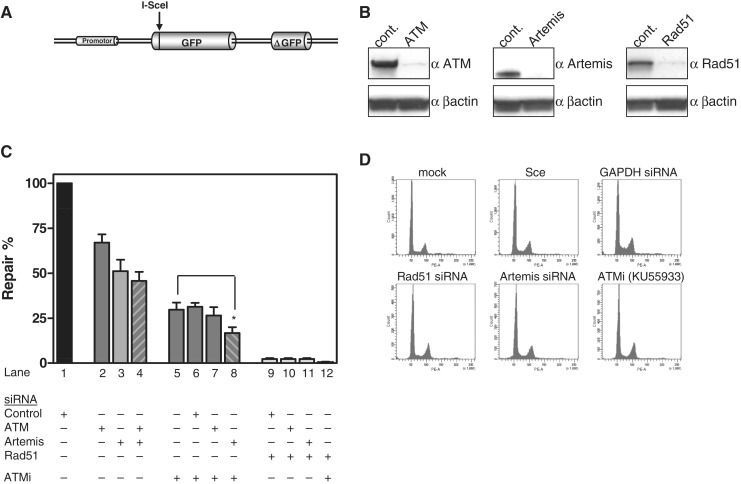
To quantify HR more directly, the pGC gene conversion reporter construct (Figure 3A, (26)) was stably integrated into HeLa cells. After DSB induction through expression of the I-SceI endonuclease, successful gene conversion led to green fluorescence in 1.9 ± 0.25% of the cells (Supplementary Figure 3A). Next, we depleted Artemis and ATM by siRNA (Figure 3B, Supplementary Figure 2A–C). Artemis siRNA reduced the HR frequency significantly to 51 ± 6% compared with control siRNA (Figure 3C). In contrast, ATM knock down diminished HR frequency to only 67 ± 5% of controls. Combining Artemis and ATM knock down only slightly enhanced the effect of Artemis siRNA alone (Figure 3C, columns 2–4). Perhaps, the siRNA treatment retained residual ATM activity, because western blots confirmed efficient, but not complete depletion of the ATM protein (Figure 3B).
Figure 3.
Reduced rate of gene conversion in the absence of ATM and Artemis. (A) Schematic structure of the GFP-based reporter construct for gene conversion (pGC). (B) Western blot of HeLa cells that have been treated with siRNA against ATM, Artemis and Rad51. siRNA anti-GAPDH or anti-CyclophyllinB were used as controls. (C) Gene conversion (GC) was determined after DSB induction using HeLa-pGC cells. Cells treated with control siRNA were transfected with the I-SceI expression vector and the fraction of GFP-positive cells was determined 72 h later by flow cytometry (control siRNA reduced GC from 1.9 to 1.4%) (Supplementary Figure 3A) and expressed as 100% relative repair efficiency (lane 1). ATM, Artemis or combined siRNA pretreatment reduced this repair efficiency (lanes 2–4). Combination of siRNAs and ATM inhibitor (10 µM of KU55933) further reduced GC efficiency (lane 5–8). Combination of Artemis siRNA and ATM inhibition (lane 8) was significantly different from ATM inhibition alone (lane 5) (Mann–Whitney two-tailed t-test, P = 0.0381). Rad51 depletion completely abrogated GC (lanes 9–12). DMSO did not affect recombination in controls (Supplementary Figure 3B). (D) Cell cycle distribution 36 h after transfection of the I-SceI expression vector did not show marked differences upon drug treatment.
The assumption was investigated by inhibiting ATM chemically using the inhibitor KU55933 (Supplementary Figure 2D–F, Supplementary Figure 3B). Inhibition of ATM decreased the HR efficiency to 29 ± 4% (Figure 3C, column 5) and this value could not be further reduced by additional ATM or control siRNA (Figure 3C, columns 6–7), supporting previous findings that the KU55933-inhibitor completely abrogated ATM function (20,32). However, combining Artemis depletion with ATM inhibition (Figure 3C, column 8) further compromised HR capacity to 17 ± 3% of controls (P = 0.038). This additive effect suggests that both proteins exert essential but partly divergent functions in HR. For comparison, we depleted Rad51, which completely abrogated gene conversion (Figure 3B and C, columns 9–12), confirming our previous observation (26). In parallel, the DNA content was monitored to exclude pure cell cycle-related effects through inhibition of ATM and Artemis (Figure 3D). Together, these results indicate that in fact ∼80% of the gene conversion activity in this system depends on ATM and Artemis.
DSBs induced during the S-phase account for different Rad51 foci numbers in AT and Artemis cells
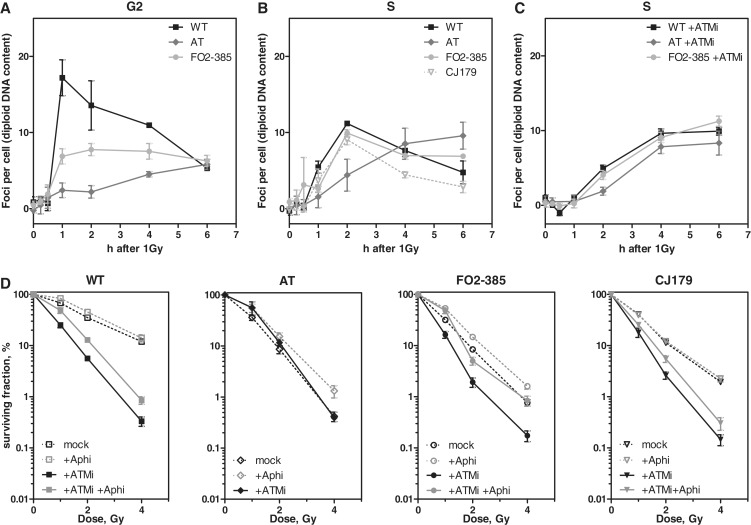
The moderate differences in gene conversion might not well match with the discrepancies of persistent Rad51 foci in AT and Artemis cells in G2 (Figure 2C). To elucidate whether Rad51 foci form during the G2-phase, we excluded S-phase cells from analysis. To this end, cells were labeled with EdU for 30 min before irradiation and a subsequent addition of aphidicolin prevented S-phase cells from entering the G2-phase (20). Rad51 focus formation in G2 was monitored in EdU-negative/Cenp-F-positive cells. WT cells showed a steep increase and a subsequent decline of the Rad51 foci number likely due to repair by HR (Figure 4A). Both AT and Artemis cells showed only a moderate increase and almost no decline of Rad51 foci with time, which confirms reduced capability of HR in both defective cell lines also observed previously (20). The increase of the foci number was remarkably delayed in AT cells. However, AT and Artemis as well as WT cells, being still in the G2-phase 6 h after IR, displayed identical numbers of Rad51 foci (Figure 4A).
Figure 4.
HR during the S-phase is dependent on ATM but not Artemis. (A) WT, AT and FO2-385 cells were EdU pulse-labeled, irradiated with 1 Gy and further incubated with aphidicoline (+Aphi) (5 µM) to block the transition of S-phase cells into G2. Kinetics of Rad51 foci numbers were monitored in Cenp-F-positive and EdU-negative G2 cells for the time interval indicated. (B) WT, AT and the Artemis cell lines FO2-385, CJ179 were EdU pulse-labeled, irradiated (1 Gy) and kinetics of Rad51 foci were evaluated in EdU-positive cells. (C) Upon ATM inhibition (10 µM KU55933), all cell lines showed similar kinetics of Rad51 foci. (D) WT, AT and both Artemis cell lines were seeded for colony formation, irradiated with 1, 2 or 4 Gy as indicated and incubated for 24 h in the presence or absence of 10 µM KU55933. Data without ATM inhibitor (black open symbols, dashed lines) were taken from Figure 1. WT and both Artemis cell lines but not AT cells were sensitized by the ATM inhibitor KU55933 (black closed symbols). Co-treatment with 5 µM aphidicolin (gray closed symbols) partially relieved this sensitization.
Different residual Rad51 foci numbers were seen in AT and Artemis cells only after they had traveled through the S-phase before being arrested in G2 (Figure 2B). Thus, we sought to elucidate whether Rad51 foci preferably appear during replication. Cells were EdU-labeled for 30 min before irradiation and nuclear Rad51 signals subsequently determined in those S-phase cells (Supplementary Figure 4A). Discrete foci were clearly detectable after 1 h reaching in WT and FO2-385 and CJ179 Artemis cells a maximum number at 2 h (11.1, 9.9 and 9.1 foci, respectively) (Figure 4B and Supplementary Figure 4A) and declined thereafter presumably again due to successful repair. Notably, the concordant Rad51 kinetics of WT and Artemis cells strongly suggest that Artemis is not required for HR during the S-phase.
Surprisingly, in AT cells the number of Rad51 foci constantly increased over the whole time period reaching 9.6 foci at 6 h (Figure 4B and Supplementary Figure 4A). This amount of foci is well comparable to the foci numbers that were recorded 24 h after IR (Figure 2B). Application of the ATM inhibitor KU55933 perfectly recapitulated the delayed but steadily increasing formation of Rad51 foci in all utilized cell lines during the S-phase (Figure 4C and Supplementary Figure 4B) as it was observed in AT cells without inhibitor. Moreover, in the presence of the ATM inhibitor, the fast increase and subsequent decline of Rad51 foci was abrogated in WT and Artemis cells indicating that HR during the S-phase is deficient in AT but not in Artemis cells. Consequently, we asked whether Artemis cells could be further sensitized by additional ATM inhibition and whether this preferably occurs in replicating cells. For this purpose, colony formation was measured in exponentially growing cultures. The ATM inhibitor decreased significantly the survival not only of WT but also of Artemis cells (Figure 4D). To verify S-phase specificity, we temporarily blocked replication progress by additional treatment with aphidicolin for 6 h after IR, corresponding to the time frame in which AT cells accumulated most Rad51 foci (Figure 4B). Strikingly, the radiosensitization through ATM inhibition was largely abrogated in WT and Artemis cells when replication was blocked (FO2-385 > CJ179) (Figure 4D). Accordingly, aphidicolin-treated AT cells also showed a moderate increase in cell survival. Together, these results indicate that HR processes during the S-phase strongly rely on ATM and that the particular deficiency during replication significantly contributes to the radiosensitive AT phenotype.
In the absence of ATM Rad51 focus formation requires functional ATR
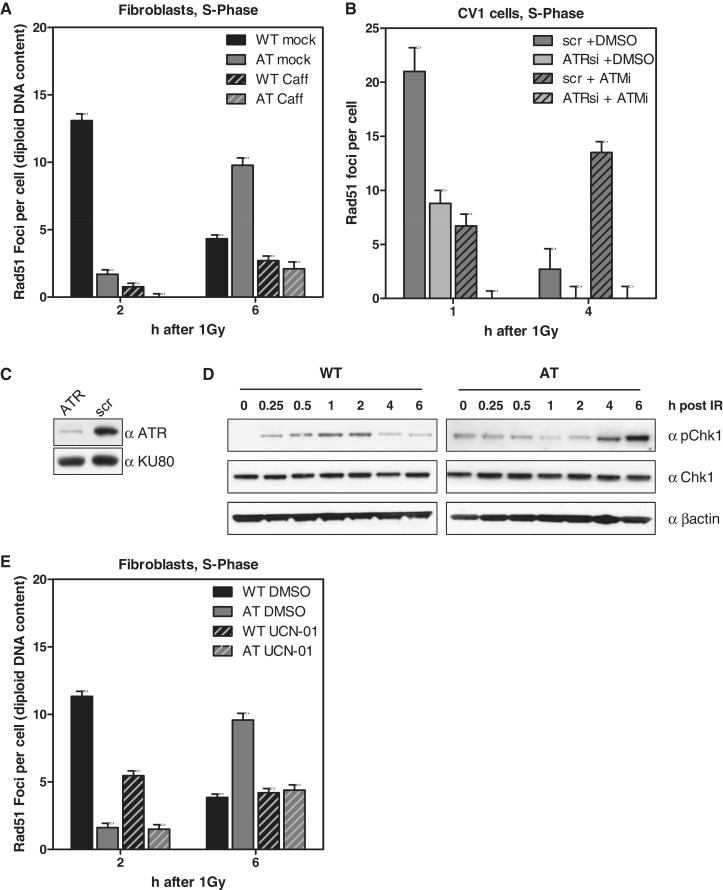
Recruitment of Rad51 to DSBs requires resection of DNA ends to generate RPA coated 3′-single-stranded overhangs. The resection process and hence the Rad51 focus formation was shown to be ATM-dependent (33–35) and may well explain that AT cells fail to accumulate Rad51 early after IR. However, the delayed but substantial generation of foci (Figure 4B) must rely on other factors. Irradiation during the S-phase induces DSBs either directly or indirectly through collision of replication forks with single-strand breaks (SSBs) leading to ‘one-ended’ DSBs. During repair after IR both types of DSBs co-exist and cannot be systematically distinguished. However, the slowly and steadily increasing number of Rad51 foci in AT cells (Figure 4B) may represent indirect DSBs accumulated during ongoing replication (i.e. RDS, see above (7,8)). To test whether this Rad51 accumulation is ATR-dependent—reminiscent of damage processing after UV-irradiation (36,37)—we treated EdU-labeled AT cells with 10 mM caffeine (38) and stained for Rad51 6 h after IR. In fact, caffeine treatment greatly diminished Rad51 focus formation to only background levels (Figure 5A, and Supplementary Figure 5A and B). Caffeine, inhibiting both the ATR and ATM kinase (38), also widely reduced Rad51 focus formation in WT cells at 2 and 6 h after IR (Figure 5A and Supplementary Figure 5A and B). To more specifically confirm that ATR is the major kinase required for Rad51 focus formation in ATM-impaired cells (34,39), we treated CV-1 cells with the ATM-specific inhibitor KU55933 and additionally depleted ATR by siRNA (Figure 5C). As expected, ATM inhibition alone slowly increased the number of radiation-induced Rad51 foci upon progression through the S-phase (Figure 5B, scr + ATMi between 1 and 4 h). Depletion of the ATR kinase alone reduced Rad51 foci to half the amount observed in mock-depleted cells (Figure 5B, i.e. 1 h after IR: light gray bar, 8.8 ± 1.2, versus dark gray bar, 21 ± 2.2 foci per cell). However, combined ATM inhibition and ATR depletion completely abolished Rad51 focus formation, well in line with the results after caffeine treatment (Figure 5A). Notably, the combined function of ATM and ATR for Rad51 focus formation during the S-phase and its inhibition by caffeine was also previously observed after thymidine-induced replication stress (40).
Figure 5.
Rad51 focus formation in AT cells depends on functional ATR. (A) WT and AT cells were pre-treated with 10 mM caffeine for 2 h and pulse-labeled with EdU shortly before IR (1 Gy). Rad51 foci were recorded 2 and 6 h later in EdU-positive cells. Not only the foci number declined upon caffeine treatment but also the size of the remaining foci was reduced (see also Supplementary Figure 5). (B) Quantification of Rad51 foci in EdU-positive CV-1 cells treated with ATR siRNA (C) and scrambled control without ATM inhibition (10 µM KU55933). (D) Western blot of Chk1 expressed in WT and AT cells irradiated with 10 Gy. (E) WT and AT cells were EdU-labeled, irradiated with 1 Gy and during repair continuously exposed to the Chk1 inhibitor UCN-01 (0.1 µM). Quantification of Rad51 foci 2 and 6 h after IR in EdU-positive cells.
ATR is the major kinase for Chk1 activation, an effector kinase, which is a critical component of the intra-S checkpoint and possibly implicated in Rad51 focus formation (41). Thus, we biochemically monitored the activation of Chk1 in both WT and AT cells (Figure 5D). AT cells displayed a late phospho-Chk1 signal, most intense at 4–6 h after IR, in parallel with the delayed formation of Rad51 foci. WT cells showed a much weaker signal with the most intense band at 1–2 h after IR, again consistent with the Rad51 focus peak of this cell line (Figure 4A and B). To more directly test whether Chk1 is involved in Rad51 focus formation during the S-phase, we inhibited Chk1 by using the specific inhibitor UCN-01 (41). Following Chk1 inhibition, EdU-positive WT and AT cells showed a reduced number of Rad51 foci (Figure 5E), indicating that not only ATR but also Chk1 is involved in Rad51 focus formation.
Taken together, during the S-phase ATR and its effector kinase Chk1 are partly required for Rad51 focus formation in ATM-proficient cells but essential in the absence of ATM. However, functional ATR and Chk1 alone appear not to be sufficient for successful repair of IR-induced DSBs during the S-phase because no decline of Rad51 foci was observed in ATM-deficient cells (Figure 4B and C). The enhanced Rad51 focus number in G2-phase AT cells 24 h after IR (Figure 2B) strongly suggests that the repair of DSBs that arose during the replicative S-phase cannot be completed in the subsequent G2-phase.
DISCUSSION
Both ATM and Artemis were shown to be equally required for NHEJ repair of a subset of IR-induced DSBs in both the G1- and G2-phase (5,12,20). ATM is also known to be involved in HR (21–25) and recently Artemis cells were shown to display a similar HR defect after IR in the G2-phase (20) (and this study Figure 4A) The remarkably diverse repair phenotype reported here demonstrates that ATM but not Artemis is essential for HR during the S-phase (Figure 4B–D). We thus conclude that the AT and Artemis repair defects are widely epistatic in G1- and G2- but not in the S-phase.
AT cells irradiated in S-phase accumulate numerous Rad51 foci gradually, which could be blocked when ATR or Chk1 are additionally inhibited. The radiation-induced Rad51 focus formation in AT cells was unexpected because ATM is known to be required for the resection of 5′-DNA ends (33–35,42,43). The free 3′-ss-tails are rapidly coated by RPA, which is the substrate for subsequent chromatin loading of Rad51 (31). ATM presumably activates through phosphorylation several essential components of the resection process, such as the MRN complex, CtIP, BRCA1, WRN, BLM and EXO1 (44–51). However, ATM’s precise role for the resection process and for subsequent recombination steps is not clear yet. In the absence of ATM, the related kinase ATR might phosphorylate several of the latter proteins with delayed kinetics (50,52). However, activation of ATR requires the presence of RPA-coated ssDNA and accordingly is considered to be a downstream event in ATM activation (43,53,54). Two explanations may reconcile these apparent contradictions. (i) A yet unknown ATM-independent resection step precedes the ATR activation. (ii) Resection is not required because the replicative MCM2-7 helicase complex may progress beyond an IR-induced SSB (i.e. when located on the lagging strand), exposing sufficient ssDNA to activate ATR–ATRIP and subsequent Rad51 loading. This scenario might perfectly recapitulate the signalling and processing of UV damage (36,37,55–57). In addition, MCM2-7 is a regular phosphorylation target of ATM (44) and ATM-catalyzed phosphorylation possibly inhibits the helicase’s activity, promoting the generation of ssDNA after DNA damage. Thus, cells lacking ATM might then expose particularly long ssDNA stretches, thereby facilitating loading of RPA and subsequently activation of ATR. The possibility that ATR is activated without preceding resection does not exclude that HR of replication-associated DSB implicates delayed secondary resection, i.e. by EXO1 (50).
How might ATR control Rad51 focus formation? Beside its hypothesized function in delayed end resection (50), ATR is the principal kinase that activates Chk1, which in turn phosphorylates Rad51 at T309. This phosphorylation step has been shown to be essential for Rad51 focus formation and HR after replication stress (41). In addition, chromatin loading of Rad51 requires c-Abl-mediated phosphorylation at Y54 and Y315 (58) and damage-induced activation of c-Abl is known to be ATM-dependent (44,59,60). Hence, it is tempting to speculate that ATR can take over in AT cells particularly during the S-phase, which has hitherto not been demonstrated because only asynchronous cell populations were investigated (59).
Although recruitment of Rad51 in AT cells leads to visible foci, its kinetics are significantly delayed during the S- and G2-phase (Figure 4A and B (61)). This delay might result from both insufficient DNA end resection and/or insufficient H2AX phosphorylation (62), which is known to be a prerequisite for timely Rad51 accumulation at sites of DNA damage (63–65). Despite successful recruitment of Rad51 to finally all unrejoined DSBs, HR appears to be widely inhibited in cells lacking functional ATM as (i) the number of IR-induced Rad51 foci never declined during the S- or G2-phase and (ii) gene conversion efficiency of I-SceI-generated DSBs was strongly reduced. Recently, it was shown that the ATM defect in G2 is restricted to HR of DSBs induced in heterochromatic regions (20), similar to NHEJ in G1 (18,19). Notably, neither I-SceI cleavage nor the majority of replication-associated DSBs likely occur in heterochromatin. Thus, we suggest that ATM has more universal but still not precisely defined functions in HR. In addition to end resection and relaxation of heterochromatin, ATM emerges as being more generally involved in chromatin remodeling through activation of RNF20 and 40 (66), as well as in damage-induced sister chromatid cohesion, presumably through activation of the cohesin component SMC1 (67–69), which facilitates homology search and strand exchange. Moreover, the accessory factors XRCC2 and 3 are also ATM phosphorylation targets (44), of which at least XRCC3 is involved in branch migration and perhaps resolution of the putative Holliday structure (70,71), two other steps downstream of Rad51 loading.
Artemis—and particularly its nuclease function (20)—is required for efficient DSB repair (γH2AX foci) in general and Rad51 focus formation in the G2-phase (Figures 1, 2, and 4). However, we show here that Artemis is dispensable for HR during the S-phase, which is clearly different from ATM. Two explanations can be advanced. The end resection process is modulated by CDKs of which CDK2 activity peaks during S-phase (47,49,72). This kinase activity thus stimulates the MRN/CtIP-mediated resection during replication to an extend where it might not depend upon supportive Artemis nuclease—in contrast to the slower and less extensive end degradation in G2 (73) where Artemis is always required. An alternative explanation for Artemis being dispensable in S-phase is the relaxed DNA structure, which may also facilitate end processing independent of the specific Artemis activity.
Based on our observation that Artemis depletion reduced the gene conversion rate in the chromosomal I-SceI-based reporter construct (Figure 3C), we propose that Artemis, similar to ATM, has HR function beyond the repair of only heterochromatic lesions. However, ATM appears to play a more pivotal role in HR (Figure 3C) because cell survival of Artemis-deficient cells could be further compromised by ATM inhibition (Figure 4D). Artemis-deficient cells have now in addition lost the G1/S checkpoint and fail to execute HR repair of direct and indirect DSBs during the S-phase. However, this additional toxicity of ATM inhibition was widely relieved when replication was temporarily blocked (Figure 4D).
In conclusion, we show that both radiosensitivity syndromes, AT and Artemis, are linked to DSB repair deficiencies affecting NHEJ and HR. The repair phenotypes are similar in the G1- and G2-phase but not fully epistatic. Importantly, only AT but not Artemis cells display an additional HR defect in S-phase. Lack of functional ATM affects repair of direct and indirect replication-associated DSBs, leading to enhanced numbers of unresolved recombination foci in G2 as corroborated by persisting Rad51 foci and a more prominent G2-arrest than it is the case for Artemis or WT cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
S.K. is supported by a grant from the German Federal Ministry of Education and Research [BMBF 02NUK001D to J.D.D.]; T.R. and G.R. are supported by a grant of the German Cancer Aid [Deutsche Krebshilfe 107889 to J.D.D. and 107980 to I.D.] and the German Federal Ministry of Environmental Protection [Bundesamt für Strahlenschutz 3610530016 to J.DD. and I.D.]. Funding for open access charge: Institutional.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Kriegs and K. Dreffke for critical reading of this article.
REFERENCES
- 1.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 2.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell. Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 3.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell. Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 4.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 2002;22:1049–1059. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, Stiff T, Jeggo P, Lobrich M. Chromosome breakage after G2 checkpoint release. J. Cell. Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotman G, Shiloh Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- 7.Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc. Natl. Acad. Sci. USA. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beamish H, Khanna KK, Lavin MF. Ionizing radiation and cell cycle progression in ataxia telangiectasia. Radiat. Res. 1994;138:S130–S133. [PubMed] [Google Scholar]
- 9.Blocher D, Sigut D, Hannan MA. Fibroblasts from Ataxia Telangiectasia (AT) and AT heterozygotes show an enhanced level of residual DNA double-strand breaks after low dose-rate gamma-irradiation as assayed by pulsed field gel electrophoresis. Int. J. Radiat. Biol. 1991;60:791–802. doi: 10.1080/09553009114552601. [DOI] [PubMed] [Google Scholar]
- 10.Dikomey E, Dahm-Daphi J, Brammer I, Martensen R, Kaina B. Correlation between cellular radiosensitivity and non-repaired double-strand breaks studied in nine mammalian cell lines. Int. J. Radiat. Biol. 1998;73:269–278. doi: 10.1080/095530098142365. [DOI] [PubMed] [Google Scholar]
- 11.Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004;64:500–508. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- 12.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 14.Geng L, Zhang X, Zheng S, Legerski RJ. Artemis links ATM to G2/M checkpoint recovery via regulation of Cdk1-cyclin B. Mol. Cell. Biol. 2007;27:2625–2635. doi: 10.1128/MCB.02072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Succi J, Feng Z, Prithivirajsingh S, Story MD, Legerski RJ. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol. Cell. Biol. 2004;24:9207–9220. doi: 10.1128/MCB.24.20.9207-9220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 17.Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3'-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J. Biol. Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 18.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat. Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 19.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signalling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. Embo. J. 2009;9:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyn MS. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- 22.Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. Embo. J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golding SE, Rosenberg E, Khalil A, McEwen A, Holmes M, Neill S, Povirk LF, Valerie K. Double strand break repair by homologous recombination is regulated by cell cycle-independent signalling via ATM in human glioma cells. J. Biol. Chem. 2004;279:15402–15410. doi: 10.1074/jbc.M314191200. [DOI] [PubMed] [Google Scholar]
- 24.Bryant HE, Helleday T. Inhibition of poly(ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo CM, Tang W, Mekeel KL, DeFrank JS, Anne PR, Powell SN. High frequency and error-prone DNA recombination in ataxia telangiectasia cell lines. J. Biol. Chem. 1996;271:4497–4503. doi: 10.1074/jbc.271.8.4497. [DOI] [PubMed] [Google Scholar]
- 26.Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohnke A, Westphal F, Schmidt A, El-Awady RA, Dahm-Daphi J. Role of p53 mutations, protein function and DNA damage for the radiosensitivity of human tumour cells. Int. J. Radiat. Biol. 2004;80:53–63. doi: 10.1080/09553000310001642902. [DOI] [PubMed] [Google Scholar]
- 28.Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, Goodarzi AA, Lobrich M, Jeggo PA. Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol. Cell. Biol. 2010;30:3371–3383. doi: 10.1128/MCB.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landberg G, Erlanson M, Roos G, Tan EM, Casiano CA. Nuclear autoantigen p330d/CENP-F: a marker for cell proliferation in human malignancies. Cytometry. 1996;25:90–98. doi: 10.1002/(SICI)1097-0320(19960901)25:1<90::AID-CYTO10>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 30.Rattner JB, Rao A, Fritzler MJ, Valencia DW, Yen TJ. CENP-F is a .ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskeleton. 1993;26:214–226. doi: 10.1002/cm.970260305. [DOI] [PubMed] [Google Scholar]
- 31.Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 33.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell. Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 34.Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. Embo. J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 39.Wang H, Boecker W, Wang H, Wang X, Guan J, Thompson LH, Nickoloff JA, Iliakis G. Caffeine inhibits homology-directed repair of I-SceI-induced DNA double-strand breaks. Oncogene. 2004;23:824–834. doi: 10.1038/sj.onc.1207168. [DOI] [PubMed] [Google Scholar]
- 40.Sirbu BM, Lachmayer SJ, Wulfing V, Marten LM, Clarkson KE, Lee LW, Gheorghiu L, Zou L, Powell SN, Dahm-Daphi J, et al. ATR-p53 restricts homologous recombination in response to replicative stress but does not limit DNA interstrand crosslink repair in lung cancer cells. PLoS One. 2011;6:e23053. doi: 10.1371/journal.pone.0023053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 42.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 43.You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T. CtIP links DNA double-strand break sensing to resection. Mol. Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 45.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, et al. Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J. Biol. Chem. 2002;277:30515–30523. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- 49.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 50.Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821–1831. doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19:5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- 52.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 54.Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaufmann G, Nethanel T. Did an early version of the eukaryal replisome enable the emergence of chromatin? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:173–209. doi: 10.1016/S0079-6603(04)77005-1. [DOI] [PubMed] [Google Scholar]
- 56.Labib K, Diffley JF. Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev. 2001;11:64–70. doi: 10.1016/s0959-437x(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 57.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popova M, Shimizu H, Yamamoto K, Lebechec M, Takahashi M, Fleury F. Detection of c-Abl kinase-promoted phosphorylation of Rad51 by specific antibodies reveals that Y54 phosphorylation is dependent on that of Y315. FEBS Lett. 2009;583:1867–1872. doi: 10.1016/j.febslet.2009.04.044. [DOI] [PubMed] [Google Scholar]
- 59.Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 60.Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan MB, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 61.Yuan SS, Chang HL, Lee EY. Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM(−/−) and c-Abl(−/−) cells. Mutat. Res. 2003;525:85–92. doi: 10.1016/s0027-5107(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 62.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 63.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 64.Dodson H, Morrison CG. Increased sister chromatid cohesion and DNA damage response factor localization at an enzyme-induced DNA double-strand break in vertebrate cells. Nucleic Acids Res. 2009;37:6054–6063. doi: 10.1093/nar/gkp684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 2002;16:571–582. doi: 10.1101/gad.970702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bauerschmidt C, Arrichiello C, Burdak-Rothkamm S, Woodcock M, Hill MA, Stevens DL, Rothkamm K. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 2010;38:477–487. doi: 10.1093/nar/gkp976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 70.Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Masson JY, Shah R, O’Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 72.Johnson N, Cai D, Kennedy RD, Pathania S, Arora M, Li YC, D’Andrea AD, Parvin JD, Shapiro GI. Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell. 2009;35:327–339. doi: 10.1016/j.molcel.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. Embo J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.