Abstract
Zfp206 (also named as Zscan10) belongs to the subfamily of C2H2 zinc finger transcription factors, which is characterized by the N-terminal SCAN domain. The SCAN domain mediates self-association and association between the members of SCAN family transcription factors, but the structural basis and selectivity determinants for complex formation is unknown. Zfp206 is important for maintaining the pluripotency of embryonic stem cells presumably by combinatorial assembly of itself or other SCAN family members on enhancer regions. To gain insights into the folding topology and selectivity determinants for SCAN dimerization, we solved the 1.85 Å crystal structure of the SCAN domain of Zfp206. In vitro binding studies using a panel of 20 SCAN proteins indicate that the SCAN domain Zfp206 can selectively associate with other members of SCAN family transcription factors. Deletion mutations showed that the N-terminal helix 1 is critical for heterodimerization. Double mutations and multiple mutations based on the Zfp206SCAN–Zfp110SCAN model suggested that domain swapped topology is a possible preference for Zfp206SCAN–Zfp110SCAN heterodimer. Together, we demonstrate that the Zfp206SCAN constitutes a protein module that enables C2H2 transcription factor dimerization in a highly selective manner using a domain-swapped interface architecture and identify novel partners for Zfp206 during embryonal development.
INTRODUCTION
Zfp206 is a transcription factor highly expressed in mouse and human embryonic stem cells (ESCs) but repressed on differentiation (1–3). Zfp206 is reported to be a regulator of pluripotency and implicated in the maintenance of the pluripotent state by jointly functioning with other plurpotency factors such as Sox2 and Oct4 (1–3). Consistently, ESCs with overexpressed Zfp206 could resist differentiation. The Zfp206 protein contains 14 C2H2 zinc fingers within its C-terminal DNA-binding domain and one SCAN domain near its N-terminus (1).
The 84 amino acid SCAN domain contains a highly conserved leucine-rich region and is found near the N-terminal end of approximately 10% of human C2H2 zinc-finger transcription factors (4). The SCAN domain was named after the first letter of some the first identified family members (SREZBP, Ctfin51, AW-1 (ZNF174) and Number 18) (5,6). The SCAN domain appears an evolutionary invention within the mammalian lineage and shows a varying degree of gene family expansions in different taxa (4). For example, the human genome contains 71 SCAN domain proteins (7), whereas the mouse genome contains only 40 (8). The primary function of the SCAN domain appears to mediate self-association and selective association with other SCAN family members. Interestingly, protein modules that mediate the dimerization of C2H2 zinc fingers have independently evolved and underwent lineage specific expansions in invertebrates (9,10). Therefore, it is possible that dimerization domains attached to C2H2 zinc fingers are important to expand the regulatory potential of the otherwise evolutionarily ancient C2H2 DNA-binding domain in a combinatorial fashion. The selective association of SCAN domain SCAN domain transcription factors (TFs) can determine which gene are bound and regulated and, as a consequence, execute specific developmental programs. Specifically, a hypothetical SCAN TF A would specify cell type X when it pairs with SCAN TF B but would specify cell type Y when SCAN TF C is expressed provided the domains are dimerization competent. However, the function of SCAN TF A would not be affected if a SCAN TF D is expressed, which is not capable of heterodimer formation. Indeed, several studies have demonstrated that the dimerization of SCAN domains is highly selective. Some SCANs form exclusive homodimers, while others form homodimers or heterodimers with a specific subset of family members (6,11–13). Hence, to understand the regulatory function of SCAN domains, it is mandatory to know its dimerization potential. Ideally, one should be able to predict the dimerization profile of a SCAN domain by analyzing the primary sequence.
The previously solved structures of the SCAN domains of ZNF174 and MZF1 revealed a domain-swapped homodimer (14,15). However, the previous structural data could not unravel a recognition code for SCAN domain dimerization and did not allow prediction of the interaction pattern from sequence.
Here, we report the first high-resolution of the SCAN domain of Zfp206 solved by X-ray crystallography. We further assessed the heterodimerization potential of Zfp206SCAN against a panel of 20 SCAN domains using a maltose-binding protein (MBP) pull-down assay. We identified Zscan4 and Zfp110 as a novel and selective interaction partners of Zfp206. Zscan4 is known to regulate telomere extension and genomic stability in ESCs (16). Zfp110 is involved in programmed cell death in the mouse embryonic neural retina (17). The interactions between the Zfp206SCAN and other embryonically expressed SCAN transcription factors suggest new functions of Zfp206. Given the important roles of Zfp206 and Zscan4 in ESCs and our demonstration of a strong dimerization between both proteins, we suggest a combinatorial action of both proteins analogous to the Sox-Oct partnerships during lineage commitment (18). Rational mutations using a Zfp206SCAN–Zfp110SCAN fusion protein showed that the N-terminal helix 1 takes parts in the heterodimerization and the domain-swapped topology is likely adopted for a Zfp206SCAN–Zfp110SCAN heterodimer.
MATERIALS AND METHODS
Molecular biology
pDEST-HisMBP-Zfp206SCAN expression vector was constructed as described previously (19). An analogous strategy was used to generate additional SCAN domains. In brief, GATEWAY BP cloning (Invitrogen) was used to produce DNA fragments coding for the mouse SCAN domains of Zscan4, Zfp110, Zfp167, ZNF75, MZF1, ZNF24, Zfp213, ZNF165, ZNF174, Zfp192, ZNF193, ZNF19, ZNF390, ZNF394, ZNF435, ZNF452, Zfp445, Zfp449 and Zfp496 (see Supplementary Table 1 for cloning primers and identifiers of the polymerase chain reaction (PCR) templates). To produce a form of SCAN proteins fused to a MBP tag, SCAN domains were transferred into pDEST-HisMBP (20) expression vector by GATEWAY LR (Invitrogen) following the manufacturers’ instructions.
Chimeric proteins were produced as follows. A DNA fragment of Zfp206 SCAN (amino acids 36–128) was amplified from the full-length Zfp206 cDNA (IMAGE: 30006755) by PCR using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAAACCTGTATTTTCAGGGCAGGCCTAGGCCTGAGGTGGCC-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTACATGTGGCTGATGTCTCTGGG-3′. DNA fragment of Zfp110 SCAN (amino acids 154–247) was amplified from the full-length Zfp110 cDNA (IMAGE: 3500984) using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAAACCTGTATTTTCAGGGCCGTTTGACTGACACTGAAGCT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTAATCGTCCTTAGACACCGAGGT-3′. The two PCR products were linked by a linker sequence 5′-CCCAGAGACATCAGCCACATGGGTGGTTCCGGTCGTTTGACTGACACTGAAGCT-3′ to produce a PCR product of Zfp206SCAN–Zfp110SCAN connected by a linker. The PCR product was cloned into the Gateway entry vector pDONR221 (Invitrogen). The DNA sequence of Zfp206SCAN–Zfp110SCAN in the produced entry vector pENTR-Zfp206SCAN–Zfp110SCAN was confirmed by sequencing. By LR reaction (Invitrogen), the Zfp206SCAN–Zfp110SCAN insert was cloned into the destination vector pTH27. The final expression vector was named as pTH27-Zfp206SCAN–Zfp110SCAN.
Deletion mutations of Zfp206SCAN–Zfp110SCAN were created using standard PCR amplifications. All point mutations of Zfp206SCAN–Zfp110SCAN in this study were created using the standard PCR-based mutagenesis method (QuikChange II Site-Directed Mutagenesis Kit, Stratagene) (see Supplementary Tables 2 and 3 for mutagenesis primers) and verified by DNA sequencing.
Protein expression and purification
The procedure for the expression and purification of Zfp206SCAN protein and other factors was carried out as described previously (19). In brief, the pDEST-HisMBP expression plasmids with different SCAN domain inserts were transformed into Escherichia coli BL21 (DE3) cells (Invitrogen). Ten milliliters of overnight culture of the cells was inoculated into 250 ml Terrific Broth (TB) medium supplemented with 100 μg/ml ampicillin. The cells were then grown at 37°C until OD600 reached 0.6. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into the culture with a final concentration of 0.5 mM to induce the expression of MBP-fused SCAN proteins. After induction, the cells were cultured overnight at 18°C. The harvested cells were lysed in buffer containing 50 mM Hepes, pH 7.3, 200 mM NaCl, 1 mM EDTA and 10 mM 2-mercaptoethanol. The MBP-fused SCAN proteins were purified by using amylose–resin (New England Biolabs (NEB)) and column chromatography as described previously (19).
The pTH27-Zfp206SCAN–Zfp110SCAN expression plasmid (21) was transformed into E. coli BL21 (DE3) cells (Invitrogen). Two hundred milliliters of overnight culture of the cells transformed with pTH27-Zfp206SCAN–Zfp110SCAN expression plasmid was used to inoculate 6 lTB medium with 100 μg/ml ampicillin. The cells were then grown at 37°C until OD600 reached 0.7. To induce protein expression, IPTG was added into the culture with a final concentration of 0.5 mM. The cells were further grown overnight at 20°C. The harvested cells were lysed in buffer containing 50 mM Tris–Cl, pH 8.0, 150 mM NaCl, 30 mM imidazole and 10 mM 2-mercaptoethanol. The initial purification was conducted by using affinity technique with Ni-NTA+ (Invitrogen). Subsequent purification of Zfp206SCAN–Zfp110SCAN protein-included cation-exchange chromatography using Resource S resin (GE Healthcare) and gel filtration using a Sephacryl S75 16/60 column (GE Healthcare). The purified Zfp206SCAN–Zfp110SCAN protein were pooled and stored in appropriate buffer for further experiments.
All deletion mutated and point mutated Zfp206SCAN–Zfp110SCAN proteins were purified by the same procedure as described for the wild-type protein.
MBP pull-down assay
Twenty micrograms of MBP or MBP-SCAN proteins (2 mg/ml) were used for each binding reaction and incubated with 100 µg of purified and tag-free Zfp206SCAN (5 mg/ml) in 250 μl binding buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol and 0.1% Triton X-100) overnight at 25°C. Then 10 μl of amylose beads was added into the respective reaction and incubated for another 3 hr at 4°C. After extensive washing with the binding buffer for three times, the proteins were eluted with the sodium dodecyl sulfate (SDS) loading buffer and analyzed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE). The identity of Zfp206SCAN protein pulled down was confirmed by mass spectrometry. Intriguingly, Zfp206SCAN was found to be capable of heterodimer formation and did so in a highly selective fashion.
Immunoprecipitation
Immunoprecipitation was performed using Pierce Crosslink IP kit (#26147) according to manufacturer’s protocol. Briefly, 10 µg of antibody was coupled and crosslinked to protein A/G plus agarose resin. A total of 1 × 108 mouse ESCs were rinsed with phosphate-buffered saline, scraped and lysed in immunoprecipitate lysis buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40 and 5% glycerol supplemented with 1× EDTA-free protease inhibitor, Roche) for an hour at 4°C. Lysate was centrifuged at 10 000 g (13 000 rpm) for 15 min. The supernatant was collected and subjected to Protein assay (Bio-rad). One milligram of whole cell protein lysate was incubated with the crosslinked antibody overnight at 4°C. Immunoprecipitated proteins were washed, eluted and resolved using 12% SDS–PAGE. Protein of interest was then detected by the corresponding antibody in western analysis. Immunoprecipitation and western analysis were performed using anti-Zfp206 antibody (1:1000 (1)) and anti-Zfp110 antibody (1:500; Abcam #ab68789).
Crystallization of Zfp206SCAN and preparation of heavy atom derivative
Crystallization of Zfp206SCAN and preparation of heavy atom derivative were done as described previously (19). In brief, crystal hits of Zfp206SCAN were obtained by high-throughput crystallization screen and manually optimized in a hanging-drop setting. The better crystals were obtained with 15 mg/ml of Zfp206 protein under the condition of 0.3 M ammonium sulfate, 0.1 M Tris–Cl, pH 8.6 and 25% PEG 3350 with 25 mM ethylenediaminetetraacetic acid disodium salt dehydrate. Mercury derivative crystals were prepared by soaking crystals in a reservoir solution containing 10 mM HgCl2. Finally, crystals were transferred to a reservoir solutions containing 10% glycerol and flash frozen directly in liquid nitrogen.
X-ray data collection, processing and structure solution
A native data set at 1.85 Å resolution and a mercury derivative data at 2.2 Å resolution were collected on the X29 beamline at the Macromolecular Crystallography Research Resource (PXRR, USA). All data set were processed using the HKL-2000 software (22) (Table 1). Five heavy atom sites of Zfp206SCAN were found by SOLVE/PHENIX (23) at a resolution range of 50–2.2 Å. The Single anomalous dispersion (SAD) phases were calculated in AUTOSOL/PHENIX (24). The model derived from the data of mercury derivative crystals was used as an initial model for molecular replacement using the higher resolution native dataset of Zfp206SCAN by AUTOMR in PHENIX. An initial model was automatically built using AUTOBUILD in PHENIX. The model was manually built into 2Fo–Fc and Fo-Fc maps in COOT (25). The refinement was carried out using PHENIX.REFINE (26). Translation/Libration/Screw (TLS) refinement was applied in the last steps of the refinement. PyMol (27) (http://www.pymol.org) was used for generating molecular figures and structural analysis.
Table 1.
Crystallographic data collection and refinement statistics
| Parameter | Native | Hg |
|---|---|---|
| Crystal data | ||
| Space group | I422 | I422 |
| Cell dimensions (Å) | ||
| a | 67.565 | 67.776 |
| c | 87.544 | 87.456 |
| Diffraction dataa | ||
| Wavelength (Å) | 1.08 | 1 |
| Resolution (Å) | 50–1.85 (1.92–1.85) | 50–2.2 (2.28–2.2) |
| Rmergeb (%) | 4.5 (46.8) | 8.5 (61.5) |
| I/I | 47.5 (6.0) | 41.1 (6.7) |
| Completeness (%) | 100 (100) | 100 (100) |
| Redundancy of data | 15.2 (14.1) | 29.9 (27.5) |
| Phasing statistics | ||
| Riso | 32.2 | |
| No. sites | 4 | |
| Rcullis (acentric/centric) | 0.673/0.339 | |
| Structure refinement | ||
| Resolution (Å) | 47.4–1.85 (1.97–1.85) | |
| No. reflections | 8692 | |
| Rwork/Rcfree (%) | 20.3/23.5 (20.5/26.3) | |
| No. atoms | ||
| Protein | 690 | |
| Water | 25 | |
| Average isotropic (or equivalent) B-factors | ||
| Macromolecule | 45.135 | |
| Main chain (85 residues) | 40.857 | |
| Side chain (74 residues) | 49.290 | |
| Solvent | 41.21 | |
| R.m.s deviations from ideal | ||
| Bond lengths (Å) | 0.008 | |
| Bond angles (°) | 0.984 | |
| Ramachandran analysis (%) | ||
| Favored | 97.5 | |
| Additionally allowed | 2.5 | |
| Disallowed | 0 |
aValues in parentheses are for the highest resolution shell.
bRmerge = Σhkl Σi |Ii(hkl) − <I(hkl)> |/Σhkl Σi Ii(hkl), where Ii(hkl) and <I(hkl)> are the intensity of measurement i and the mean intensity for the reflection with indices hkl, respectively.
cRwork = Σhkl[||Fobs| − k|Fcalc||]/Σhkl[|Fobs|]; Rfree = ΣhklCT [||Fobs| − k|Fcalc||]/ΣhklCT[|Fobs|]; hklCT – test set.
Model for Zfp206SCAN–Zfp110SCAN heterodimer
Currently, a 3D structure is unavailable for Zfp110SCAN. We built a three-dimensional model for Zfp110SCAN using homology modeling software, MODELLER (28). Zfp206SCAN was used as a structural template for modeling. The accurate alignment between query and template protein is important for homology modeling. We performed the pairwise alignment between Zfp110SCAN and Zfp206SCAN using CLUSTALW software (29). The sequence identity between the two domains is 42.7%. We generated 20 homology models, and the model with lowest energy was selected as a final model for further analysis. To assess the quality of the model to check whether the model has any local or global error, we utilized HARMONY server, which assesses the compatibility of an amino acid sequence with a proposed three-dimensional structure (30).
We employed two steps to create Zfp206SCAN–Zfp110SCAN heterodimer. In the first step, Zfp206SCAN model was superimposed on top of Zfp110SCAN homodimer using LSQMAN program (31). Zfp206SCAN homodimer consists of two identical subunits namely chain A and chain B. LSQMAN program superimposed Zfp110SCAN model on top of chain B of Zfp206SCAN. In the second step, chain B of Zfp206SCAN was removed from the superimposed coordinates. This gave us a heterodimer complex containing Zfp110SCAN and Zfp206SCAN. Molecular dynamics (MD) simulations were performed to analyze the stability of the model (32) (see Supplementary Method).
RESULTS
Overall structure of Zfp206SCAN
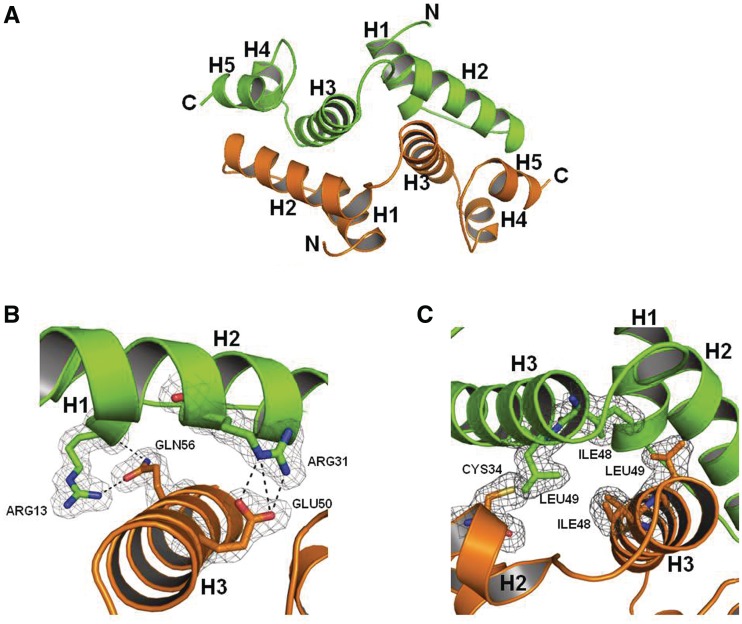
The 1.85 Å structure of Zfp206SCAN was solved by molecular replacement using a lower resolution model derived from the data of a mercury-derived Zfp206SCAN crystal, and the structure was finally refined with a Rfree value of 23.5% including data to 1.85 Å resolution (Table 1). The backbone dihedrals are found in ‘allowed’ regions of Ramachandran space. The deviations between the bond distances in the model and ‘ideal’ bond distances is small (0.008 Å) and so are the bond angle deviations (0.984°). Except for the C-terminal eight residues ‘VPRDISHM’, all the other amino acids are visible in the electron density map. Zfp206SCAN is a domain-swapped antiparallel homodimer arranged around a crystallographic 2-fold axis with five α helices in each monomer (Figure 1A). The loop between helix 2 and helix 3 connects the two subdomains in each monomer. Helix 2 and helix 3 form the core of the dimer interface with helix 2 packing against helices 3 and helix 5 of the opposing monomer and vice versa. The amino-terminal helix 1 provides additional dimer contact with helix 3 of the other monomer (Figure 1B). However, helix 4 does not contribute to the dimer interface contacts (Figure 1A). The dimer interface contains several polar interactions (Figure 1B), but the majority of the contact interface is composed of hydrophobic amino acids (Figure 1C). The electron density is very clear at most regions of the model enabling unbiased modeling, and the dimer interface, in particular, is very well defined (Figure 1B and C) (33).
Figure 1.
Overall structure of Zfp206SCAN. (A) The Zfp206SCAN domain-swapped dimer is formed by packing helix H2 of one monomer (green) against helices H3 and helix H5 of the opposing monomer (orange). (B) The (2FO-FC) map of the dimer interface involving residues Arg13 and Arg31 in molecule 1 and Gln56 and Glu50 (as indicated in Figure 2A) in molecule 2 of Zfp206SCAN contacted by hydrogen bond. The electron density (2Fo-Fc) is displayed at the 0.5σ level. (C) The (2FO-FC) map of the dimer interface involving residues Ile48 and Leu49 in molecule 1 and Cys34, Ile48 and Leu49 of molecule 2 (as indicated in Figure 2A).
Selective dimerizations of Zfp206SCAN with other SCAN domains of family members
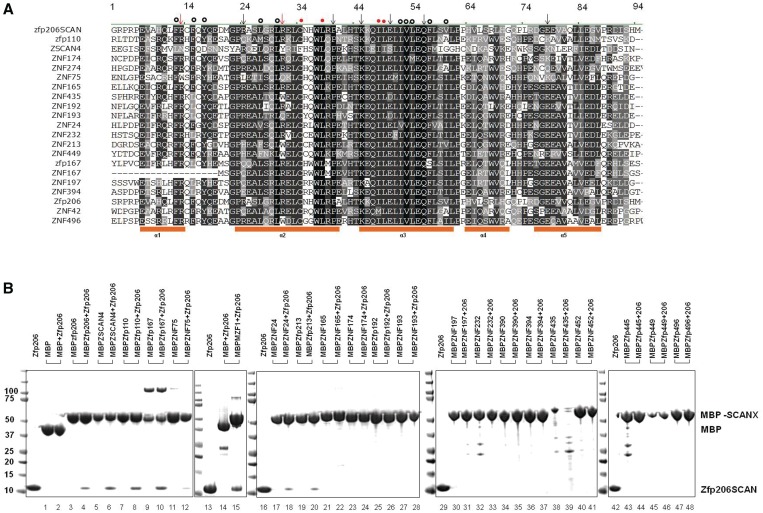
Next, we asked whether the Zfp206SCAN is able to form heterodimers with other SCAN family members. To this end, we performed multiple sequence alignments using CLUSTAL W (34) and selected 20 SCAN domains (details of functions shown in Supplementary Table 4) TFs based on the distances to Zfp206 on a phylogenetic tree based on multiple sequence alignments of 52 SCAN domain for interaction studies (Figure 2A and Supplementary Figure 1). We expressed and purified those 20 SCAN family members with N-terminal MBP tags and conducted MBP pull-down assays to identify partners of the Zfp206SCAN. Zfp206SCAN was purified from MBP-Zfp206SCAN by TEV digestion to remove MBP tag. MBP-Zfp206SCAN is a dimer in solution (Supplementary Figure 2). To increase the chance of heterodimer formation, excess tag-free Zfp206SCAN (100 µg) was mixed with MBP-SCAN proteins (20 µg). Higher incubation temperature (25°C) other than 4°C was chosen to favor the formation of heterodimers. In the previous study (35), high concentration and temperature were adopted for heterodimerization of SCAN domains in pull-down assay too. Zfp206SCAN was pulled down by MBP-fused SCAN domains of ZSCAN4, Zfp110, Zfp167, ZNF24, ZNF75 and MZF1 (Figure 2B). However, the SCAN domains of ZNF165, ZNF174, Zfp192, ZNF193, ZNF197, ZNF390, ZNF394, ZNF435, ZNF452, Zfp445, Zfp449 and Zfp496 did not interact with Zfp206SCAN at all (Figure 2B). This suggests that the Zfp206SCAN selectively interacts with a subset of SCAN domains amongst the 20 selected candidates. In addition, this indicates that ZSCAN4, Zfp110, Zfp167, ZNF24, ZNF75 and MZF1 have the potential to be novel biological partners for Zfp206. The binding was also observed between Zfp206SCAN and MBP-Zfp206SCAN, which further supports the homodimerization of the SCAN domain of Zfp206.
Figure 2.
In vitro MBP pull-down assay of Zfp206SCAN and other SCAN domains. (A) Multiple sequence alignments of the SCAN domains used in MBP pull-down assay. Secondary structure elements of SCAN domains are shown under the alignments as orange blocks. Arrows are for polar interface residues, and dots are for hydrophobic residues seen in the structure of the Zfp206SCAN homodimer. The numbers are shown above the alignment mark residues with the conserved portion of the SCAN domain for easier comparison. The red arrows and dots are the mutated residues described in the later study (Figure 4). (B) Pull-down of non-tagged Zfp206SCAN using amylose beads pre-coated with recombinant MBP or MBP-SCAN fusion proteins. The prey proteins were detected by SDS–PAGE and Coomassie stain, and identified to be Zfp206SCAN by mass spectrometry. The upper bands in lanes 9 and 10 are the dimer of MBP167SCAN. The additional bands seen in lanes 14, 15, 32, 39 and 43 are non-specific proteins produced during purification.
We also tried to co-express and purify Zfp206SCAN (with C-terminal His-tag)-Zfp110SCAN (with C-terminal Strep-tag) heterodimer, which was cloned in pCOLA plasmid. The complex was first purified by Ni-NTA affinity column and then by streptactin beads. After two steps of affinity purifications, Zfp206SCAN–Zfp110SCAN heterodimer was purified (Supplementary Figure 3).
Domain-swapped topology is a possible preference of Zfp206SCAN–Zfp110SCAN heterodimer
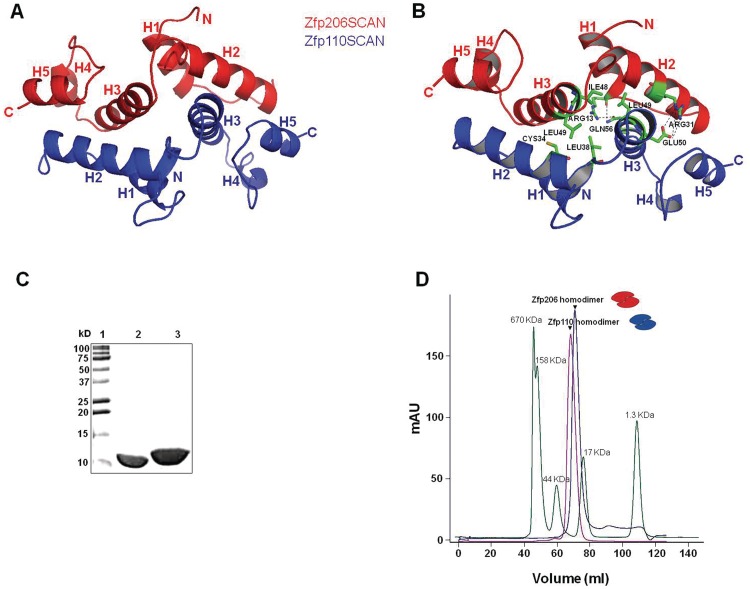
Next, we wanted to understand the structural basis for the selective heterodimerization of Zfp206 and Zfp110. To this end, we generated a structural model of Zfp110SCAN, a homodimer, using MODELLER (28) based on the atomic coordinates of Zfp206SCAN homodimer. We then superimposed one single chain of Zfp110SCAN on one chain of Zfp206SCAN to create the model of Zfp206SCAN–Zfp110SCAN heterodimer. Next, we performed MD simulations to confirm the stability and the molecular contacts of the heterodimer model. In our model, Zfp206SCAN–Zfp110SCAN heterodimer resembles Zfp206SCAN homodimer with a domain-swapped topology (Figure 3A). The interface interactions to stabilize the Zfp206SCAN–Zfp110SCAN heterodimer involve a certain number of hydrophobic interactions and hydrogen bonds (Figure 3B). Next, we attempted to introduce interface mutations to selectively disrupt homodimer or heterodimer formation.
Figure 3.
Model for Zfp206SCAN-Zfp110SCAN heterodimer. (A) Model for Zfp206SCAN-Zfp110SCAN created by superimposing Zfp110SCAN monomer on Zfp206SCAN molecule of the homodimer followed by MD simulations. In the model, the Zfp206SCAN monomer is presented in red and the Zfp110SCAN monomer is presented in blue. (B) Based on the model of Zfp206SCAN and Zfp110SCAN heterodimer, interface residues (shown in green) including Arg 13, Arg 31, Ile 48 and Leu 49 in Zfp206SCAN monomer and Cys 34, Leu 38 and Leu 49 in Zfp110SCAN were chosen for mutations to disrupt interactions between the two monomers. (C–D) Homodimerization of Zfp206SCAN and Zfp110SCAN. (C) Fifteen percent SDS–PAGE showing purified Zfp206 in lane 2 and Zfp110 in lane 3 and molecular weight standards in lane 1 in kilo Daltons. (D) Size-exclusion chromatogram showing that the Zfp206SCAN as well as the Zfp110SCAN elute as a single symmetric peaks corresponding to the molecular weight of the homodimeric forms of the proteins (21 and 24.8 kDa). The cartoons represent the protein conformations.
Construction of Zfp206SCAN–Zfp110SCAN heterodimer connected by a linker
Next, we wanted to further study the basis for the selective heterodimerization of Zfp206 and Zfp110. The Zfp206SCAN protein could be expressed and purified from E. coli in a soluble form as shown in the SDS–PAGE analysis (Figure 3C). When analyzed by size-exclusion chromatography, the Zfp206SCAN elutes as a single symmetric peak reminiscent of the molecular weight of the dimeric form of the protein (21 kDa, Figure 2E). Likewise, the purified Zfp110SCAN elutes at a size corresponding to a homodimeric form (Figure 3C and D). We next asked whether the homodimers are preferentially formed when the Zfp206SCAN and Zfp110SCAN are mixed. However, we are unable to distinguish homodimers and heterodimers in size-exclusion chromatograms. Therefore, we resorted to the stochiometric analysis of a Zfp206SCAN–Zfp110SCAN fusion protein.
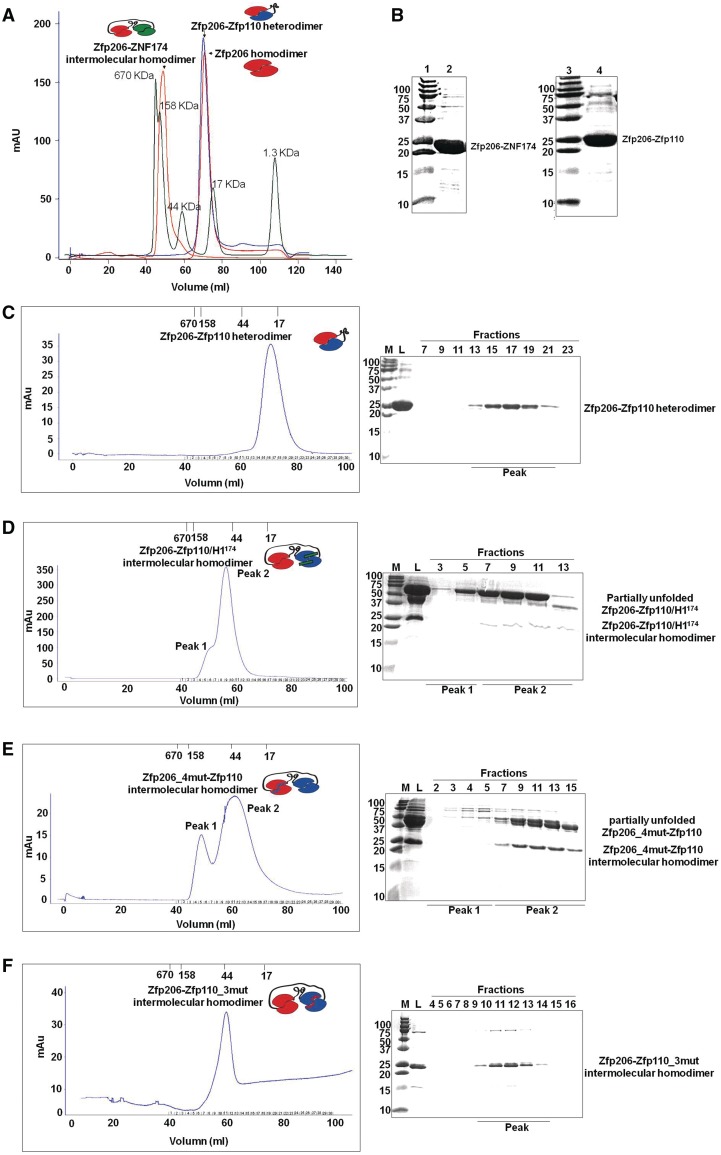
To design the fusion protein, we connected the Zfp206SCAN and Zfp110SCAN with a flexible 21 amino acid linker that would permit the formation of either intramolecular or intermolecular complex (Figure 4A). By size-exclusion analysis, we observed a single peak reminiscent of the theoretical molecular weight of a single Zfp206SCAN–Zfp110SCAN fusion protein of 23 kDa forming an intramolecular heterodimer (Figure 4A–C). This peak perfectly superimposes with a peak obtained from unlinked homodimers of the Zfp206SCAN. Because both of Zfp206SCAN and Zfp110SCAN can self-associate in the absence of a linker, the absence of an intermolecular interaction between fusion proteins strongly suggests that heterodimeric interactions are preferred between the Zfp206SCAN and the Zfp110SCAN, while homodimers would be formed only in the absence of a heterodimerization partner. The Zfp206–Zfp110 heterodimer shows α-helical characters in CD spectra (Supplementary Figure 4A). By contrast, an analogous chimeric Zfp206SCAN–ZNF174SCAN fusion protein eluted at a volume suggesting a molecular weight higher than 46 kDa (Figure 4A). This molecular weight suggests intermolecular interactions reminiscent of a homodimeric interaction in the absence of a linker. Consistently, Zfp206SCAN and the ZNF174SCAN were found to not interact in our pull-down assay (Figure 2B). Thus, our size exclusion-based assay allows us to robustly distinguish between SCAN homodimers and heterodimers and verifies that Zfp206 and Zfp110 form heterodimers, whereas Zfp206 and ZNF174 do not.
Figure 4.
Role of the amino-terminal helix 1 for Zfp206SCAN and Zfp110SCAN heterodimerization and mutations based on the Zfp206SCAN-Zfp110SCAN heterodimer. (A) The intramolecular linker-mediated Zfp206-Zfp110 heterodimer elutes at the volume corresponding to the theoretical molecular weight of a monomeric fusion protein 23 KDa, which is virtually identical with the elution profile of un-connected Zfp206SCAN homodimer (21 KDa). Oppositely, the Zfp206-ZNF174 elutes at the volume corresponding to a higher molecular weight indicating the formation of an intermolecular homodimer. The profile of a molecular weight standard is overlaid as green curve. (B) 15% SDS–PAGE gel shows purified Zfp206-ZNF174 (lane 2) and Zfp206-Zfp110 (lane 4) and molecular standards (lane 1 and 3; weights in kilo Daltons). (C) Elution profile of the linker mediated intramolecular Zfp206-Zfp110 heterodimer (left) and SDS–PAGE of the eluted fractions (right). (D) Elution profile of the Zfp206-/Zfp110_H1174 protein suggesting the formation of an intermolecular homodimer instead of the linker-mediated heterodimer seen in (C). Cartoon drawing of the various complexes are shown as inset of the chromatograms with the Zfp206SCAN in red, the Zfp110SCAN in blue and the ZNF174SCAN in green. The linker is illustrated with a black line and the helix1 of the ZNF174SCAN that was introduced into the Zfp110SCAN is shown as green bar. (E) Zfp206-4mut-Zfp110 intermolecular homodimer (Zfp206SCANI48AL49AR13AR31A-Zfp110SCAN) and (F) Zfp206-Zfp110-3mut intermolecular homodimer (Zfp206SCAN-Zfp110SCANC36AL40AL51A).
The amino-terminal helix 1 plays a role in heterodimerization of Zfp206SCAN and Zfp110SCAN
The amino-terminal helix 1 of the SCAN domain shows the most diverse sequence and a variable pattern of hydrophobic residues (Figure 2A). Hence, helix1 is a candidate to encode selectivity determinants that underlie dimerization preferences of SCAN domains (36). Therefore, we decided to investigate the role of amino-terminal helix 1 for the dimerization of Zfp206SCAN and Zfp110SCAN. As indicated before, ZNF174SCAN did not interact with Zfp206SCAN, so we swapped the helix 1 of Zfp110SCAN with the helix 1 of ZNF174SCAN and constructed a chimeric Zfp206–Zfp110/H1174 protein. Interestingly, Zfp206–Zfp110/H1174 resulted in an intermolecular homodimer (Figure 4D) and some large partially unfolded products (Figure 4D and Supplementary Figure 4B) indicated by size-exclusion chromatography, suggesting the disruption of Zfp206–Zfp110 heterodimer in the presence of H1174. Next, helix 1 of ZNF174SCAN was swapped with the helix 1 of Zfp110SCAN to construct a chimeric ZNF174/H1110SCAN protein. The ZNF174/H1110SCAN exhibited an enhanced ability to interact with Zfp206SCAN when compared with wild-type ZNF174SCAN (Supplementary Figure 5D, and Supplementary Figure 5A shows the control for heterodimer.). In addition, deletion of the helix 1 of Zfp206SCAN and Zfp110SCAN, respectively, resulted in the formation of intermolecular homodimers of Zfp206SCAN–Zfp110SCAN ΔH1 and Zfp206SCANΔH1–Zfp110SCAN (peak 1 shown in Supplementary Figure 5B and C).
To further dissect determinants for the selective dimerization, we chose to mutate two charged residues (Zfp206SCANR13AR31A) and two other hydrophobic residues (Zfp206SCANI48AL49A) of the dimer interface based on our heterodimer models (Figure 3). Individual point mutations did not result in the disruption of the interaction between Zfp206SCAN–Zfp110SCAN (data not shown). However, double mutants Zfp206SCANI48AL49A and Zfp206SCANR13AR31A lead to the formation of a heterogeneous elution profile with intermolecular complexes indicating a loss of a stable intramolecular complex as proxy for a heterodimer (Supplementary Figure 6B and C, and Supplementary Figure 6A is shown as a control for heterodimer). Furthermore, combining all four mutations (Zfp206SCANI48AL49AR31AR13A) revealed one peak at the position of dimerization of Zfp206SCAN–Zfp110SCAN protein by gel filtration (Figure 4E) and some large partially unfolded products (Figure 4E and Supplementary Figure 4C). Likewise, mutations on corresponding Zfp110SCAN interface residues (Zfp110SCANC34AL38AL49A and Zfp110SCANE50AQ56A) also resulted in upper shifted peak compared with the chromatography of the native Zfp206SCAN–Zfp110SCAN (Figure 4F and Supplementary Figure 6D).
Zfp206 interacts with Zfp110 in embryonic stem cells
Zfp206 is known to play a role in maintaining pluripotency (1). Zfp110 was reported to participate in apoptosis in the mouse embryonic neural retina (17). To test the endogenous interaction between Zfp206 and Zfp110 in the ESCs, we assayed for their interaction via co-immunoprecipitation. The anti-Zfp110 antibody was used to pull down the endogenous Zfp110 protein. Anti-Zfp206 antibody was then used to probe the pull-down products to test whether Zfp110 interacted with Zfp206 (Figure 5). In the reciprocal experiment, anti-Zfp206 antibody was used to immunoprecipitate the endogenous Zfp206 protein. The pulled down products were probed with anti-Zfp110 antibody to examine whether Zfp206 interacted with Zfp110 (Figure 5). The results of both experiments showed that Zfp206 co-immunoprecipitated with Zfp110.
Figure 5.
Zfp206 interacts with Zfp110 in vivo. (A) Anti-Zfp110 antibody was used to immunoprecipitate (IP) Zfp110 and the co-IP products were analyzed by western blot (WB) with an anti-Zfp206 antibody to test whether Zfp110 interact with Zfp206. Vice versa, (B) anti-Zfp206 antibody was used to immunoprecipitate Zfp206, and co-IP products were analyzed by WB with an anti-Zfp110 antibody to check whether Zfp206 interacts with Zfp110.
DISCUSSION
Transcription factor proteins often form molecular complexes on cis-regulatory DNA to combinatorially regulate the expression of nearby genes. In many cases, those partnerships are mediated by the DNA-binding domains through direct protein contacts or via an indirect, DNA-mediated, crosstalk between the partnering proteins (37–41). We have previously demonstrated that subtle modifications at the dimerization interface of TFs can have profound consequences and dramatically swap the potential of TFs to trigger cellular specification events (40). However, some TFs contain additional domains that can affect their dimerization potential and thereby control which genes are regulated. Therefore, we decided to study the SCAN that is found in many transcription factors with critical roles in early mammalian development.
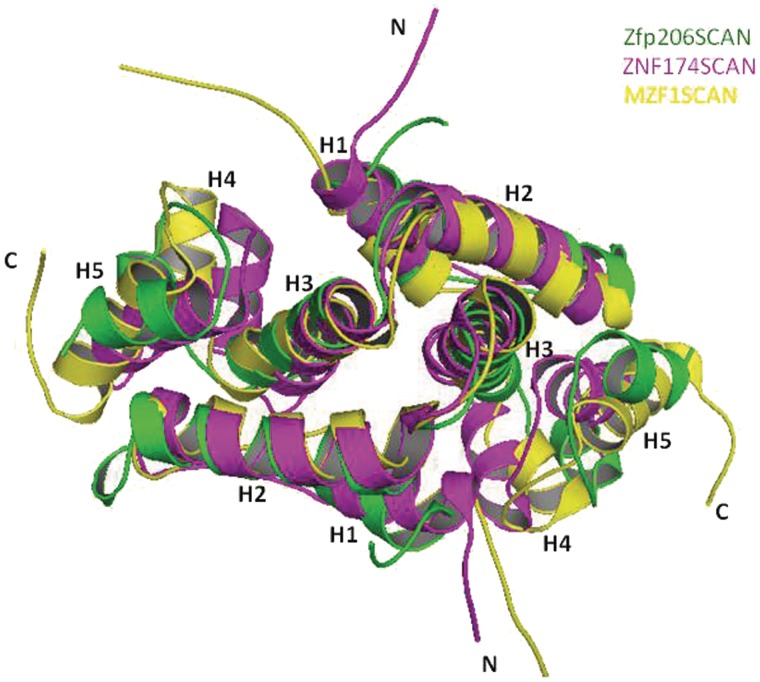
To shed light on the complex formation of C2H2 zinc finger proteins, we report the structure of the SCAN domain of Zfp206 transcription factor to high resolution. Zfp206SCAN has a domain-swapped topology similar to two other family members, which were solved by NMR (14,15) (Figure 6). The presence of the SCAN domain in Zfp206 transcription factor and the homodimerization of this SCAN domain suggest that Zfp206 might bind to DNA as a homodimer. This is consistent with the finding that Zfp206 binds to a consensus palindrome motif GCGCATGCGC (3). It is assumed that the members from the SCAN family of transcription factors form a transcription network by self-association and selective association with other members mediated by SCAN domain (11,13,36). The results of our MBP pull-down assay suggest that Zfp206 can selectively interact with other SCAN-containing transcriptional regulators by SCAN-mediated heterodimerization. Zfp206 is reported to play an important role in maintaining the pluripotent stage of ESCs (1,3). The newly found biological partners for Zfp206 give us more clues about the functions of this transcription factor. For instance, as reported, Zscan4 promotes telomere elongation and maintains genomic stability in ESCs (16). Zfp206 might collaborate with Zscan4 or regulate its functions by heterodimerization. Another partner ZNF24 (ZNF191) is known to be necessary for the maintenance of the undifferentiated stage of neural progenitors (42). This suggests possible role of Zfp206 in inhibiting neuronal differentiation and the maintenance of progenitor characteristics. This is consistent with the finding that Zfp206 functionally teams up with Sox2 (3), another key component of maintaining neural progenitor identity (43). However, till now there is little knowledge about the functions of some other partners indicated in our results, such as ZNF75, Zfp167 and Zfp213. The interactions of these transcription factors with Zfp206 might hint at their possible roles involved in the transcription pathways of ESCs.
Figure 6.
Structural alignment of Zfp206SCAN homodimer with ZNF174SCAN homodimer (PDB entry 1Y7Q) and MZF1SCAN homodimer (PDB entry 2FI2). The Zfp206SCAN is shown in green; the ZNF174SCAN is shown in pink and MZF1SCAN is shown in yellow.
Zfp110, also known as neurotrophin receptor interacting factor, is reported to be involved in p75(NTR)/NGF-mediated developmental cell death in the mouse embryonic neural retina. Zfp110 is ubiquitously expressed at higher and constant levels in the embryo than in the adult. Overexpressed GFP-Zfp110 alone is primarily located in nucleus but could trans-localize outside the nucleus on co-expression of p75NTR (17). The heterodimerization of Zfp206 and Zfp110 was indicated in our in vitro MBP pull-down assay and in vivo co-immunoprecipitation experiments. It is possible that Zfp110 is retained in the nucleus to conduct functions of gene regulation by interacting with Zfp206 through SCAN dimerization, but not translocate outside the nucleus to interact with p75NTR to induce apoptosis. Apoptosis is known to be associated with differentiation of ESCs (44). This indicates another potential function of Zfp206 for maintaining pluripotency in ESCs.
To study the heterodimerization of Zfp206SCAN and Zfp110SCAN, we constructed a fusion protein Zfp206SCAN–Zfp110SCAN by connecting the two monomers with a flexible linker. As shown in the results of size-exclusion chromatography, the interaction between Zfp206SCAN and Zfp110SCAN make this Zfp206SCAN–Zfp110SCAN protein favor a compact topology of a heterodimer (23 kDa) rather than the formation of homodimer or multimer. This construct also facilitated the evaluation of mutation effects on the heterodimer. A previous study had shown that the monomer status of the SCAN domain is unfolded. The SCAN domain is in equilibrium between the domain-swapped folding status and the unfolded monomer (12). Domain swapping is a kind of oligomerization in which each monomer is interchanged with another identical partner by interactions that are important for stabilizing the protein from its unfolded monomeric form (45). One recent study showed that the overall monomer topology is a crucial factor that determines the preference for the domain-swapped topology (46). The sequence alignment (Figure 2A) and structural alignment (Figure 6) of SCAN domains indicate high conservation between each member. We superimposed the Zfp110SCAN monomer on one monomer of Zfp206SCAN to create the Zfp206SCAN–Zfp110SCAN heterodimer. Similar to the homodimer, the heterodimer interface interactions involve a number of hydrophobic interactions and hydrogen bonds (Figure 3B). Based on the analysis of the model interface, double mutations or multiple mutations on Zfp206SCAN monomer or comparable Zfp110SCAN monomer, which disrupt the intramolecular hydrophobic interactions or hydrogen bonds between these two monomers, resulted in the loss of compact topology of the Zfp206SCAN–Zfp110SCAN protein, and gave rise to intermolecular interactions resulting in the formation of Zfp206SCAN–Zfp110SCAN intermolecular homodimer. The mutations not only disrupt the interactions between Zfp206SCAN and Zfp110SCAN but also prevent the formation of intermolecular homodimer. This explains the formation of partially unfolded products due to the flexible SCAN monomer failing to form homodimer in the linker protein (12). This composite data thus suggest the possibility of a domain-swapped topology for Zfp206SCAN-Zfp110SCAN heterodimer. Deleting the N-terminal helix1 of either Zfp206SCAN monomer (Supplementary Figure 5B) or Zfp110SCAN monomer (Supplementary Figure 5C) and swapping amino terminal helix 1 of Zfp110SCAN with that of ZNF174SCAN (Figure 4D), which is not a partner for Zfp206SCAN, resulted in disruption of the interaction between Zfp206SCAN and Zfp110SCAN and the formation of Zfp206SCAN–Zfp110SCAN intermolecular homodimer or partially unfolded products. This indicated that the amino-terminal helix 1 plays a role in heterodimerization of Zfp206SCAN and Zfp110SCAN. However, there is still a population of protein with the same character of the native Zfp206SCAN–Zfp110SCAN protein. This is consistent with our model that other parts of SCAN domain such as helix 2 and helix 3 also enable heterodimerization. Based on our model, the N-terminal helix 1 appears to not participate within the core of the dimer interface. During the initial studies, we had difficulties in getting soluble Zfp206SCAN protein without N-terminal helix 1 (data not shown). It has been demonstrated that N-terminal motif participates in the initial folding stage of a protein (47,48). The SCAN monomer is in unfolded state (12). From energy landscape analysis, it is inferred that the most favorable mechanism of domain swapping is native monomers ↔ partially folded monomers ↔ unfolded monomers ↔ open-end domain-swapped dimers ↔ domain-swapped dimers (49). The intermediate folding states of SCAN monomers might determine the final topology of heterodimer. The amino-terminal helix 1 of SCAN domain shows the most diverse pattern in hydropathy. Hydropathy is an important force for protein folding. It is possible that difference in N-terminal helix 1 or different contacts between helix 1 and other parts of SCAN domain enable different folding intermediates of the monomers while they encounter each other. This might explain the possible route to selective heterodimerization between SCAN proteins. It is probable that the selectivity may be determined by combinational features in original SCAN monomer topology such as hydropathy, hydrophobicity, buried surface area or isoelectric effects. However, only the structural analysis of heterodimeric SCAN complexes can ultimately resolve the basis for the selective heterodmierziation between members of the SCAN family.
ACCESSION NUMBERS
Coordinate and Structure Factors have been submitted to the protein databank with the accession number 4E6S.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–6, Supplementary Methods and Supplementary References [50–61].
FUNDING
Diffraction data were collected at beamline X29 of the National Synchrotron Light Source (NSLS) supported by the US Department of Energy, Office of Science, Office of Basic energy Sciences. Funding for open access charge: Agency for Science, Technology and Research (A*STAR), Singapore.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Robert Robinson (IMCB, Singapore) for helping with diffraction data collection. They thank Dr Howard Robinson for access to the X29 beamline at the Macromolecular Crystallography Research Resource (PXRR, USA) and for providing assistance in data collection.
REFERENCES
- 1.Wang ZX, Kueh JL, Teh CH, Rossbach M, Lim L, Li P, Wong KY, Lufkin T, Robson P, Stanton LW. Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells (Dayton, Ohio) 2007;25:2173–2182. doi: 10.1634/stemcells.2007-0085. [DOI] [PubMed] [Google Scholar]
- 2.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 3.Yu HB, Kunarso G, Hong FH, Stanton LW. Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. J. Biol. Chem. 2009;284:31327–31335. doi: 10.1074/jbc.M109.016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pengue G, Calabro V, Bartoli PC, Pagliuca A, Lania L. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 1994;22:2908–2914. doi: 10.1093/nar/22.15.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee PL, Gelbart T, West C, Adams M, Blackstone R, Beutler E. Three genes encoding zinc finger proteins on human chromosome 6p21.3: members of a new subclass of the Kruppel gene family containing the conserved SCAN box domain. Genomics. 1997;43:191–201. doi: 10.1006/geno.1997.4806. [DOI] [PubMed] [Google Scholar]
- 7.Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi: 10.1016/s0378-1119(03)00509-2. [DOI] [PubMed] [Google Scholar]
- 8.Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Chung HR, Schafer U, Jackle H, Bohm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002;3:1158–1162. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jauch R, Bourenkov GP, Chung HR, Urlaub H, Reidt U, Jackle H, Wahl MC. The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure. 2003;11:1393–1402. doi: 10.1016/j.str.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis CM, Wang LL, Ballinger D, Bowen BR, et al. The SCAN domain mediates selective oligomerization. J. Biol. Chem. 2000;275:17173–17179. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- 12.Stone JR, Maki JL, Blacklow SC, Collins T. The SCAN domain of ZNF174 is a dimer. J. Biol. Chem. 2002;277:5448–5452. doi: 10.1074/jbc.M109815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams AJ, Blacklow SC, Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell Biol. 1999;19:8526–8535. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov D, Stone JR, Maki JL, Collins T, Wagner G. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol. Cell. 2005;17:137–143. doi: 10.1016/j.molcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Peterson FC, Hayes PL, Waltner JK, Heisner AK, Jensen DR, Sander TL, Volkman BF. Structure of the SCAN domain from the tumor suppressor protein MZF1. J. Mol. Biol. 2006;363:137–147. doi: 10.1016/j.jmb.2006.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casademunt E, Carter BD, Benzel I, Frade JM, Dechant G, Barde YA. The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. EMBO J. 1999;18:6050–6061. doi: 10.1093/emboj/18.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng CK, Li NX, Chee S, Prabhakar S, Kolatkar PR, Jauch R. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012;40:4933–4941. doi: 10.1093/nar/gks153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Y, Choo SH, Rossbach M, Baburajendran N, Palasingam P, Kolatkar PR. Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:443–447. doi: 10.1107/S1744309112006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nallamsetty S, Austin BP, Penrose KJ, Waugh DS. Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 2005;14:2964–2971. doi: 10.1110/ps.051718605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammarstrom M, Woestenenk EA, Hellgren N, Hard T, Berglund H. Effect of N-terminal solubility enhancing fusion proteins on yield of purified target protein. J. Struct. Funct. Genomics. 2006;7:1–14. doi: 10.1007/s10969-005-9003-7. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta. Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta. Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta. Crystallogr. D Biol. Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 28.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2006 doi: 10.1002/0471250953.bi0506s15. Chapter 5, Unit 5 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugalenthi G, Shameer K, Srinivasan N, Sowdhamini R. HARMONY: a server for the assessment of protein structures. Nucleic Acids Res. 2006;34:W231–W234. doi: 10.1093/nar/gkl314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleywegt GJ, Jones TA. A super position. Joint CCP4/ESF-EACBM Newslett. Protein Crystallography. 1994;31:9–14. [Google Scholar]
- 32.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 33.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2, Unit 2 3. [DOI] [PubMed] [Google Scholar]
- 35.Noll L, Peterson FC, Hayes PL, Volkman BF, Sander T. Heterodimer formation of the myeloid zinc finger 1 SCAN domain and association with promyelocytic leukemia nuclear bodies. Leukemia Res. 2008;32:1582–1592. doi: 10.1016/j.leukres.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Nam K, Honer C, Schumacher C. Structural components of SCAN-domain dimerizations. Proteins. 2004;56:685–692. doi: 10.1002/prot.20170. [DOI] [PubMed] [Google Scholar]
- 37.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 38.Muller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 39.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 40.Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, Kolatkar PR. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells. 2011;29:940–951. doi: 10.1002/stem.639. [DOI] [PubMed] [Google Scholar]
- 41.Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 42.Khalfallah O, Ravassard P, Lagache CS, Fligny C, Serre A, Bayard E, Faucon-Biguet N, Mallet J, Meloni R, Nardelli J. Zinc finger protein 191 (ZNF191/Zfp191) is necessary to maintain neural cells as cycling progenitors. Stem Cells (Dayton, Ohio) 2009;27:1643–1653. doi: 10.1002/stem.88. [DOI] [PubMed] [Google Scholar]
- 43.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 44.Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Apoptosis and differentiation of human embryonic stem cells induced by sustained activation of c-Myc. Oncogene. 2007;26:5564–5576. doi: 10.1038/sj.onc.1210353. [DOI] [PubMed] [Google Scholar]
- 45.Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc. Natl Acad. Sci. USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho SS, Levy Y, Onuchic JN, Wolynes PG. Overcoming residual frustration in domain-swapping: the roles of disulfide bonds in dimerization and aggregation. Phys. Biol. 2005;2:S44–S55. doi: 10.1088/1478-3975/2/2/S05. [DOI] [PubMed] [Google Scholar]
- 47.Feng H, Zhou Z, Bai Y. A protein folding pathway with multiple folding intermediates at atomic resolution. Proc. Natl Acad. Sci. USA. 2005;102:5026–5031. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishna MM, Englander SW. The N-terminal to C-terminal motif in protein folding and function. Proc. Natl Acad. Sci. USA. 2005;102:1053–1058. doi: 10.1073/pnas.0409114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang S, Cho SS, Levy Y, Cheung MS, Levine H, Wolynes PG, Onuchic JN. Domain swapping is a consequence of minimal frustration. Proc. Natl Acad. Sci. USA. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP. JOY: protein sequence-structure representation and analysis. Bioinformatics. 1998;14:617–623. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 51.Tina KG, Bhadra R, Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zverev VV, Khmel’ IA. Nucleotide sequence of the replicative region of the plasmid ColA-CA31. Mol. Gen. Mikrobiol. Virusol. 1985:18–24. [PubMed] [Google Scholar]
- 53.Tan S, Guschin D, Davalos A, Lee YL, Snowden AW, Jouvenot Y, Zhang HS, Howes K, McNamara AR, Lai A, et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc. Natl Acad. Sci. USA. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong XY, Yang XA, Wang YD, Chen WF. Zinc-finger protein ZNF165 is a novel cancer-testis antigen capable of eliciting antibody response in hepatocellular carcinoma patients. Br. J. Cancer. 2004;91:1566–1570. doi: 10.1038/sj.bjc.6602138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirata RD, Hirata MH, Strufaldi B, Possik RA, Asai M. Creatine kinase and lactate dehydrogenase isoenzymes in serum and tissues of patients with stomach adenocarcinoma. Clin. Chem. 1989;35:1385–1389. [PubMed] [Google Scholar]
- 56.Shi H, Ren J, Xu H, Pan J, Hao X, Barlow LL, Dong W. Upregulated expression of hITF in Crohn's disease and screening of hITF Interactant by a yeast two-hybrid system. Dig. Dis. Sci. 2010;55:2929–2939. doi: 10.1007/s10620-010-1227-0. [DOI] [PubMed] [Google Scholar]
- 57.Gonsky R, Knauf JA, Elisei R, Wang JW, Su S, Fagin JA. Identification of rapid turnover transcripts overexpressed in thyroid tumors and thyroid cancer cell lines: use of a targeted differential RNA display method to select for mRNA subsets. Nucleic Acids Res. 1997;25:3823–3831. doi: 10.1093/nar/25.19.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang C, Wang Y, Li D, Li Y, Luo J, Yuan W, Ou Y, Zhu C, Zhang Y, Wang Z, et al. Inhibition of transcriptional activities of AP-1 and c-Jun by a new zinc finger protein ZNF394. Biochem. Biophys. Res. Commun. 2004;320:1298–1305. doi: 10.1016/j.bbrc.2004.06.080. [DOI] [PubMed] [Google Scholar]
- 59.Gu X, Zheng M, Fei X, Yang Z, Li F, Ji C, Xie Y, Mao Y. ZNF435, a novel human SCAN-containing zinc finger protein, inhibits AP-1-mediated transcriptional activation. Mol. Cells. 2007;23:316–322. [PubMed] [Google Scholar]
- 60.Luo K, Yuan J, Shan Y, Li J, Xu M, Cui Y, Tang W, Wan B, Zhang N, Wu Y, et al. Activation of transcriptional activities of AP1 and SRE by a novel zinc finger protein ZNF445. Gene. 2006;367:89–100. doi: 10.1016/j.gene.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 61.Lu Z, Poliakov E, Redmond TM. Identification of a KRAB-zinc finger protein binding to the Rpe65 gene promoter. Curr. Eye Res. 2006;31:457–466. doi: 10.1080/02713680600678059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.