Abstract
To better define the roles of assembly factors required for eukaryotic ribosome biogenesis, we have focused on one specific step in maturation of yeast 60 S ribosomal subunits: processing of 27SB pre-ribosomal RNA. At least 14 assembly factors, the ‘B-factor’ proteins, are required for this step. These include most of the major functional classes of assembly factors: RNA-binding proteins, scaffolding protein, DEAD-box ATPases and GTPases. We have investigated the mechanisms by which these factors associate with assembling ribosomes. Our data establish a recruitment model in which assembly of the B-factors into nascent ribosomes ultimately leads to the recruitment of the GTPase Nog2. A more detailed analysis suggests that this occurs in a hierarchical manner via two largely independent recruiting pathways that converge on Nog2. Understanding recruitment has allowed us to better determine the order of association of all assembly factors functioning in one step of ribosome assembly. Furthermore, we have identified a novel subcomplex composed of the B-factors Nop2 and Nip7. Finally, we identified a means by which this step in ribosome biogenesis is regulated in concert with cell growth via the TOR protein kinase pathway. Inhibition of TOR kinase decreases association of Rpf2, Spb4, Nog1 and Nog2 with pre-ribosomes.
INTRODUCTION
Eukaryotic ribosome biogenesis initiates in the nucleolus, where ribosomal RNA (rRNA) is transcribed, folded, bound by ribosomal proteins (r-proteins) and assembly factors, modified and processed to begin to form mature ribosomal subunits. Subsequent steps in maturation of pre-ribosomal particles (pre-rRNPs) occur on their release from the nucleolus to the nucleoplasm and finally, on export to the cytoplasm. This assembly pathway requires a dynamic series of remodeling steps in which protein–protein, RNA–protein and RNA–RNA interactions are established, disrupted and reconfigured (1–6).
Ribosome biogenesis is best studied in the yeast Saccharomyces cerevisiae, where more than 200 phylogenetically conserved trans-acting ribosome assembly factors have been discovered (2,3). Genetic analyses led to the discovery of many of these factors and pinpointed specific steps in ribosomal precursor RNA (pre-rRNA) processing for which each of these proteins is required (Figure 1B). Affinity purifications of pre-ribosomes containing pre-rRNA processing intermediates enabled identification of many other assembly factors and determined with which pre-rRNAs each factor associates. Although this approach establishes a timeline of association of factors with nascent ribosomes, no doubt assembly involves more steps than those defined by the known pre-rRNA processing reactions. Thus, we need to understand recruitment at higher resolution.
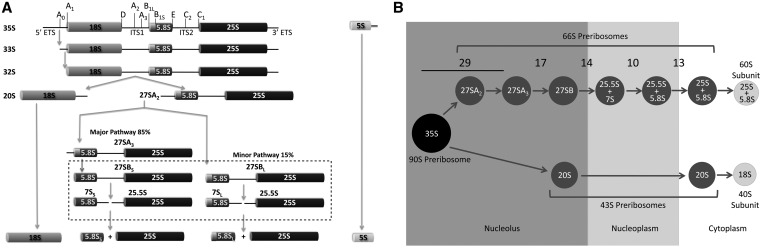
Figure 1.
Pathway for processing of pre-rRNA and maturation of pre-rRNPs in Saccharomyces cerevisiae. (A) rRNA processing pathway. The 35S primary transcript, synthesized by RNA polymerase I, contains sequences for mature 18S, 25S and 5.8S rRNA flanked and separated by external and internal transcribed spacers (ETS/ITS). The 5S rRNA is transcribed from separate linked genes by RNA polymerase III and contains a 3′ETS. The spacers are removed by a series of endonucleolytic and exonucleolytic processing steps in assembling ‘pre-rRNPs’. Processing of the 27SB pre-rRNA to form 25.5S and 7S pre-rRNAs involves endonucleolytic cleavage at the C2 site in ITS2, as shown (boxed). (B) Assembly intermediates in the biogenesis of 60S ribosomal subunits. ‘pre-rRNPs’ that mature into 60S ribosomal subunits are shown, with the pre-rRNAs contained within them indicated. Known numbers of assembly factors required for each of the consecutive steps in subunit maturation are shown.
Having discovered most of the factors required for ribosome biogenesis, it is now critical to determine how these molecules facilitate rearrangement of pre-rRNAs and pre-rRNPs. Transition between different conformations may create the structures necessary for protein binding, pre-rRNA processing and export from the nucleus to produce functional ribosomes. To understand these rearrangements, we are studying one specific step in assembly: processing of 27SB pre-rRNA to generate 25.5S and 7S pre-rRNAs. The clearest landmark for this processing step is endonucleolytic cleavage at the C2 site of internal transcribed spacer 2 (ITS2) (Figure 1A). This step is believed to require major conformational rearrangements of pre-rRNAs and pre-rRNPs. For example, structural and mutational analysis supports two possible secondary structures of the ITS2 RNA, the ‘ring structure’ (7) and the ‘hairpin structure’ (8). Processing of 27SB pre-rRNA may be an important control point in ribosome biogenesis; nascent ribosomes are released from the nucleolus into the nucleoplasm once this step is completed (9).
Of the ∼75 different assembly factors reproducibly found in 66S precursors to 60S ribosomal subunits (Figure 1B and Supplementary Table S1), 14 assembly factors, hereafter referred to as the ‘B-factors’, have been shown to be necessary for processing of 27SB pre-rRNA (10–26). Yeast strains mutant for each of these B-factors exhibit a diagnostic pre-rRNA processing phenotype-increased amounts of 27SB pre-rRNA relative to 27SA2 and 27SA3 pre-rRNAs, indicating a failure to cleave the C2 site within ITS2 (Figure 1A). The B-factors fall into several functional categories: RNA-binding proteins such as Tif6, Nip7, Rpf2, Rlp24 and Nsa2; GTPases such as Nog1 and Nog2; DEAD box proteins (DBPs)/ATPases such as Spb4, Dbp10, Drs1 and Has1 and the scaffolding protein Mak11. Nop2 is thought to function as an RNA methyltransferase, and Rrs1 has no predicted function. Because Drs1 and Has1 appear to also function during earlier steps of pre-rRNA processing (10,22) (data not shown and J. Dembowski, personal communication), we have not included them in this study. This work focuses on the order by which these B-factors are recruited into nascent ribosomes, by systematically examining their interdependence for assembly into pre-rRNPs. Defining the recruiting pathway for the proteins involved in this step in assembly will enable identification of cofactors that function with each B-factor and the substrates on which they act. These are important steps to more precisely determine the roles of these proteins in ribosome assembly.
Fromont-Racine and coworkers showed that Mak11, Rlp24, Nog1 and Nsa2 form a network of genetic and physical interactions to generate a recruiting pathway ending with Nog2 (17,21,23,24,27). They showed that Nsa2 and Nog2 associate with pre-ribosomes only after 27SB pre-rRNA is generated and that Nsa2 is required to recruit Nog2. Here, we show that all B-factor proteins are required to recruit Nog2. Interestingly, we found that only a subset of the B-factors are required to recruit Nsa2 and that Nsa2 alone is not sufficient to recruit Nog2.
We further define the order in which the B-factors bind to pre-ribosomes, by assaying their association in yeast strains depleted of each protein. We show that a newly defined subcomplex containing Nop2 and Nip7 is necessary for the stable incorporation of the previously characterized Rpf2 subcomplex containing the assembly factors Rpf2 and Rrs1 and the r-proteins L5 and L11 (25). The Rpf2 subcomplex is required for association of the DBP Spb4, which is necessary, but not sufficient to load the GTPase Nog2 into nascent ribosomes. Furthermore, we found that the Nop2/Nip7 subcomplex is necessary for stable association of Rlp24, Nog1 and Tif6 with pre-ribosomes. Dbp10 also seems to be important for the recruitment of Rlp24; it is involved in association of Rlp24 with pre-ribosomes but is not necessary for recruiting Rpf2, Tif6 or Nop2/Nip7 subcomplex. Taken together, our data lead to a recruitment model in which the B-factors assemble into nascent ribosomes in a hierarchical manner via two largely independent linear recruiting pathways that converge on Nog2. Finally, we also identified a means by which this step in ribosome biogenesis is regulated in concert with cell growth via the TOR protein kinase pathway.
MATERIALS AND METHODS
Construction of yeast strains
Yeast strains conditional for expression of RPF2, RLP24, SPB4, NOG1, TIF6 or NOG2 were constructed as described by Longtine et al. (28), as follows. The sequences containing a selectable marker, plus the GAL1 promoter sequence followed by an ATG and codons encoding three copies of the hemagglutinin epitope (3HA), were amplified by polymerase chain reaction (PCR). The PCR products were transformed into yeast. Transformants were screened for correct integration of the GAL1 promoter and the triple hemagglutinin (3HA) tag upstream and in-frame with the respective genes, by western blotting with anti-HA antisera. Strains conditional for expression of Nip7, Nop2 or Dbp10 were obtained from other laboratories (11,12,14) and contain a genomic knockout of the respective genes plus a plasmid bearing a GAL promoter fusion of each gene. For NIP7, we used the temperature-sensitive allele nip7-1 fused to the GAL promoter. Because the Nip7-1 protein is functional at 30°C but is less stable than wild-type Nip7 (12), Nip7-1 can be more rapidly depleted than with wild-type NIP7 fused to the GAL1 promoter.
Yeast strains expressing C-terminal TAP-tagged Nop7 or C-terminal 3HA-tagged proteins were created by PCR of the tag sequence and a selectable marker (URA3 or TRP1 for the TAP tag and HIS3 or kanMX6 for the 3HA tag), transformation and selection, as described in the study by Rigaut et al. (29) and Longtine et al. (28), respectively. Transformants were screened by western blotting to identify those expressing the tagged proteins, and, in most cases, by polysome gradients for defects in ribosome assembly, which would indicate the effects of the tag on protein function. Sequences of oligonucleotides used as PCR primers are available on request.
Growth of yeast strains and depletion of factors
Yeast strains used in this study are listed in Supplementary Table S1. Unless otherwise noted, yeast was grown at 30°C in YEPGlu medium (2% dextrose, 2% peptone and 1% yeast extract) or YEPGal medium (2% galactose, 2% peptone and 1% yeast extract). Cells were harvested during mid-logarithmic phase growth, at 3–5 × 107 cells/ml, except where otherwise indicated. The strains containing GAL1 promoter fusions of B-factor genes were grown at 30°C in YEPGal liquid medium to 3–5 × 107 cells/ml or grown in YEPGal medium and shifted to YEPGlu for indicated times, to 3–5 × 107 cells/ml, to deplete the proteins in vivo. Depletion was assayed by western blotting of whole-cell extracts with anti-HA antiserum, to measure amounts of the 3HA-tagged assembly factors (data not shown).
Sucrose gradient assays of ribosomes and polyribosomes
Pre-ribosomes, ribosomes and polyribosomes were fractionated from 40 OD254 units of whole-cell extracts on 7–47% (wt/vol) sucrose gradients as described by Deshmukh et al. (30), with the following modifications. Cycloheximide (5 mg) was added to cultures 20 min before harvesting cells. A Teledyne ISCO Foxy R1 density gradient fractionator was used to fractionate and analyze gradients.
Affinity purification of pre-ribosomes or pre-ribosome subcomplexes
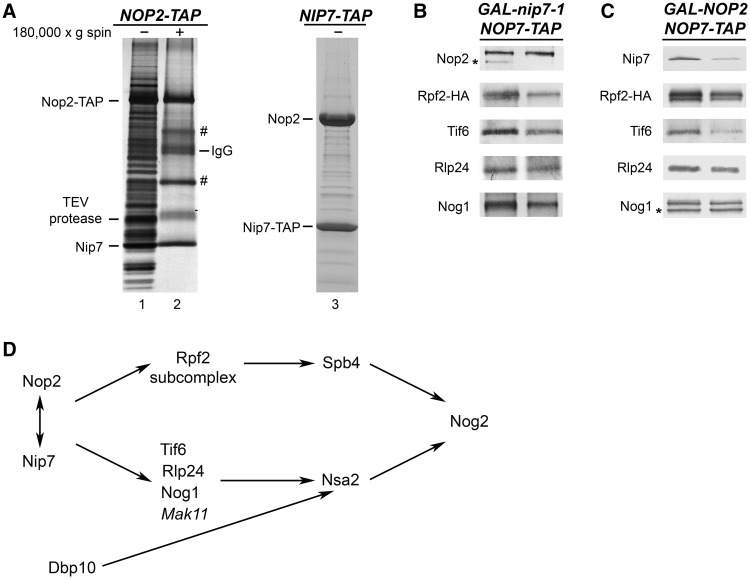
Ribosome assembly intermediates were affinity purified from whole-cell extracts with magnetic Dynabeads (Invitrogen), using TAP-tagged assembly factor Nop7, as described in the study by Sahasranaman et al. (31). The Nop2/Nip7 subcomplex was purified as follows: extracts were prepared from a NOP2-TAP strain, and pre-ribosomes and ribosomes were pelleted by centrifugation of whole-cell extracts at 180 000g for 2 h at 4°C, as described by Krogan et al. (32). The supernatant was subjected to a second centrifugation at 180 000g for 45 min at 4°C. TAP-tagged Nop2 was used for affinity purification of the Nop2/Nip7 subcomplex from the supernatant as described previously (29).
Protein extractions, SDS–PAGE and western blot analysis
Proteins in whole-cell extracts were prepared for gel electrophoresis by dissolving the extract in sodium dodecyl sulfate (SDS) sample buffer. Proteins were recovered from sucrose gradient fractions or from eluates during affinity purification by precipitation with 10% trichloroacetic acid (TCA) and were subsequently suspended in SDS sample buffer. Proteins were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) on 4–20% Novex precast gels (Invitrogen) and stained with silver by standard methods. To assay Nog2 protein by western blotting, NuPage 4–12% Bis-Tris gels (Invitrogen) were used, because Nog2 comigrates with immunoglobulin G (IgG) on 4–20% Novex gels. Proteins from whole-cell extracts, gradient fractions or purified pre-ribosomes were assayed by western blot analysis (33), with the following modification. To enable detection of multiple different proteins on one blot and to conserve antiserum, after electroblotting, we cut nitrocellulose membranes into smaller sections based on the known mobility of the different proteins. Edges of cut membranes, when visible, are indicated by ‘<’ in the figures. TAP-tagged proteins were detected using alkaline phosphatase conjugated to IgG (Pierce). 3HA-tagged proteins were identified with mouse monoclonal antibody 12CA5. Otherwise, antibodies specific for r-proteins or ribosomal assembly factors were used. Alkaline phosphatase (AP)-conjugated anti-mouse or anti-rabbit secondary antibodies (Promega) were used, and colorimetric detection was performed using NBT and BCIP (Promega). Often, two bands were seen in western blotting with Nog2, Nop2 or Nog1. The band corresponding to the bona fide protein was confirmed by comigration on SDS–PAGE with silver-stained protein identified by mass spectrometry or by decrease on turning off expression of the corresponding gene and is indicated with ‘#’.
RESULTS
Pre-ribosomes are largely intact in the absence of B-factors
To determine the interdependence of B-factors for assembly into pre-ribosomes, we assayed association of these proteins with pre-rRNPs in the absence of each B-factor. Because this is a first attempt to understand the order in which all B-factors assemble and there are few if any missense alleles for each of the genes encoding these proteins, we chose to assay interdependence in conditional null strains. To do so, we replaced the promoter of each B-factor gene with the GAL1 promoter. In galactose, the GAL1 promoter is active, but in the presence of glucose, GAL1 is repressed. We used TAP-tagged assembly factor Nop7 for affinity–purification of pre-ribosomes from these strains containing or lacking each B-factor. Nop7 is present in 90S particles and each of the 66S pre-RNPs, but does not depend on B-factors for association with pre-ribosomes (data not shown), and is not known to be required for processing of 27SB pre-rRNA (34). In addition, the population of pre-rRNAs with which Nop7 is associated does not change significantly in the absence of the B-factors (Supplementary Figure S1 and data not shown). Thus, any changes that we observe in any B-factor mutant most likely would not result from Nop7 failing to associate with pre-ribosomes or being associated with a different population of assembly intermediates.
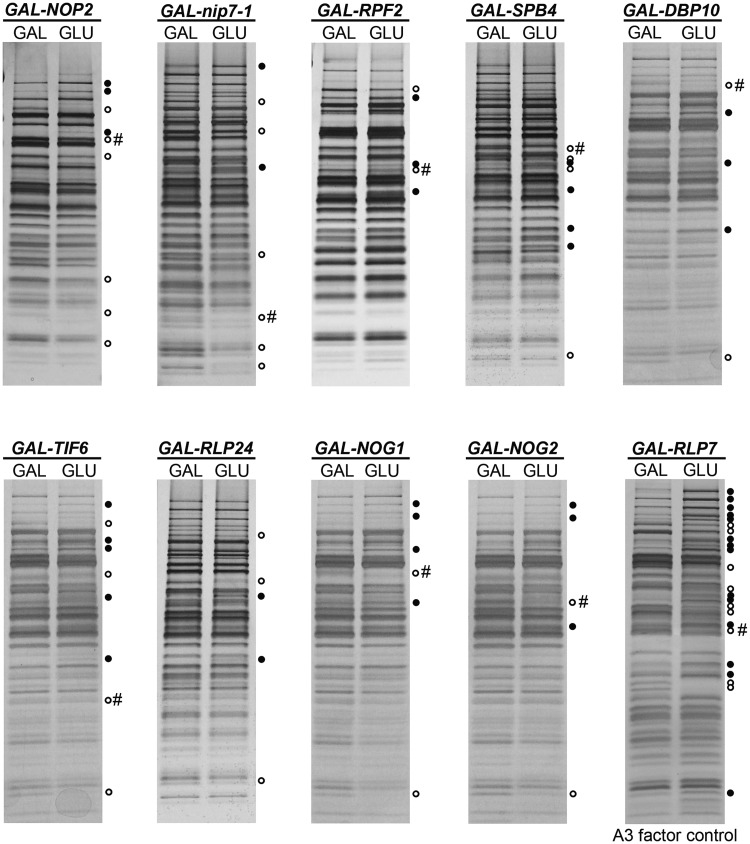
Pre-ribosomes were purified on depletion of each of nine B-factors, and pre-ribosomal proteins were resolved by SDS–PAGE (Figure 2). Typically we observed the changes in the levels of 4–8 assembly factors associated with pre-ribosomes in each of the B-factor mutants. Many of these changes were the same in each mutant, but there were observable differences. The fact that we see only a small number of changes in each mutant suggests that pre-ribosomes are largely intact. In contrast, depletion of proteins that block the preceding step in ribosome assembly, processing of 27SA3 pre-rRNA, causes a greater number of changes in pre-ribosome composition (Figure 2, A3 factor control). The proteins required for this earlier step are interdependent of their association with pre-ribosomes, and in their absence, pre-ribosomes are unstable and turned over (31). Thus, the phenotype that we observe in B-factor mutants is specific of blocking this step of assembly and is different from that seen when previous steps are blocked.
Figure 2.
Depletion of B-factors causes few changes in the SDS–PAGE profile of proteins contained in pre-ribosomes. Nine of the 12 B-factors necessary for processing of 27SB pre-rRNA were individually depleted using the conditional GAL promoter. TAP-tagged ribosome assembly factor Nop7 was used to purify 90S and 66S pre-ribosomes from yeast grown in YEPGal (left lane in each pair), and from strains grown in YEPGal and shifted to YEPGlu for 16 h to deplete each B-factor (right lane in each pair). Proteins present in purified pre-ribosomes were separated by SDS–PAGE and stained with silver. TAP-tagged Rpf2 was used to purify pre-ribosomes from the GAL-RLP7 strain in which Rlp7 was depleted. Rlp7 is required for processing of 27SA3 pre-rRNA. Thus, this strain serves as a control to demonstrate specificity of the gel profiles for B-factor-depleted strains. Silver-stained bands that increased in the absence of each assembly factor are indicated with solid circles. Bands that decreased are indicated with hollow circles. When able to be identified, the protein under control of the GAL promoter is indicated with #.
Nog2 is the last B-factor to associate with pre-ribosomes and is dependent on the other B-factors
Previously, two sets of interacting assembly factors, the Rpf2 subcomplex as well as Rlp24, Mak11 and Nog1, were shown to be required to recruit Nog2 to pre-ribosomes (21,24,25). These proteins assemble into early pre-rRNPs containing 35S or 27SA2 pre-rRNAs (21,24,25). However, these proteins do not appear to be required for subunit maturation until much later, for processing of 27SB pre-rRNA within 66S pre-rRNPs. In contrast, Nog2 assembles into pre-ribosomes just before 27SB pre-rRNA is cleaved at the C2 site (17). Thus, the B-factors may serve as a recruiting scaffold or help create pre-rRNP structures in early precursor particles, to enable proper loading of Nog2 and other B-factors.
To test the requirement of the other B-factors for the recruitment of Nog2 into pre-ribosomes, we assayed levels of Nog2 in pre-ribosomes purified from strains in which each B-factor was depleted. Consistent with previous results, we saw that the levels of Nog2, associated with pre-ribosomes, were severely decreased, and in some cases undetectable, in the absence of Rpf2, Rlp24 and Nog1 (Figure 3A). In addition, we saw that Nog2 levels were strongly diminished in pre-ribosomes in the absence of Nop2, Nip7, Spb4, Tif6 or Dbp10. This was not simply a result of protein turnover, as Nog2 was stable in whole-cell extracts on depletion of other B-factors (Supplementary Figure S2A). Thus, all of the known B-factors are required to recruit and/or maintain stable association of Nog2 with pre-ribosomes.
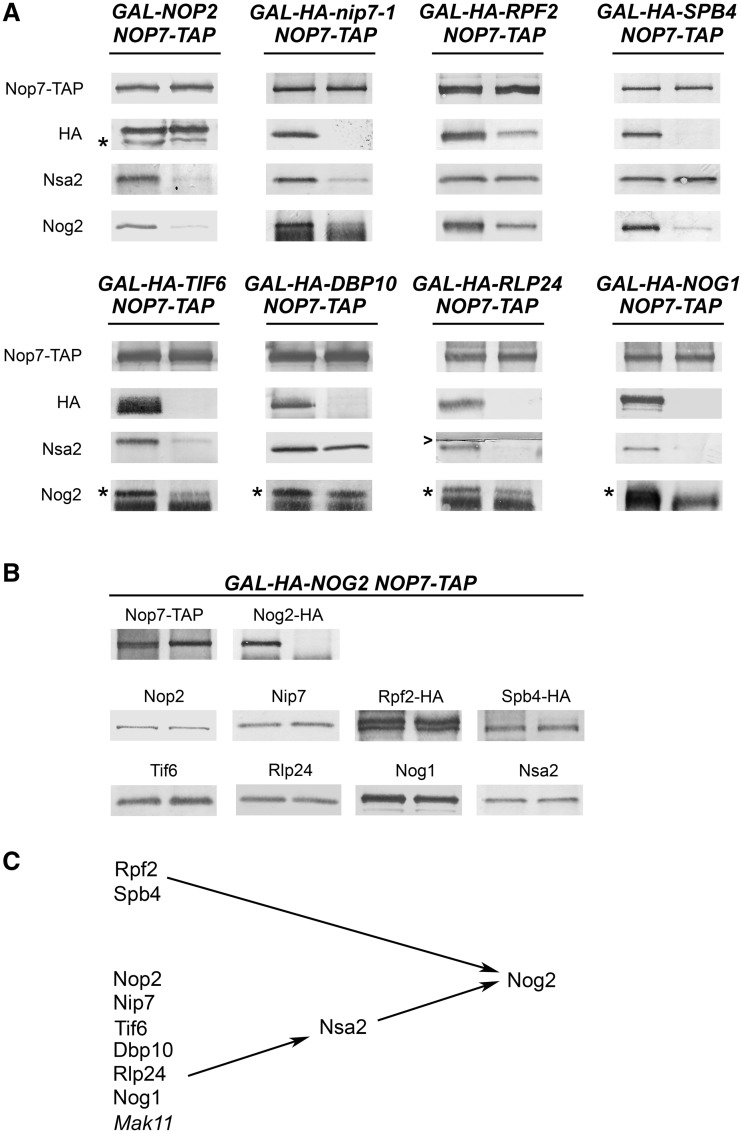
Figure 3.
All the B-factors are necessary to recruit Nog2 to pre-ribosomes; only a subset of B-factors are required to recruit Nsa2 into pre-ribosomes. (A) The changes in the levels of pre-ribosome-associated Nsa2 and Nog2 in the absence of each of the B-factors. Pre-ribosomes were purified from cells grown in YEPGal or shifted to YEPGlu to deplete each B-factor. Proteins from pre-rRNPs were resolved by SDS–PAGE and assayed by western blotting. In each pair of samples shown in this figure and in Figures 4–7, protein from the undepleted strain is on the left and protein from the depleted strain is on the right. In this blot and all subsequent blots, where two bands are detected, the bona fide protein is indicated with an asterisk. Nop7-TAP serves as the loading control. (B) Nog2 is not required to recruit any of the B-factors. Proteins in pre-ribosomes purified from GAL-NOG2 yeast grown in YEPGal, or shifted to YEPGlu to deplete Nog2 were assayed by western blotting. Nop7-TAP serves as a loading control. Each pair of samples is shown as described in Figure 3A. (C) All of the B-factors are required to recruit Nog2 to pre-ribosomes. A subset of B-factors is required to recruit Nsa2 and Nog2.
Because Nog2 assembles into pre-ribosomes after 27SB pre-rRNA is generated, whereas the other B-factors assemble earlier (excluding Nsa2), we reasoned that Nog2 would not be required to recruit these proteins. Semiquantitative assays of pre-ribosomes in Nog2-depleted yeast using SILAC mass spectrometry suggested that few changes occur in its absence (27). To test this idea in more detail, we assayed by western blotting for the presence of eight other B-factor proteins in pre-ribosomes when Nog2 was depleted (Figure 3B). We observed no changes in the level of these proteins in pre-ribosomes, consistent with Nog2 associating with pre-ribosomes after the other B-factors.
Nsa2 is recruited to pre-ribosomes by a subset of B-factors
Like Nog2, Nsa2 assembles into pre-ribosomes only after 27SB pre-rRNA is generated (23). Nsa2 depends on the B-factors Rlp24, Mak11 and Nog1, and its recruitment is a prerequisite for association of Nog2 (23,24). Therefore, we tested the association of Nsa2 with pre-ribosomes in the absence of the eight remaining B-factors (Figure 3A). Western blotting showed that Nsa2 fails to associate with pre-ribosomes in the absence of Nop2, Nip7, Tif6, Dbp10, Rlp24 and Nog1. However, Nsa2 did not depend on the Rpf2 subcomplex or the DBP Sbp4. Thus, there are B-factor mutants in which both Nsa2 and Nog2 fail to associate with pre-ribosomes as well as mutants in which only Nog2 fails to associate. These results show that Nsa2 alone is not sufficient to recruit Nog2 to pre-ribosomes and that recruitment of Nog2 requires the concerted assembly of additional proteins. Because Nog2 is the last B-factor recruited to pre-ribosomes, it may use GTP hydrolysis as a signal or trigger for cleavage of 27SB pre-rRNA at the C2 site. Thus, its recruitment may be highly regulated by the cooperation of a number of assembly factors to ensure that cleavage of the C2 site does not occur prematurely. Thus, our results, with those of the Fromont-Racine group, describe a recruiting pathway among the B-factor proteins that culminates with the association of the GTPase Nog2 (Figure 3C).
The Rpf2 subcomplex is required to recruit Spb4 into pre-ribosomes and Spb4 is required for subsequent recruitment of Nog2
Next, we tested the interdependence of the B-factors that associate with pre-ribosomes before Nsa2 and Nog2. To do so, we assayed their association with pre-ribosomes when each of the other B-factors was depleted. We began by investigating the requirement of the Rpf2 subcomplex for association of other B-factors. Previous work showed that Rpf2, Rrs1, L5 and L11 are mutually interdependent for assembly of the subcomplex and association with the pre-ribosome (25). Thus, depleting Rpf2 is analogous to depleting the entire Rpf2 subcomplex. To test whether the Rpf2 subcomplex is required to load any of the other B-factors into nascent ribosomes, we assayed the amounts of B-factors in purified pre-ribosomes when Rpf2 was depleted. The levels of Nog2 and HA-tagged Spb4 in pre-ribosomes were significantly decreased when Rpf2 was depleted, but amounts of Nog1, Nop2-HA, Nip7, Nsa2 and Tif6-HA remained largely the same (Figures 3A and 4A). There was a reproducible but very slight decrease of Rlp24. These results, plus our previous experiments with Rpf2 depletions (Figure 3A and (25)), suggest that association of Spb4 with pre-ribosomes occurs early during assembly and depends on the Rpf2 subcomplex. Conversely, Spb4 recruits Nog2 much later.
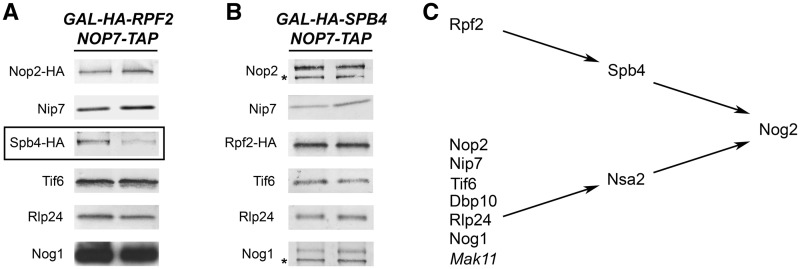
Figure 4.
Spb4 depends on Rpf2 to dock with pre-ribosomes. (A) Rpf2 is necessary to load Spb4 into nascent ribosomes. Pre-ribosomes were purified from a GAL-RPF2 strain grown in YEPGal or shifted to YEPGlu to deplete Rpf2. Pre-ribosomes were purified from whole cell lysates of undepleted and depleted cells using TAP-tagged Nop7. Proteins from pre-rRNPs were resolved by SDS–PAGE and assayed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Rpf2 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. B-factors that decrease in the absence of Rpf2 are boxed. (B) No other B-factors are recruited into pre-ribosomes by Spb4. Proteins in pre-ribosomes purified from GAL-SPB4 yeast grown in YEPGal or shifted to YEPGlu to deplete Spb4 were assayed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Spb4 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. (C) Hierarchy for recruiting Rpf2, Spb4 and Nog2 into pre-ribosomes.
We then tested whether the other B-factors depend on Spb4 to associate with pre-ribosomes. To do so we assayed their association on depletion of Spb4. Of eight B-factors, only Nog2 was decreased (Figures 3A and 4B). We did not directly test the association of Dbp10 in the absence of Spb4. Because Nsa2 depends on Dbp10 (Figure 3A and C), and Nsa2 does not change in the absence of Spb4, we infer that Dbp10 does not depend on Spb4. We conclude that the Rpf2 subcomplex is necessary for association of Spb4 with pre-ribosomes and that Spb4 recruits Nog2 (Figure 4C). No other B-factors depend on Rpf2, Spb4 or Nog2 for association with pre-ribosomes.
Nop2 and Nip7 form a subcomplex important for recruitment of the other B-factors into pre-ribosomes
Previously, Nop2 and Nip7 were found to be necessary for processing of 27SB pre-rRNA (11,12). Both proteins cosedimented with 66S pre-ribosomes when whole-cell extracts were fractionated by sucrose gradient centrifugation (Supplementary Figure S3A), and both are commonly found in affinity-purified 66S pre-ribosomes (Supplementary Table S2). To investigate Nop2 and Nip7 in more detail, we TAP-tagged each of them and assayed which pre-ribosomal proteins copurified with them.
When affinity purification was carried out with whole-cell extracts from a Nip7-TAP strain, large amounts of Nop2 copurified with Nip7-TAP, and much smaller amounts of other pre-ribosomal proteins were present (Figure 5A, lane 3). This may be caused by the C-terminal TAP-tag interfering with the C-terminal PUA RNA-binding domain of Nip7, weakening its association with pre-ribosomes, but not with Nop2. Consistent with this potentially deleterious effect of the TAP-tag on Nip7, we observed that the NIP7-TAP strain grew more slowly than wild-type, untagged NIP7 strains (data not shown). In addition, extracts prepared from the NIP7-TAP strain contain halfmer polyribosomes (Supplementary Figure S3B), indicative of a defect in production or function of 60S subunits (35,36). Affinity purification of Nop2-TAP using whole-cell extracts revealed the typical SDS–PAGE profile of proteins present in early and middle 66S pre-rRNPs (Figure 5A, lane1 and Supplementary Table S2). However, when whole-cell extracts from a NOP2-TAP strain were subjected to high-speed centrifugation, to pellet ribosomes and pre-ribosomes before affinity purification with Nop2-TAP, primarily Nop2 and Nip7 were recovered (Figure 5A, lane 2). Taken together, these results demonstrate that Nop2 and Nip7 are tightly associated and form a heteromeric subcomplex.
Figure 5.
Nop2 and Nip7 form a subcomplex, are interdependent for assembly into pre-ribosomes and function in recruiting of other B-factors. (A) Nop2 and Nip7 form a heteromer in vivo. Yeast strains expressing TAP-tagged Nop2 or Nip7 were grown in YEPGlu medium to 3 × 107 cells/ml. Whole-cell extracts from the NOP2-TAP strain (lane 1) or the supernatants after high-speed centrifugation (lane 2) were subjected to affinity purification using Nop2-TAP, to purify Nop2-containing pre-ribosomes or subcomplexes, respectively. Purified proteins were identified by mass spectrometry. Proteins labeled with ‘#’ are common contaminants in purified samples. Whole-cell extract from the NIP7-TAP strain was subjected to affinity purification using Nip7-TAP (lane 3). Nop2 and Nip7 in these purified samples were identified by mass spectrometry. (B and C) Nop2 and Nip7 are interdependent for their assembly into pre-rRNPs and important for incorporation of other B-factors into pre-ribosomes. GAL-nip7-1 or GAL-NOP2 yeast were grown in YEPGal or shifted to YEPGlu. Whole-cell extracts were subjected to affinity purification, using Nop7-TAP to isolate pre-ribosomes. Amounts of B-factors present in the purified pre-ribosomes were analyzed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Nop2 or Nip7 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. (D) Nop2 and Nip7 are important for the stable association of all the B-factors.
Another indication that Nop2 and Nip7 form a subcomplex is the observation that both proteins were detected in fractions 3–5 near the top of gradients (as well as in fractions containing 66S pre-ribosomes), when wild-type whole-cell extracts were subjected to centrifugation (Supplementary Figure S3A). Furthermore, much greater amounts of both Nop2 and Nip7-TAP sedimented near the top of gradients when NIP7-TAP extracts were subjected to centrifugation (Supplementary Figure S3B).
Because Nip7 and Nop2 form a subcomplex, we assayed whether they depend on each other to assemble into pre-ribosomes, as well as whether any other B-factors depend on either of them. Indeed, Nop2 was not present in pre-ribosomes purified from strains in which Nip7 was depleted (Figure 5B). Likewise, pre-ribosomes from Nop2-depleted cells contained diminished amounts of Nip7 (Figure 5C). In both Nop2- and Nip7-depleted strains, decreased amounts of Rpf2, Tif6, Rlp24, Nog1, Nsa2 and Nog2 were associated with pre-ribosomes (Figures 3A and 5B and C). Spb4, Mak11, Rrs1 and Dbp10 were not tested. We assume that when Rpf2 is absent, both Rrs1 and Spb4 are also unable to assemble into pre-rRNPs (Figure 4A and (25)). The inability of Nop2 or Nip7 to assemble with pre-rRNPs in the absence of the other was not simply a result of protein turnover when the Nip7/Nop2 subcomplex could not form. Each protein was present in whole-cell extracts at wild-type levels when the other protein was depleted (Supplementary Figure S2B). We conclude that Nop2 and Nip7 form a subcomplex and that both are important for association of at least six other B-factors, as well as each other, with nascent ribosomes (Figure 5D).
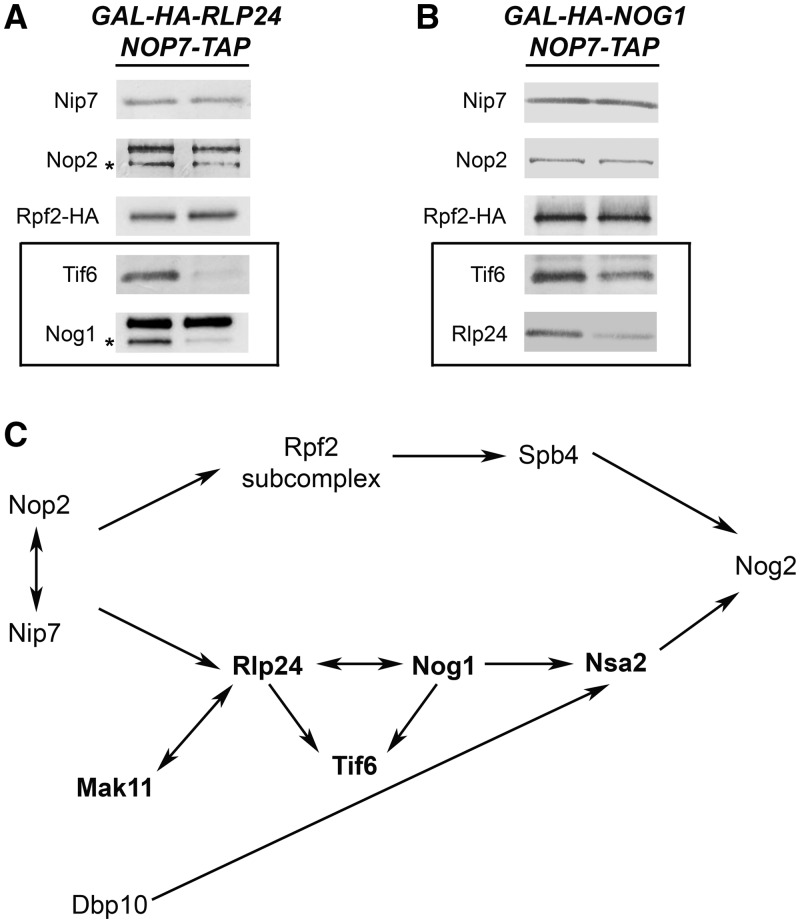
Nog1, Rlp24 and Tif6 are interdependent for recruitment into pre-ribosomes
Fromont-Racine and coworkers (17,21,23,24,27) defined a hierarchy of recruitment among Mak11, Rlp24, Nog1, Nsa2 and Nog2 (boldface in Figure 6C). To first investigate the relationship of Rlp24 and Nog1 with other B-factors for entry into nascent ribosomes, we surveyed the effects of depleting either of these two proteins on ribosome assembly. As previously observed by Saveanu et al. (21), we found that amounts of Nsa2 and Nog2 in pre-ribosomes were diminished when expression of either RLP24 or NOG1 was turned off (Figure 3A). Saveanu and coworkers had also found that Nog1 was absent from pre-rRNPs when Rlp24 was depleted, but that Rlp24 did not depend on Nog1 for stable association with pre-ribosomes. However, when we depleted Nog1, we saw a decrease in levels of Rlp24 associated with purified pre-ribosomes (Figure 6B). We also found that pre-ribosome-associated Tif6 decreased when either Nog1 or Rlp24 was depleted, but Rpf2, Nop2 and Nip7 were not affected (Figure 6A and B). It is likely that the effect on Tif6 that we observe on depleting Nog1 or Rlp24 is mediated through these two proteins rather than via downstream factors Nsa2 or Nog2, because we found that Nog2 is not necessary for docking of Tif6 with pre-rRNPs (Figure 3B), and Lebreton et al. (23) showed that Tif6 is not affected by depletion of Nsa2. This is also consistent with the observations that Nsa2 and Nog2 associate with pre-rRNPs several steps after Tif6 (17,23). Together, our results and those of Fromont-Racine and coworkers demonstrate a network of physical and functional interactions among Tif6, Nog1 and Rlp24. These three proteins exhibit interdependence for association with pre-ribosomes and are required for later recruitment of Nsa2 and Nog2 (Figure 6C).
Figure 6.
Nog1 and Rlp24 are important for association with pre-ribosomes of Tif6, as well as each other. (A and B) The changes in amounts of pre-ribosomal proteins when either Rlp24 or Nog1 is depleted. GAL-RLP24 or GAL-NOG1 yeast were grown in YEPGal or shifted to YEPGlu for 16 h to deplete either assembly factor. Pre-ribosomes were purified from whole-cell lysates of undepleted and depleted cells using TAP-tagged Nop7. Western blotting was used to assay amounts of pre-ribosomal proteins. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Nog1 or Rlp24 from preribosomes, and the Nop7-TAP loading control are shown in Figure 3A. (C) Interdependence among Rlp24, Nog1, Tif6, Nsa2 and Nog2 for assembly into pre-ribosomes. Proteins in boldface were previously studied by Fromont-Racine and coworkers.
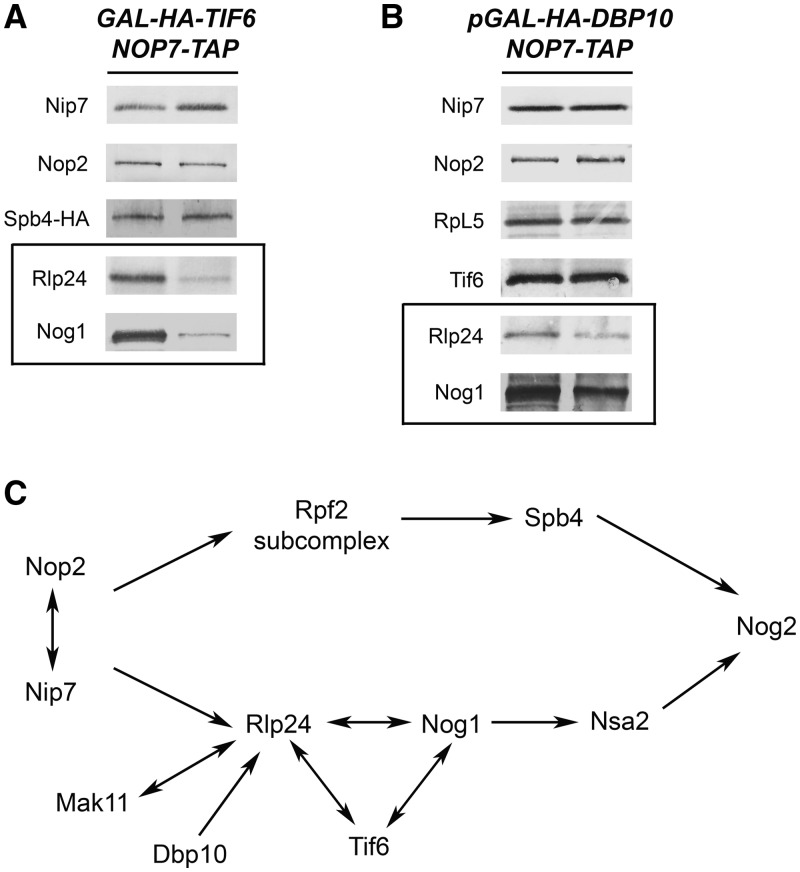
Tif6 and Dbp10 are important for association with pre-rRNPs of Rlp24, Nog1, Nsa2 and Nog2
Tif6 is associated with pre-ribosomes containing 27SA2, 27SB, 25.5S and 7S pre-rRNAs and is found in both the nucleus and the cytoplasm (37). Tif6 binds to r-protein L23 on the subunit interface of pre-ribosomes and therefore prevents premature entry of nascent ribosomes into the translation pathway, by blocking association with 40S subunits (38). Assembly factors such as Sdo1 and Efl1 release Tif6 from pre-rRNPs, to enable recycling of Tif6 from the cytoplasm back to the nucleus (39,40). However, Tif6 also functions earlier in 60S subunit biogenesis. Depletion of Tif6 causes accumulation of 27SB pre-rRNA, by blocking pre-rRNA processing in the nucleolus (16). To investigate where Tif6 fits into the hierarchy of B-factor assembly into pre-ribosomes, we assayed the effects of depleting Tif6 on pre-ribosome constituents. We found that Rlp24, Nog1, Nsa2 and Nog2 were diminished, whereas Nop2, Nip7 and the Rpf2 subcomplexes were not affected (Figures 3A and 7A). Western blotting of whole-cell extracts from the Tif6 and Rlp24 depleted strains also revealed that in the absence of Tif6, Rlp24 undergoes turnover, whereas Tif6 remains stable when Rlp24 is depleted (Supplementary Figure 2C). Therefore, our results (Figures 6A and B and 7A), together with those of Saveanu et al. (21), show that association of Tif6, Rlp24 and Nog1 are mutually interdependent, but independent of assembly of Rpf2 and Spb4.
Figure 7.
Both Tif6 and Dbp10 are required for assembly of Rlp24 and Nog1 into pre-ribosomes.(A) Tif6 is required to recruit Rlp24 and Nog1 pre-rRNPs. Proteins present in pre-rRNPs purified from a GAL-TIF6 strain grown in YEPGal or shifted to YEPGlu for 16 h were resolved by SDS–PAGE and subjected to western blot analysis. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Tif6 from preribosomes and the Nop7-TAP loading control are shown in Figure 3A. (B) Dbp10 is important for association of Rlp24 and Nog1 with pre-rRNPs. The GAL-DBP10 strain was grown in YEPGal or shifted to YEPGlu for 16 h to deplete Dbp10. Pre-ribosomes were purified from extracts prepared from each strain, and proteins were resolved by SDS–PAGE. Pre-ribosomal proteins were assayed by western blotting. Each pair of samples is shown as described in Figure 3A. Western blots showing the depletion of Dbp10 from preribosomes, and the Nop7-TAP loading control are shown in Figure 3A. (C) Position in the association hierarchy for Tif6 and Dbp10.
Dbp10 is a DEAD-box protein that routinely copurifies with 66S pre-ribosomes (Supplementary Table S2). Depletion of Dbp10 or growing the cold-sensitive dbp10-1 mutant at 18°C leads to accumulation of 27SB pre-rRNA (14). We investigated the role of Dbp10 in recruiting proteins to nascent ribosomes, by purifying pre-ribosomes from a strain in which Dbp10 was depleted. Western blotting revealed that levels of Rlp24, Nog1, Nsa2 and Nog2 were decreased, whereas Tif6, Nop2, Nip7 and r-protein L5 (representative of the Rpf2 subcomplex) were unaffected (Figures 3A and 7B). The effects observed on depletion of Dbp10 were relatively mild. This may reflect the transient nature of DBPs. Although DBPs undoubtedly play a major role in ribosome assembly, they may not always be intimately associated with pre-ribosomes.
Taken together, our studies of the recruiting pathway of 10 different assembly factors required for processing of 27SB pre-rRNA are summarized in Figure 7C. Our data establish that many of the B-factors are recruited into early assembly intermediates in two independent pathways. These two pathways ultimately converge later in assembly to recruit the last factor, the GTPase Nog2.
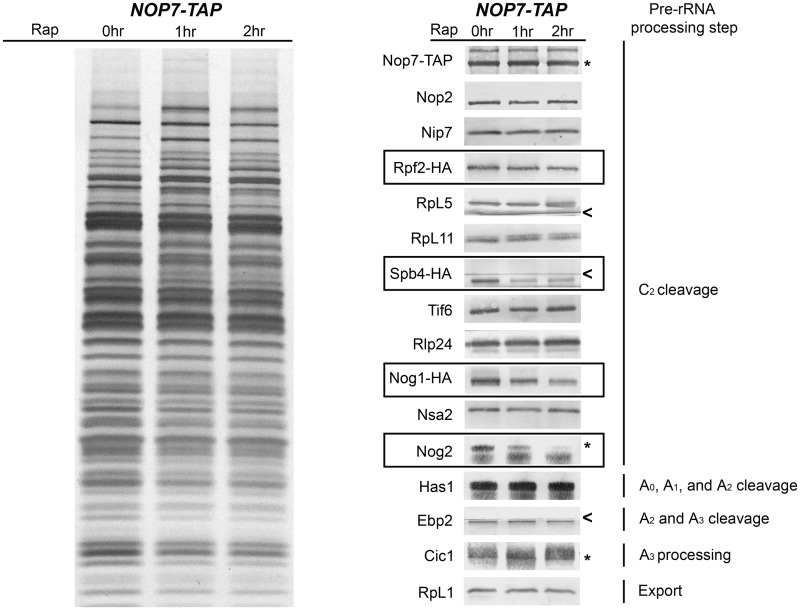
The TOR protein kinase pathway regulates biogenesis of 60S ribosomal subunits via targeting the B-factors Rpf2, Spb4, Nog1 and Nog2
The number of ribosomes in a cell as well as their function are adjusted in response to cellular growth rates, mediated in part by the protein kinase TOR (target of rapamycin) (41,42). TOR inactivation on treatment of cells with rapamycin results in decreased transcription of rRNAs and nucleolar entrapment of pre-rRNPs, possibly via targeting Nog1, one of the B-factors (43–45). To test whether other B-factors serve as targets of TOR kinase, we examined their levels in pre-ribosomes from cells treated with rapamycin. After treatment with rapamycin, pre-ribosomes remained largely intact, but levels of Rpf2, Spb4, Nog1 and Nog2 associated with pre-rRNPs decreased, whereas amounts of other B-factors remained the same (Figure 8). As controls, we assayed levels of assembly factors and r-proteins that do not function in 27SB pre-rRNA processing. These proteins remained stably associated with pre-rRNPs after treatment with rapamycin (Figure 8). When amounts of B-factors in whole-cell extracts were tested, we found that levels of Rpf2, Spb4, Nog2 and Nog1 decreased (Supplementary Figure S4). These data suggest that the TOR kinase pathway may regulate ribosome biogenesis in response to cellular growth rate via targeting a subset of B-factors. Failure to recruit or maintain Rpf2, Spb4, Nog2 and Nog1 in pre-rRNPs may block maturation of 60S ribosomal subunits, entrapping them within the nucleolus.
Figure 8.
The TOR protein kinase pathway regulates assembly of 60S ribosomal subunits via targeting the B-factors Rpf2, Spb4, Nog1 and Nog2. The Nop7-TAP strain was grown at 30°C in YEPGlu medium to 3 × 107 cells/ml. Two more cultures were grown to 2 × 107 cells/ml, and 200 ng/ml rapamycin was added to each culture. These two cultures were then grown for another 1 or 2.5 h to 3 × 107 cells/ml. Whole-cell extracts were subjected to single-step purification using TAP-tagged Nop7 to isolate 66S pre-ribosomes. Proteins present in the affinity-purified pre-ribosomes were resolved by SDS–PAGE and stained with silver (lanes 1–3) or subjected to western blot analysis (lanes 4–6). Proteins from untreated cells are in lanes 1 and 4, those from cells treated with rapamycin for 1 h are in lanes 2 and 5 and those from cells treated for 2.5 h are in lanes 3 and 6. Nop7-TAP serves as the loading control. Proteins with decreased association with pre-ribosomes upon treatment with rapamycin are boxed. The pre-rRNA processing step in which each protein tested functions is indicated.
DISCUSSION
Our working hypothesis is that the numerous assembly factors associate with nascent ribosomes in a hierarchical and cooperative manner, through protein–protein or protein–RNA interactions, to drive forward the formation of consecutive assembly intermediates. Therefore, our approach to begin to uncover the precise roles of these factors is to systematically investigate physical or functional interactions among them.
Stable interactions among some assembly factors and r-proteins are evident by isolating assembly subcomplexes containing them (25,32,34,46–52). These subcomplexes might serve as smaller building blocks to minimize the complexity of assembly of these enormous ribonucleoprotein particles containing so many components. Importantly, identifying and characterizing these physical clusters of assembly factors will enable discovery of cofactors and substrates of these factors, thus unraveling their functions, and will also help define the local topology within pre-rRNPs.
Rearrangements of local structures within assembling ribosomes may be propagated by allosteric mechanisms to alter the global architecture of pre-rRNPs. Such reorganizations might affect recruitment or activity of assembly factors working in the same step of subunit biogenesis, even though these proteins may not physically interact. Thus, to obtain a comprehensive picture of ribosome assembly, it is necessary to ‘walk out’ of neighborhoods defined by physical subcomplexes within pre-ribosomes and to investigate interactions among all assembly factors that function together in one specific step of pre-rRNA processing and assembly. Studying molecules in one ‘functional cluster’ will help to understand how these molecules coordinate their precise functions with each other in one assembly step.
We previously discovered that six assembly factors required for processing of 27SA3 pre-rRNA assemble early into pre-ribosomes, are mutually interdependent for association with pre-ribosomes and are important for assembly of r-proteins with domain I and II of 25S/5.8S rRNAs (31) (Jakovljevic et al., accepted with minor revisions). Here, we have studied recruitment of the factors required for the next step in pre-rRNA processing, removal of ITS2 from 27SB pre-rRNA. Previous experiments to map the relative timing of association of B-factors with pre-ribosomes demonstrated that seven of these proteins assembled rather early, with 90S pre-ribosomes or 66S particles containing 27SA2 pre-rRNA (21,24–26,37). In contrast, Nsa2 and Nog2 assembled later, with 27SB pre-rRNA (17,23). The entry points of Dbp10, Nip7 and Nop2 have not been studied. Experiments described in this article are consistent with and extend previous results to generate a higher resolution picture of the assembly hierarchy.
Building on work from the Fromont-Racine group, we began by assaying the association of the late-entering B-factors, Nsa2 and Nog2, in the absence of the other B-factors. We show that all of the B-factors are required to recruit the GTPase Nog2, but only a subset (Nop2/Nip7, Dbp10, Tif6, Rlp24 and Nog1) are required to recruit Nsa2. Fromont-Racine and coworkers showed that Nsa2 is necessary to recruit Nog2. Here, we show that in the absence of Rpf2 or Sbp4, Nsa2 is still recruited into pre-ribosomes, but Nog2 is not. Thus, Nsa2 alone is not sufficient to recruit the GTPase Nog2.
We then extended our model by systematically testing the interdependence of all the B-factors. We discovered that these B-factors are loaded into pre-ribosomes sequentially via two largely separate assembly routes that converge on Nog2 (Figure 7C). Association of Nop2 and Nip7 with each other and with pre-ribosomes is necessary for stable association of downstream B-factors, beginning with the Rpf2 subcomplex (Rpf2, Rrs1, r-proteins L5 and L11 and 5S rRNA), and Rlp24, Tif6 and Nog1. Members of each of these three sets of proteins are mutually interdependent for assembly and exhibit significant physical and genetic interactions with each other (Supplementary Table S3 and Figure 9A). Furthermore, many contain RNA binding motifs and thus are likely to bind pre-rRNA. Taken together, these results suggest that processing of 27SB pre-rRNA requires formation of one or more assembly platforms of B-factor subcomplexes within pre-ribosomes, several steps before the processing event. In contrast, the last steps in recruiting of the B-factors occur later in assembly, after formation of 27SB pre-rRNA.
Figure 9.
Physical and genetic interactions provide clues about the location of the B-factors in pre-ribosomes. (A) Predicted locations of B-factors in assembling 60S subunits. Binding sites of proteins were predicted using a combination of genetic and physical interactions and then superimposed on a schematic of the 60S subunit viewed form the intersubunit side. Grey circles indicate assembly factors and ribosomal proteins that were used as landmarks to help position the B-factors. Genetic interactions listed in Supplementary Table S3 are indicated. PCA, protein complementation assay; H.C. sup, high-copy suppressor; Sup, extragenic suppressor; sl−, synthetic lethal. (B) r-proteins such as L4, L15 and L18 are shown in the crystal structure of the 60S subunit as viewed from the crown view of the intersubunit face rotated right 90°. Domains I and III of 25S rRNA are indicated in red and blue, respectively. The Nop7 binding site determined by Granneman et al. (56) is shown in cyan. Pymol images of the ribosome structure were generated using PDB files 3U5H and 3U5I.
In the absence of each B-factor, other B-factors associate less well with pre-ribosomes, but to varying extents. Western blotting showed that the B-factors are usually not completely absent from pre-ribosomes, but rather diminished. The strongest effect that we observed in our mutants was the association of Nsa2 and Nog2. This probably reflects blocking assembly at a step before these proteins associate with pre-ribosomes. Conversely, effects on the association of other B-factors were often not as strong. One possibility is that we observe small changes in the levels of these proteins because we purify a different population of assembly intermediates in cells depleted of B-factors. We believe that this is not the case because TAP-tagged Nop7 does not co-IP significantly different pre-rRNA intermediates in B-factor mutants (Supplementary Figure S1 and data not shown). In fact, Nop7 co-IPs slightly greater amounts of 27SB pre-rRNA in the absence of the B-factors. Unlike Nsa2 and Nog2, the other B-factors associate with 35S or 27SA2 containing pre-ribosomes and remain associated throughout the lifetime of 27SB pre-rRNA. Because 27SB-containing pre-ribosomes are the longest-lived intermediates and these intermediates accumulate in B-factor mutants, we predict that the small changes that we see might in fact be slightly underrepresented.
The experiments performed in this study fail to distinguish two possibilities that could occur in the absence of each B-factor: (i) The other B-factors fail to associate with pre-ribosomes. (ii) The other B-factors associate with pre-ribosomes, but fail to be stably incorporated. We believe the former to be true regarding Nsa2 and Nog2 and the later regarding the other B-factors. Each B-factor in our model (Figure 7C) does not necessarily recruit the next through direct interaction, but rather may form a structure or niche in the pre-ribosome that allows stable incorporation of the next B-factor. Thus, each B-factor only stably associates with pre-ribosomes after stable incorporation of the preceding B-factors.
Our model shows that the B-factors associate with pre-ribosomes through two converging pathways that result in recruiting Nog2. Parallel assembly pathways that converge to a single intermediate have been observed in ribosome biogenesis and are proposed to be quality control mechanisms (53–55). Johnson and coworkers (55) showed that cytoplasmic release of assembly factors occurs via parallel pathways that converge to trigger the release of Tif6. This convergence of pathways may represent a quality control checkpoint to ensure Tif6 is not released before the assembling 60S subunit is competent for translation. Lamanna and Karbstein (56) speculate that parallel assembly pathways may converge before irreversible steps in assembly. They reason that irreversible steps should be tightly regulated and it is easier to survey a single intermediate rather than a number of different parallel intermediates before proceeding to the next step of assembly. This may be reflected in the recruitment of Nog2 to pre-ribosomes. As the last known B-factor to associate with pre-ribosomes, Nog2 may be a key factor in triggering C2 cleavage. Recruitment of Nog2 by two parallel pathways may ensure that it is not loaded onto pre-ribosomes prematurely, thus preventing premature cleavage at the C2 site.
Of the 12 assembly factors required for processing of 27SB pre-rRNA, 4 are thought to exert their action by using the power of NTP hydrolysis. Involvement of this many NTPases during this step suggests substantial rearrangements may occur. Consistent with this, 27SB pre-rRNA is one of the most abundant pre-rRNA intermediates destined for 60S subunits, with a lifetime substantially longer than other pre-rRNAs (57). The early assembling protein Spb4, a DBP and potential RNA-dependent ATPase, and the late assembling factor Nsa2, are necessary to recruit the GTPase Nog2. Neither Spb4 nor Nsa2 alone is sufficient to recruit Nog2. Spb4 associates early with pre-ribosomes, whereas Nsa2 associates later. Temporal separation of the association of these two proteins may act as a proofreader of ribosome assembly, ensuring that Nog2 is recruited to the pre-ribosome only at the proper time. Because Sbp4 and Nog2 are at the end of the recruiting pathway (Figure 7C), it is tempting to speculate that removal of ITS2 is triggered by the concerted action of these NTPases. Spb4 might be regulated by Nog2, analogous to regulation of the spliceosomal ATPase Brr2 by the GTPase Snu114 (58). During spliceosome assembly, GTP-bound Snu114 activates Brr2, whereas GDP-bound Snu114 represses Brr2 function. Alternatively, Nog2 alone could harness the energy of GTP hydrolysis to cause a conformational switch within the pre-ribosome. In particular, the A3 assembly factors Nop15 and Cic1 that bind to ITS2 may need to be removed or reorganized (59). GTP hydrolysis may cause a structural change within ITS2 that makes binding of Nop15 and Cic1 less favorable in the new conformation (60). Consistent with Nog2 being a member of the myosin/kinesin superfamily of GTPases, Nog2 could also act in a mechanical fashion, using GTP hydrolysis to physically displace Cic1 or Nop15 (60,61).
Rearrangements before and during processing of 27SB pre-rRNA may also occur as a result of posttranslational modification of assembly factors. We show that the B-factors Rpf2, Spb4, Nog1 and Nog2 are specifically reduced in pre-ribosomes when the protein kinase TOR is inactivated. On the basis of our recruiting model, we would predict that if Nog1 was diminished in pre-ribosomes, Nsa2 would also be diminished. Contrary to this idea, we do not see a change in Nsa2 on inactivation of TOR signaling. This may indicate that these proteins are not failing to be recruited to pre-ribosomes, but rather are recruited normally, but then prematurely dissociate from pre-ribosomes on inactivation of TOR signaling. Interestingly, three of these B-factors are the putative NTPases Spb4, Nog1 and Nog2. These NTPases may fail to stably assemble into pre-ribosomes in the absence of posttranslational modifications. This could be analogous to r-protein S3, which must undergo a series of phosphorylation and dephosphorylation events to stably associate with pre-40S intermediates and promote formation of the 40S beak (62).
Alternatively, inactivation of TOR signaling may repress expression of a subset of assembly factors. Nog1 was previously shown to be regulated by TOR kinase (43). Inactivation of TOR caused decreased transcription of NOG1. It was also shown that Nog1 is rapidly turned over, although independently of TOR signaling. We show that on inactivation of TOR, levels of Rpf2, Spb4, Nog1 and Nog2 are diminished in whole-cell extracts. Expression of the genes encoding these proteins, as well as other assembly factors and r-proteins, is controlled and coordinated by a set of core promoter sequences termed the Ribi regulon (Ribosome biogenesis) and RP regulon. In addition, inactivation of TOR signaling represses expression of genes controlled by the Ribi and RP regulons (45,63–69). Thus, Rpf2, Spb4 and Nog2 may be rapidly turned over, similarly to Nog1.
A current challenge in understanding ribosome biogenesis is determining where and how assembly factors contact the pre-ribosome. Recent studies on crosslink proteins to RNA with UV light have revealed the rRNA-binding sites of some assembly factors (59,70,71). In addition, advances in cryo-EM have provided information about the locations of assembly factors in pre-ribosomes (38,72,73), and higher resolution crystal structures of eukaryotic ribosomes are allowing better visualization of how r-proteins contact rRNA (74). Although these techniques are powerful for determining how and where assembly factors contact the pre-ribosome, their application is limited in ribosome assembly. For example, not all assembly factors contact rRNA and thus cannot be cross-linked to determine their binding sites. Of those proteins that do bind RNA, crosslinking may not capture all RNA-binding sites in these dynamic particles. In addition, structural analyses often require a fairly homogenous purified sample. Because most assembly factors that function in 60S subunit biogenesis are present in more than one consecutive intermediate, obtaining homogenous samples for cryo-EM or generating crystals is difficult. Therefore, other methods need to be employed to help predict and determine the binding sites of assembly factors.
We used known genetic and physical interactions among the B-factors to generate a model of the locations of some of these assembly factors (Figure 9A and Supplementary Table S3). Nop2 and Nip7 form a subcomplex and are synthetically lethal with deletion of genes encoding r-proteins L26B or L17A, respectively (75). These two r-proteins are adjacent to each other in domain I of 25S/5.8S rRNA, lying near the polypeptide exit tunnel (74). Consistent with the synthetic lethal interactions observed between these two r-proteins and the B-factors Nop2 and Nip7, L17 and L26 function in processing of 27SB pre-rRNA (M. Gamalinda, personal communication). This suggests that Nop2 and Nip7 might dock pre-ribosomes near these two r-proteins, close to the exit tunnel (Figure 9A).
Cryo-EM studies show that Tif6 binds L23 and L24 on the intersubunit side of assembling 60 subunits and functions to prevent premature association of the 60S and 40S subunits (38). Tif6 is one of the last assembly factors to be released from the assembling ribosome. Its removal is dependent on the Shwachman-Bodian-Diamond syndrome protein homologue Sdo1 and the GTPase Efl1 (40). Sdo1 was shown to interact with r-protein L3 in a two-hybrid screen, positioning Sdo1 near Tif6. Interestingly, Sdo1 was also shown to bind directly to Nip7 (76). Thus, there may be a physical connection between Nip7, Sdo1 and Tif6. This provides a testable model that Sdo1 may be involved in releasing not only Tif6 but also other B-factors.
L5, L11 and 5S rRNA, which are members of the Rpf2 subcomplex, form the central protuberance (CP) near the top of 60S subunits (74). Thus, it is likely that when Rpf2 and Rrs1 deliver L5, L11 and 5S rRNA to assembling ribosomes, they are located in a comparable position. We speculate that Rpf2 and Rrs1 are located on the intersubunit side of the CP based on the following evidence: (i) Rrs1 was shown to interact with Ebp2 in both two-hybrid and protein complementation assay (PCA) screens (77,78); (ii) Ebp2 exhibits two hybrid interactions with Brx1 (79) and (iii) Brx1 interacts with Tif6 by PCA (78). These three pieces of evidence suggest a chain of interactions beginning with Tif6, perpetuated through Brx1 and Ebp2, and ending with the Rpf2 subcomplex.
Nsa2 exhibits two-hybrid interactions with r-proteins L4, L15 and L18 (23) (Figure 9B), which are located adjacent to each other on the left edge of the 60S subunit when viewed from the intersubunit face. Nsa2 shares a number of both physical and genetic interactions with Rlp24, Nog1 and Mak11, suggesting that these proteins may associate with pre-ribosomes in close proximity to each other (21,23). Consistent with this idea, our results, with those of Saveanu et al., show that Rlp24, Nog1 and Mak11 are interdependent for their assembly into pre-ribosomes and all three are required to recruit Nsa2. Interestingly, overexpression of the A3 factor Nop7 suppresses mutations in nog1 (43). Nop7 is a member of a group of mutually interdependent proteins required for processing 27SA3 pre-rRNA (31). UV crosslinking has shown most of the A3 factors bind rRNA in domain I of 25S/5.8S rRNA or in ITS2, and in their absence, these regions are not properly assembled (31,59). Consistent with genetic interactions between Nog1 and Nop7, in the absence Nop7 and mutually interdependent A3 factors, Nog1 and Rlp24 fail to associate with pre-ribosomes, suggesting that they may also bind near the A3 factors (31) (data not shown). Specifically, Nop7 was shown to bind rRNA near helix 54 in domain III (59) (Figure 9B). This region of rRNA is in close proximity to the rRNA-binding sites of L4, L15 and L18 further supporting the idea Rlp24, Nog1, Mak11 and Nsa2 bind the left edge of the 60S subunit. It is also interesting that inactivation of TOR signaling causes both Nog1 and Nop7 to associate less well with pre-ribosomes (43), suggesting that this neighborhood could be a target of TOR regulation.
Although we were unable to predict the binding sites of all B-factors, our model provides a number of important insights. First, we predict that a large majority of the B-factors are binding on the intersubunit face. Could some of these factors prevent premature subunit joining analogous to Tif6? Could Nog2 remove or reorganize these proteins analogous to other GTPases that function on the subunit interface (55)? Second, we predict that Rlp24, Nog1 and Nsa2 are binding near r-proteins L4, L15 and L18, respectively. These are in close proximity to the ITS2-proximal stem, where ITS2 would be predicted to be located in assembling 60S subunits. Our predictions for the location of these B-factors put them in prime position to facilitate the removal of ITS2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–4 and Supplementary References.
FUNDING
Funding for open access charge: National Institutes of Health [GM28301 to J.L.W.]; Richard King Mellon Foundation Presidential Graduate Fellowship in the Life Sciences and the Semon H. Stupakoff Scholarship (to J.T.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the following people for generously providing strains: John Aris (GAL-NOP2), David Goldfarb and Nilson I.T. Zanchin (GAL-nip7-1) and Jesus de la Cruz (GAL-DBP10). The authors are grateful to the following people for generously providing antibodies: John Aris (Nop2), David Goldfarb (Nip7), Janine Maddock (Nog1), Cosmin Saveanu and Micheline Fromont-Racine (Rlp24, Nog2, Nsa2 and Tif6) and Jeff Brodsky (Sec61). They also thank Susan Dowd for assistance with mass spectrometry.
REFERENCES
- 1.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karbstein K. Inside the 40S ribosome assembly machinery. Curr. Opin. Chem. Biol. 2011;15:657–663. doi: 10.1016/j.cbpa.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joseph N, Krauskopf E, Vera MI, Michot B. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999;27:4533–4540. doi: 10.1093/nar/27.23.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh LC, Lee JC. Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 1990;211:699–712. doi: 10.1016/0022-2836(90)90071-S. [DOI] [PubMed] [Google Scholar]
- 9.Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002;157:941–951. doi: 10.1083/jcb.200111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ripmaster TL, Vaughn GP, Woolford JL., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl Acad. Sci. USA. 1992;89:11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong B, Brockenbrough JS, Wu P, Aris JP. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanchin NI, Roberts P, DeSilva A, Sherman F, Goldfarb DS. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell Biol. 1997;17:5001–5015. doi: 10.1128/mcb.17.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Cruz J, Kressler D, Rojo M, Tollervey D, Linder P. Spb4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA. 1998;4:1268–1281. doi: 10.1017/s1355838298981158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burger F, Daugeron MC, Linder P. Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res. 2000;28:2315–2323. doi: 10.1093/nar/28.12.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuno A, Miyoshi K, Tsujii R, Miyakawa T, Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu U, Si K, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wehner KA, Baserga SJ. The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell. 2002;9:329–339. doi: 10.1016/s1097-2765(02)00438-0. [DOI] [PubMed] [Google Scholar]
- 19.Morita D, Miyoshi K, Matsui Y, Toh EA, Shinkawa H, Miyakawa T, Mizuta K. Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27 SB Pre-rRNA to 25S rRNA and the 60S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:28780–28786. doi: 10.1074/jbc.M203399200. [DOI] [PubMed] [Google Scholar]
- 20.Kallstrom G, Hedges J, Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2004;52:141–158. doi: 10.1111/j.1365-2958.2003.03973.x. [DOI] [PubMed] [Google Scholar]
- 23.Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 2006;281:27099–27108. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- 24.Saveanu C, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M. The p21-activated protein kinase inhibitor Skb15 and its budding yeast homologue are 60S ribosome assembly factors. Mol. Cell Biol. 2007;27:2897–2909. doi: 10.1128/MCB.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Gomez JJ, Lebaron S, Froment C, Monsarrat B, Henry Y, de la Cruz J. Dynamics of the putative RNA helicase Spb4 during ribosome assembly in Saccharomyces cerevisiae. Mol. Cell Biol. 2011;31:4156–4164. doi: 10.1128/MCB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebreton A, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M, Saveanu C. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008;36:4988–4999. doi: 10.1093/nar/gkn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 30.Deshmukh M, Tsay YF, Paulovich AG, Woolford JL., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL., Jr Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. EMBO J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 33.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons Inc; 1994. [Google Scholar]
- 34.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 35.Helser TL, Baan RA, Dahlberg AE. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol. Cell Biol. 1981;1:51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rotenberg MO, Moritz M, Woolford JL., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- 37.Graindorge JS, Rousselle JC, Senger B, Lenormand P, Namane A, Lacroute F, Fasiolo F. Deletion of EFL1 results in heterogeneity of the 60S GTPase-associated rRNA conformation. J. Mol. Biol. 2005;352:355–369. doi: 10.1016/j.jmb.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 38.Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J. Biol. Chem. 2010;285:14848–14851. doi: 10.1074/jbc.C109.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 40.Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 41.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 42.Crespo JL, Hall MN. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002;66:579–591. doi: 10.1128/MMBR.66.4.579-591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honma Y, Kitamura A, Shioda R, Maruyama H, Ozaki K, Oda Y, Mini T, Jeno P, Maki Y, Yonezawa K, et al. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 2006;25:3832–3842. doi: 10.1038/sj.emboj.7601262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanrobays E, Leplus A, Osheim YN, Beyer AL, Wacheul L, Lafontaine DL. TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export. RNA. 2008;14:2061–2073. doi: 10.1261/rna.1176708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granneman S, Gallagher JE, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJ. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 2003;31:1877–1887. doi: 10.1093/nar/gkg300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 48.Karbstein K, Doudna JA. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J. Mol. Biol. 2006;356:432–443. doi: 10.1016/j.jmb.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 49.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL., Jr Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 51.Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol. Cell Biol. 2007;27:1207–1221. doi: 10.1128/MCB.01523-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, Tschochner H. Analysis of ribosome biogenesis factor-modules in yeast cells depleted from pre-ribosomes. Nucleic Acids Res. 2010;38:3068–3080. doi: 10.1093/nar/gkp1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;186:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamanna AC, Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J. Mol. Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Granneman S, Petfalski E, Tollervey D. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karbstein K. Role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. [DOI] [PubMed] [Google Scholar]
- 61.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 62.Schafer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 63.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 66.Miyoshi K, Shirai C, Mizuta K. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:1969–1973. doi: 10.1093/nar/gkg278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, Beyer AL, Nomura M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wade CH, Umbarger MA, McAlear MA. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast. 2006;23:293–306. doi: 10.1002/yea.1353. [DOI] [PubMed] [Google Scholar]
- 70.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, III, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 75.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luz JS, Georg RC, Gomes CH, Machado-Santelli GM, Oliveira CC. Sdo1p, the yeast orthologue of Shwachman-Bodian-Diamond syndrome protein, binds RNA and interacts with nuclear rRNA-processing factors. Yeast. 2009;26:287–298. doi: 10.1002/yea.1668. [DOI] [PubMed] [Google Scholar]
- 77.Tsujii R, Miyoshi K, Tsuno A, Matsui Y, Toh-e A, Miyakawa T, Mizuta K. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells. 2000;5:543–553. doi: 10.1046/j.1365-2443.2000.00346.x. [DOI] [PubMed] [Google Scholar]
- 78.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 79.Shimoji K, Jakovljevic J, Tsuchihashi K, Umeki Y, Wan K, Kawasaki S, Talkish J, Woolford JL, Jr, Mizuta K. Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:4574–4588. doi: 10.1093/nar/gks057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.