Abstract
p21 is a well-established regulator of cell cycle progression. The role of p21 in DNA repair, however, remains poorly characterized. Here, we describe a critical role of p21 in a replication-coupled DNA double-strand break (DSB) repair that is mechanistically distinct from its cell cycle checkpoint function. We demonstrate that p21-deficient cells exhibit elevated chromatid-type aberrations, including gaps and breaks, dicentrics and radial formations, following exposure to several DSB-inducing agents. p21−/− cells also exhibit an increased DNA damage-inducible DNA-PKCS S2056 phosphorylation, indicative of elevated non-homologous DNA end joining. Concomitantly, p21−/− cells are defective in replication-coupled homologous recombination (HR), exhibiting decreased sister chromatid exchanges and HR-dependent repair as determined using a crosslinked GFP reporter assay. Importantly, we establish that the DSB hypersensitivity of p21−/− cells is associated with increased cyclin-dependent kinase (CDK)-dependent BRCA2 S3291 phosphorylation and MRE11 nuclear foci formation and can be rescued by inhibition of CDK or MRE11 nuclease activity. Collectively, our results uncover a novel mechanism by which p21 regulates the fidelity of replication-coupled DSB repair and the maintenance of chromosome stability distinct from its role in the G1-S phase checkpoint.
INTRODUCTION
Of the various forms of DNA damage, the physical severing of the sugar-phosphate backbone poses the greatest threat to genome integrity. DNA double-strand breaks (DSBs) can arise both exogenously, as a consequence of exposure to ionizing radiation and certain chemicals, and endogenously, as a result of collapsed replication forks or reactive oxygen species generated as a by-product of normal metabolic processes. If inappropriately repaired, DSBs have the potential to lead to cell death or cellular transformation, the first step in cancer formation (1). The highly unstable karyotypes of the majority of cancer cells are a testament to the pervasive role of DSBs in the genesis of cancer.
To safeguard against the deleterious effects of DSBs, prokaryotic and eukaryotic organisms have evolved two highly regulated DSB repair pathways: homologous recombination (HR) and nonhomologous DNA end joining (NHEJ). HR is an error-free pathway that requires a homologous DNA sequence, either the sister chromatid or the homologous chromosome, to function as a template for re-synthesis of the severed DNA strand. In humans, the principal effector proteins of HR are homologs of the Saccharomyces cerevisiae RAD52 epistasis group (2). Of this group, the RAD51 protein catalyzes the central step of strand capture and invasion of the homologous DNA template (3). In contrast, NHEJ does not require a homologous DNA sequence and often re-ligates broken DNA ends with disregard for previous contiguity. Thus, NHEJ is often error prone and mutagenic. The principal effector proteins of NHEJ include KU70, KU80, DNA-PKCS, DNA ligase IV and XRCC4 (4). Despite the critical importance of accurate DSB repair for the prevention of cellular transformation and cancer, our understanding of the factors that determine how DSBs are repaired is incomplete. The cell cycle stage is one factor known to have a strong bearing on DSB repair.
The p21Cip1/Waf1 protein is a member of a family of cyclin-dependent kinase (CDK) inhibitors and is a major regulator of cell cycle progression (5). To our knowledge, no studies have systematically addressed the role of p21 in DSB repair. In this study, we have examined the role of p21 in the cellular response to the DSB-inducing agents mitomycin C (MMC), a DNA crosslinking agent, and camptothecin (CPT) and etoposide (VP-16), topoisomerase I and II poisons. We demonstrate that p21 plays a major role in the regulation of replication-coupled DSB repair that is mechanistically distinct from its role in the G1-S checkpoint. Both human and murine p21−/− cells are hypersensitive to the clastogenic effects of MMC, CPT and VP-16. Elevated chromatid-type aberrations and DNA-PKCS pS2056 nuclear foci formation indicate the increased activation of NHEJ in the absence of p21. Concomitantly, DNA replication-coupled HR, as assessed by measuring the frequency of sister chromatid exchanges (SCEs) and the repair of a crosslinked GFP reporter substrate, is reduced in the absence of p21. Furthermore, we demonstrate that p21−/− cells exhibit increased MRE11 nuclear foci and CDK-mediated BRCA2 S3291 phosphorylation. Importantly, we establish that the DSB hypersensitivity of p21−/− cells can be rescued by both the inhibition of CDK activity and MRE11 nuclease activity. Collectively, our results strongly suggest that p21 plays a central role in the coordination of early steps of replication-coupled DSB repair and promotes error-free HR.
MATERIALS AND METHODS
Cells and cell culture conditions
HCT116 wild-type, p21−/− and p53−/− cells (6,7) were grown in McCoy’s 5A medium. B6-129/Sv p21+/+ and p21−/− mouse embryonic fibroblasts (MEFs) and HeLa cells were grown in Dulbecco’s Modified Eagle’s Medium. All growth media was supplemented with 12% v/v fetal bovine serum, l-glutamine and penicillin/streptomycin.
Chromosome breakage analysis
Cells were incubated in the absence or presence of DNA-damaging agents for 16 h. Before harvesting, cells were treated with 0.1 µg/ml Colcemid (Gibco/Invitrogen) for 2 h. Cell pellets were incubated in 0.075 M KCl at 37°C for 18 min, followed by fixation in Carnoy’s fixative (3:1 methanol:glacial acetic acid) with multiple changes. Cells were dropped onto chilled slides and air-dried before staining with 2.5% w/v Giemsa solution (Sigma). Metaphases were analyzed using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software.
siRNA experiments
HeLa cells were plated in six-well dishes at a density of 200 000 cells per well and transfected the following day with p21-specific (sip21) or control, non-targeting siRNA (siControl [siCtrl]) using Lipofectamine 2000 (Invitrogen). Fifty hours following transfection, cells were incubated in the absence or presence of 50 and 100 nM MMC for 16 h and harvested for cytogenetic analysis as described earlier. The following sense and anti-sense siRNA sequences were used: sip21 (Qiagen) 5′-GACCAUGUGGACCUGUCACTT-3′ and 5′-GUGACAGGUCCACAUGGUCTT-3′ and non-targeting siCtrl (Applied Biosystems/Ambion) 5′-UAACGACGCGACGUAATT-3′ and 5′-UUACGUCGUCGCGUCGUUATT-3′.
Sister chromatid exchange analysis
Cells were incubated for two cell cycles in the presence of 12 μM bromodeoxyuridine (Sigma) and incubated in the absence or presence of DNA-damaging agents for one cell cycle before harvest. Metaphase chromosome spreads were prepared as described earlier. Slides were allowed to air dry for 48 h, stained with 10 µg/ml Hoechst 33528 (Sigma) and then irradiated with 120 mJ/cm2 UV-C irradiation for 10 min using a XL-1000 Spectrolinker UV Crosslinker (Spectronics Corporation). Slides were stained with 3% w/v Giemsa in Sorensen’s buffer (67 mM Na2HPO4 × 2H2O, 67 mM KH2PO4, pH 6.8) for 25 min and analyzed as described earlier.
Replication-coupled, interstrand crosslink homologous recombination assay
The assay involves transient transfection of the TR-OriP-GFP HR reporter substrate with pCEP4 (Invitrogen, V044-50), allowing the expression of Epstein-Barr nuclear antigen 1 (EBNA1) for replication of TR-OriP-GFP, adapted from (8,9). Before transfection, triplexes were formed in vitro by incubating 10 µM of triplex forming oligonucleotide (TFO) or psoralen-TFO (pso-TFO) with 20 µg TR-OriP-GFP for 30 min in 40 µl TFO buffer (50 mM HEPES, pH 7.2, 50 mM NaCl, 0.5 mM spermine and 10 mM MgCl2). Triplex or control DNA and 20 µg pCEP4 were added to 5 × 106 cells in 800 µl Opti-MEM (Gibco) in a 0.4 cm gap cuvette (Bio-Rad) and electroporated by pulsing at 250 V, 950 µF. After electroporation, cells were placed in a 10-cm tissue culture dish with tissue culture media and incubated at 37°C. After 1 h, the media was replaced with 1 ml phosphate-buffered saline (PBS), and then cells were irradiated with ultraviolet radiation A (UV-A) (0.15 J cm−2 at 365 nm) in a Stratalinker (Stratagene) with the lid removed. After irradiation, 10 ml of fresh tissue culture media was added to the cells. GFP+ cells were quantified by flow cytometry 48 h after transfection using a Becton Dickinson FACScan.
Immunoblotting and antibodies
For immunoblotting analysis, cell suspensions were washed in ice-cold PBS and lysed in 1% v/v Triton X-100, 0.5% w/v sodium deoxycholate, 0.1% w/v sodium dodecyl sulphate in PBS supplemented with protease and phosphatase inhibitors (Sigma). Proteins were resolved on NuPAGE 3–8% w/v Tris-Acetate or 4–12% w/v Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride membranes. Membranes were immunoblotted with mouse monoclonal antisera against BRCA2 (OP95; Calbiochem/EMD Chemicals), DNA-PKCS (Ab-2/25-4; Thermo Scientific), γH2AX (JBW301; Millipore) and α-tubulin (Ab-2; LabVision), and rabbit polyclonal antisera BRCA2 pS3291 (10), DNA-PKCS pS2056 (ab18192; Abcam), p21 (N-20; Santa Cruz Biotechnology) and MRE11 (NB100-142; Novus Biologicals).
Immunofluorescence microscopy
For immunofluorescence (IF) analyses, cells were seeded in four-well tissue culture slides (BD Biosciences) and treated with MMC for 16 hr. Cells were fixed in 4% w/v paraformaldehyde in PBS for 20 min at 4°C, followed by permeabilization for 5 min in 0.3% v/v Triton X-100 in PBS. Fixed cells were incubated with primary antibodies in 5% v/v goat serum, 0.1% v/v NP40, in PBS for 1 hr, washed three times with PBS and then incubated with alexafluor 488-conjugated anti-mouse or anti-rabbit secondary antibodies (Invitrogen) for 45 min. Cells were then counterstained and mounted in vectashield plus 4′6-diamidine-2-phenylindole dihydrochloride (Vector Laboratories) and visualized using a Zeiss AxioImager.A1 upright epifluorescent microscope with AxioVision LE 4.6 image acquisition software. Primary antibodies used for IF were anti-53BP1 (H300; Santa Cruz Biotechnology), anti-BRCA1 (D9; Santa Cruz Biotechnology), anti-CtIP (a kind gift from Richard Baer, Columbia University) and anti-MRE11 (PC388; Calbiochem/EMD Chemicals).
RESULTS
Functional requirement for p21 in the cellular response to the DNA crosslinking agent mitomycin C
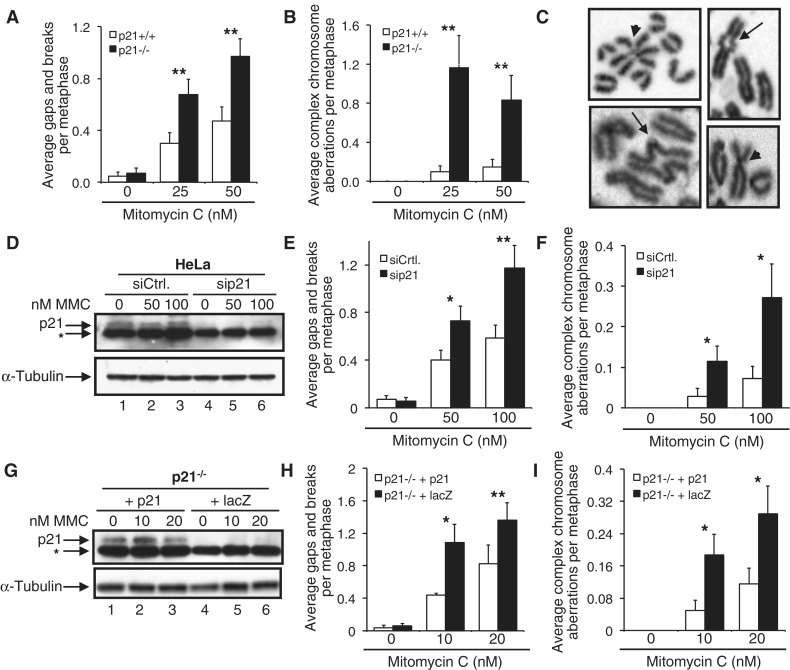
We recently demonstrated that p21 is required for the activation of the Fanconi anemia (FA) pathway: p21 promotes DNA damage-inducible FANCD2 and FANCI monoubiquitination via transcriptional regulation of the USP1 deubiquitinating enzyme (11). Therein, we also reported that HCT116 p21−/− cells exhibit elevated levels of MMC-induced chromosome aberrations, compared with p21+/+ cells. In this study, we sought to expand on these observations and to uncover the molecular basis of this phenotype. To examine the role of p21 in the maintenance of chromosome stability in a non-transformed cell model, we exposed B6-129/Sv p21+/+ and p21−/− primary MEFs to MMC and examined metaphase chromosomes for the presence of chromosome and chromatid aberrations. Similar to that previously observed for HCT116 p21−/− cells (11), p21−/− MEFs were more susceptible to the clastogenic effects of MMC (Figure 1A–C). For example, a >2-fold increase in the average number of chromosome gaps and breaks was observed for the p21−/− MEFs, compared with the p21+/+ MEFs, at both concentrations of MMC examined (P < 0.01, in both cases) (Figure 1A). Furthermore, a ∼12-fold increase in complex chromosome aberrations was observed for 25 nM MMC-treated p21−/− MEFs, compared with the p21+/+ MEFs (P < 0.01) (Figure 1B). These complex chromosome aberrations included Robertsonian-like metacentric chromosomes as well as arrangements involving multiple chromosomes (Figure 1C). Using an alternative approach, depletion of p21 in HeLa cells using siRNA also sensitized cells to the clastogenic effects of MMC; a ∼2-fold increased frequency of chromosome gaps and breaks was observed for p21-depleted cells compared with cells transfected with a control, non-targeting siRNA (p < 0.01) (Figure 1D and E). Similarly, a ∼4-fold increased frequency of MMC-inducible complex chromosome aberrations was observed for p21-depleted cells (P < 0.05) (Figure 1F). Importantly, transient transfection of HCT116 p21−/− cells with the p21 cDNA rescued their MMC hypersensitivity, resulting in a significant reduction in MMC-inducible chromosome gaps and breaks and complex chromosome aberrations (Figure 1G–I). Furthermore, spectral karyotyping (SKY)/multicolor fluorescence in situ hybridization (M-FISH) analysis of metaphase chromosomes from MMC-treated p21−/− cells confirmed the presence of illegitimate chromatid-type interchanges resulting in the formation of chromosome structural aberrations including unbalanced chromosome translocations (Supplementary Figure S1). Taken together these findings clearly establish an important function for p21 in the maintenance of chromosome stability following exposure to the DNA interstrand crosslinking agent MMC.
Figure 1.
Functional requirement for p21 in the cellular response to the DNA crosslinking agent mitomycin C. (A–C) p21+/+ and p21−/− MEFs were incubated in the absence or presence of 25 and 50 nM MMC for 18 h, and the numbers of chromosome gaps and breaks (A), and complex chromosome aberrations (B), including dicentric chromosomes and radial formations, were scored. At least 50 metaphases were scored per treatment. (C) Representative chromosome aberrations detected in p21−/− MEFs. Arrows point to radial chromosomes; arrowheads point to chromatid fusions. (D–F) HeLa cells were transfected with control, non-targeting (siCtrl) and p21 (sip21) siRNAs. Fifty-four hours post-transfection, cells were incubated in the absence or presence of 50 and 100 nM MMC for 16 h. (D) Whole-cell lysates were prepared, and resolved proteins were immunoblotted with anti-p21 and anti-α-tubulin antibodies. The asterisk denotes a non-specific band. (E and F) Metaphase spreads were prepared, and chromosome gaps and breaks (E) and complex chromosome aberrations (F) were scored. (G–I) HCT116 p21−/− cells were transiently transfected with pLenti6.2-p21 or pLenti6.2-lacZ and treated with 0, 10 and 20 nM MMC for 16 h. (G) Whole-cell lysates were prepared, and resolved proteins were immunoblotted with anti-p21 and anti-α-tubulin antibodies. The asterisk denotes a non-specific band. (H and I) Metaphase spreads were prepared, and chromosome gaps and breaks (H) and complex chromosome aberrations (I), including dicentric chromosomes and radial formations, were scored. At least 80 metaphases were scored per treatment. Error bars represent the standard errors of the means from independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
p21−/− cells are hypersensitive to the clastogenic effects of type I and II topoisomerase poisons
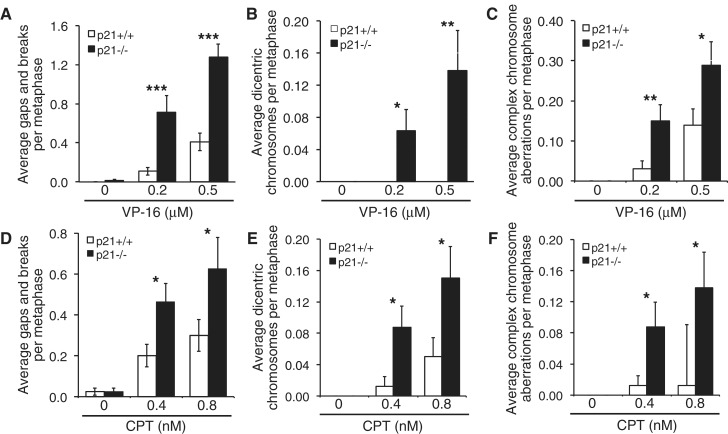
The high frequency of dicentric chromosomes and complex chromosome aberrations observed in MMC-treated p21-deficient cells suggested a more general defect in DSB repair. DSBs have been demonstrated to arise during the DNA replication-coupled repair of DNA interstrand crosslinks (ICLs) (12,13). To determine whether p21 plays a role in the cellular response to other agents known to induce the formation of DSBs, p21+/+ and p21−/− cells were exposed to the type I and II topoisomerase poisons CPT and VP-16, respectively, and metaphase chromosomes were analyzed for the presence of chromosome and chromatid aberrations. Similar to that observed for MMC, markedly elevated levels of chromatid-type aberrations were observed for VP-16-treated p21−/− cells, compared with VP-16-treated p21+/+ cells (Figure 2A–C). For example, a 6-fold increase in chromatid gaps and breaks (p < 0.001) was observed for the p21−/− cells, compared with p21+/+ cells, after exposure to 0.2 µM VP-16 (Figure 2A). Importantly, dicentric chromosomes were frequently observed for the VP-16-treated p21−/− cells and were never observed for p21+/+ cells (Figure 2B). In addition, a ∼5-fold increase in complex chromosome aberrations (P = 0.005), including radial chromosome formations, was observed for the p21−/− cells, compared with p21+/+ cells, following exposure to 0.2 µM VP-16 (Figure 2C). We also monitored γH2AX nuclear foci formation following treatment with VP-16. We observed an increased spontaneous γH2AX nuclear foci formation in the p21−/− cells. In addition, persistently elevated levels of γH2AX nuclear foci were observed for p21−/− cells up to 7 h following exposure to VP-16, at which point levels had returned to normal in wild-type cells (Supplementary Figure S2). Moreover, similar to that observed for MMC and VP-16, elevated levels of chromatid-type aberrations were observed for p21−/− cells treated with CPT (Figure 2D–F). For example, we detected an ∼11-fold increase (P < 0.05) in complex chromosome aberrations in 0.8 nM CPT-treated p21−/− cells, compared with the p21+/+ cells (Figure 2F). Taken together, our results demonstrate that p21-deficient cells exhibit pronounced chromosome instability following exposure to several DSB-inducing agents, most likely as a result of defective DSB repair.
Figure 2.
p21−/− cells are hypersensitive to the clastogenic effects of etoposide and camptothecin. (A–C) HCT116 p21+/+ and p21−/− cells were treated with 0.2 and 0.5 µM etoposide (VP-16) for 16 h and chromosome gaps and breaks (A), dicentric chromosomes (B) and complex chromosome aberrations (C) were scored. (D–F) p21+/+ and p21−/− cells were treated with 0.4 and 0.8 nM CPT, and chromosome gaps and breaks (D), dicentric chromosomes (E) and complex chromosome aberrations (F) were scored. *P < 0.05; **P < 0.01; ***P < 0.001.
The role of p21 in the maintenance of chromosome stability is independent of the G1-S checkpoint
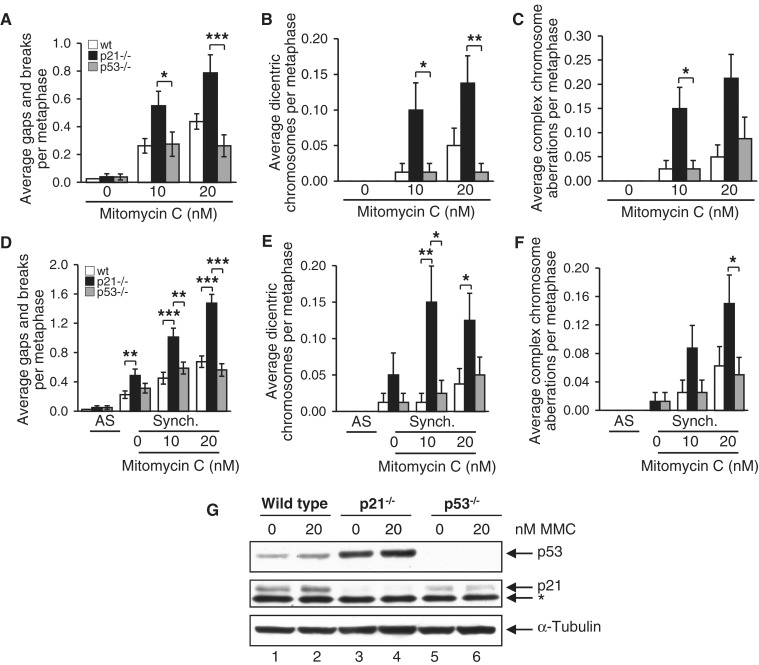
We next sought to determine whether the increased DNA damage-inducible chromosome instability of the p21−/− cells was a consequence of a defective G1-S phase checkpoint or altered cell cycle progression in general. Thus, we performed a series of experiments with both p21−/− and p53−/− cells, reasoning that if the effect of p21 was merely related to altered cell cycle dynamics, then the same phenotype would be observed for p53−/− cells (7). However, this is not what we observed. Instead, p21−/− cells were markedly more sensitive to the clastogenic effects of MMC than p53−/− cells (Figure 3A–C). For example, a ∼3-fold increase (P < 0.001) in MMC-induced gaps and breaks was observed for p21−/− cells, compared with p53−/− cells, following exposure to 20 nM MMC (Figure 3A). Under the same conditions, an ∼11-fold increase (P < 0.01) in dicentric chromosome formations was observed for p21−/− cells, compared with p53−/− cells (Figure 3B). Similar results were obtained for wild-type, p21−/− and p53−/− cells following treatment with CPT (results not shown).
Figure 3.
The role of p21 in the maintenance of chromosome stability is independent of the G1-S checkpoint. (A–C) HCT116 wild-type, p21−/− and p53−/− cells were incubated in the absence or presence of MMC for 16 h, and chromosome gaps and breaks (A), dicentric chromosomes (B) and complex chromosome aberrations (C) were scored. (D–F) HCT116 wild-type, p21−/− and p53−/− cells were synchronized via double-thymidine block, released into thymidine-free medium, and treated with 0, 10 and 20 nM MMC for 11 h. Metaphase chromosome gaps and breaks (D), dicentric chromosomes (E) and complex chromosome aberrations (F) were then scored. AS, asynchronous; Synch., synchronous. *P < 0.05; **P < 0.01; ***P < 0.001. (G) Whole-cell lysates were prepared from wild-type, p21−/− and p53−/− cells and immunoblotted with anti-p53, anti-21 and anti-α-tubulin antibodies. The asterisk denotes a non-specific band.
In an alternative approach, we synchronized wild-type, p21−/− and p53−/− cells in early S-phase by double-thymidine block, incubated cells in the absence or presence of MMC and examined metaphase chromosome damage. The cell cycle stage profiles of wild-type, p21−/− and p53−/− cells were very similar following double-thymidine arrest (Supplementary Figure S3A). Although the p21−/− cells appeared to progress to G2-M at a faster rate than wild-type and p53−/− cells, cell cycle stage profiles and mitotic indices following MMC exposure were indistinguishable (Supplementary Figure S3B). Double-thymidine arrest, by itself, resulted in a measurable increase in chromosome aberrations for all three cell types (Figure 3D–F). However, in the case of gaps and breaks and dicentric chromosome formations, the p21−/− cells exhibited the greatest degree of sensitivity (Figure 3D and E). Furthermore, following exposure of synchronized cells to MMC, a pronounced increase in gaps and breaks, dicentric chromosome formations and complex chromosome aberrations were observed for the p21−/− cells compared with wild-type and p53−/− cells (Figure 3D–F). For example, a ∼6-fold increase (P < 0.05) in dicentric chromosome formations was observed for the p21−/− cells compared with p53−/− cells, following synchronization and treatment with 10 nM MMC (Figure 3E). p53−/− cells express appreciable levels of p21 protein, albeit at a reduced level compared with wild-type cells (Figure 3G). Collectively, these results strongly suggest that the role of p21 in the maintenance of chromosome stability is independent of G1-S-phase transition and not an indirect consequence of altered cell cycle progression.
Elevated nonhomologous DNA end-joining activity in p21−/− cells
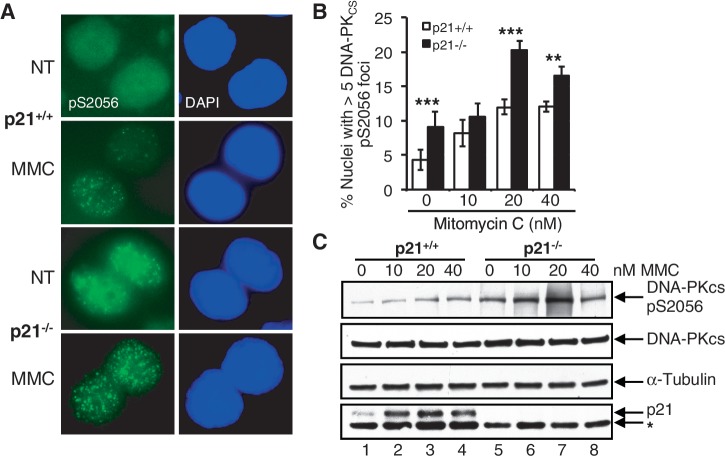
Our observations of increased MMC-, VP-16- and CPT-inducible chromatid-type interchanges in the absence of p21 suggested the increased activity of the typically error-prone NHEJ DSB repair pathway. To test this hypothesis, we examined the recruitment of DNA-PKCS to discrete nuclear foci in the p21+/+ and p21−/− cells, using an antibody raised specifically against the DNA-PKCS protein phosphorylated on S2056 (14,15). An increased DNA-PKCS pS2056 nuclear foci formation was observed in the p21−/− cells both in the absence and presence of MMC (Figure 4A and B). For example, a ∼2-fold increased DNA-PKCS pS2056 nuclear foci formation was observed in the p21−/− cells, compared with the p21+/+ cells, both spontaneously and following exposure to 20 nM MMC (P < 0.0001) (Figure 4A and B). These IF findings were corroborated by immunoblotting whole-cell lysates prepared from untreated and MMC-treated p21+/+ and p21−/− cells, with the same DNA-PKCS pS2056 antibody. Substantially increased protein levels of DNA-PKCS pS2056 were observed in both untreated and MMC-treated p21−/− cells, compared with the p21+/+ cells (Figure 4C). In addition, in a pulse-chase experiment, we observed persistently elevated levels of both γH2AX and DNA-PKCS pS2056 nuclear foci in p21−/− cells up to 7 h following exposure to MMC, at which point levels had begun to decrease in wild-type cells (P < 0.001 in both cases) (Supplementary Figure S4A and B). These findings demonstrate elevated levels of unrepaired DSBs and increased NHEJ activity in the absence of p21 following treatment with MMC, suggesting that p21 may negatively regulate this typically error-prone DSB repair pathway.
Figure 4.
An increased phosphorylation of DNA-PKCS S2056 in p21-deficient cells. (A–C) HCT116 p21+/+ and p21−/− cells were incubated in the absence or presence of 10, 20 and 40 nM MMC for 16 h. (A and B) The number of nuclei displaying >5 discrete DNA-PKCS pS2056 foci were quantified using immunofluorescence microscopy and plotted. Representative images of DNA-PKCS pS2056 nuclear foci are shown in (A). At least 300 nuclei were scored per treatment per experiment, and this experiment was performed at least three times with similar results. Error bars represent standard errors of the means from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Whole-cell lysates were prepared, and resolved proteins were immunoblotted with anti-DNA-PKCS pS2056, anti-DNA-PKCS, anti-p21 and anti-α-tubulin antibodies. The asterisk denotes a non-specific band.
p21 is required for DNA replication-coupled homologous recombination
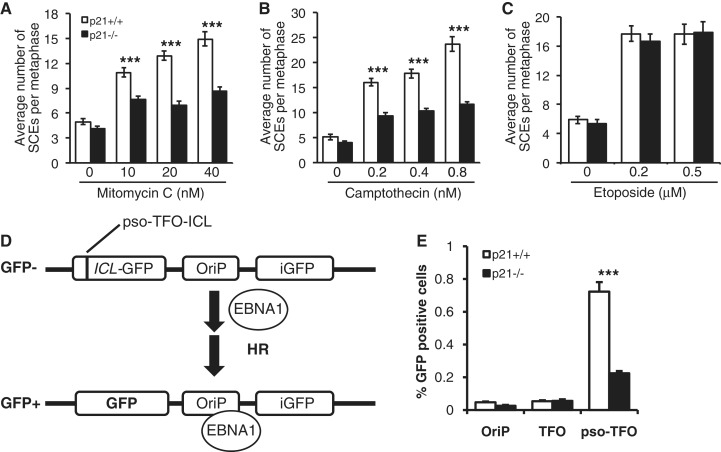
Next, to determine whether p21 plays a role in the regulation of HR DSB repair, we measured the frequencies of both spontaneous and DNA damage-inducible SCEs in p21+/+ and p21−/− cells. SCEs are a cytogenetically visible manifestation of HR between sister chromatids (16). No differences in the spontaneous frequency of SCE between p21+/+ and p21−/− cells were observed, with an average of approximately four SCEs per metaphase for both cell types (Figure 5A–C). However, we observed significant differences in the frequencies of both MMC- and CPT-inducible SCEs between the p21+/+and p21−/− cells (Figure 5A and B). For example, a ∼2-fold increase (P < 0.0001) in the numbers of SCEs per metaphase chromosome was observed for p21+/+ cells, compared with p21−/− cells, following exposure to MMC and CPT (Figure 5A and B). However, in direct contrast, no differences in the frequencies of VP-16-inducible SCEs were observed between the p21+/+ and p21−/− cells (Figure 5C).
Figure 5.
Impaired DNA replication-coupled homologous recombination in p21−/− cells. HCT116 p21+/+ and p21−/− cells were incubated in the presence of bromodeoxyuridine for two cell cycles and in the absence or presence of mitomycin C (A), camptothecin (B) or etoposide (C) for the second cell cycle period. Metaphase spreads were prepared, and the numbers of SCEs were scored. Error bars represent the standard errors of the means. At least 30 metaphases were scored per treatment per experiment. (D) HR reporter assay for the measurement of replication-dependent HR repair of a site-specific DNA ICL (9). (E) p21+/+ and p21−/− cells were transiently transfected with the TR-OriP-GFP vector alone (OriP), TR-OriP-GFP without an ICL (TFO) and psoralen-UVA crosslinked TR-OriP-GFP (pso-TFO), in the presence of EBNA1. Error bars represent standard errors of the means from three independent experiments.*P < 0.05; **P < 0.01; ***P < 0.001.
To directly test the role of p21 in DNA replication-coupled ICL HR repair we utilized the recently developed TR-OriP-GFP assay, which measures HR repair of a psoralen-UVA crosslinked triplex oligonucleotide (9). The TR-OriP-GFP substrate contains the Epstein-Barr virus (EBV) origin of replication, allowing it to replicate in the presence of the EBV nuclear antigen EBNA1 (Figure 5D). A marked reduction in replication-dependent ICL-induced HR repair was observed in the absence of p21 (P < 0.001), compared with wild-type p21 cells (Figure 5E). p21 did not affect DNA replication per se, as transfection of an OriP vector containing an intact GFP gene led to a similar increase in GFP+ cells in the absence or presence of p21 (data not shown).
To determine whether defective replication-coupled HR repair in the p21−/− cells was a result of a defect in the localization of the RAD51 recombinase, we examined RAD51 nuclear foci formation in the p21+/+ and p21−/− cells. Although we observed a >2-fold increase (P < 0.0001) in the percentage of p21−/− nuclei exhibiting RAD51 nuclear foci in the absence of DNA damage, no differences in the percentage of nuclei exhibiting RAD51 nuclear foci were observed between p21+/+ and p21−/− cells up to 48 h following exposure to MMC (Supplementary Figure S5).
Elevated MRE11 nuclease activity contributes to the increased chromosome instability of p21−/− cells
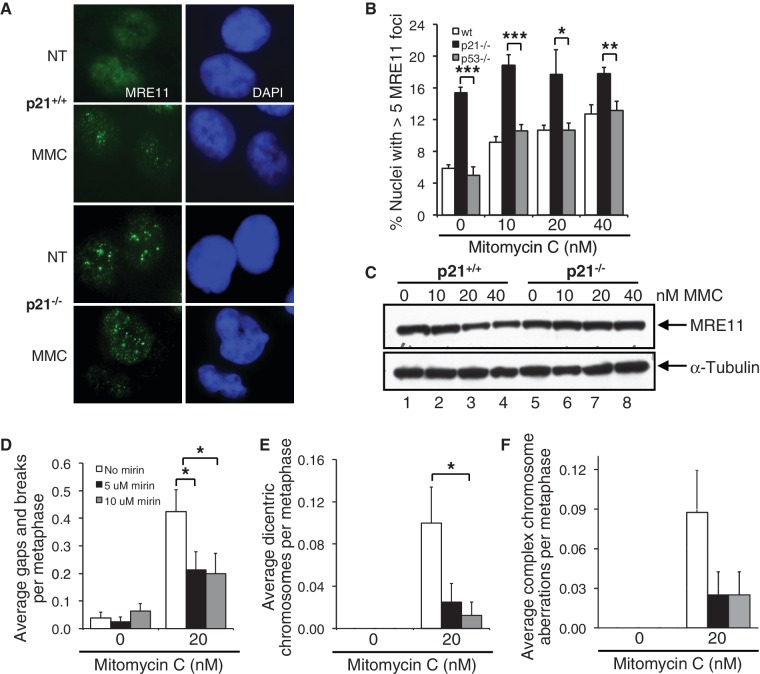
To gain mechanistic insight into the origins of increased NHEJ and decreased HR activity in p21-deficient cells, we next examined the recruitment of several DNA repair proteins known to act early in the DSB response. First, we examined the recruitment of the MRE11 nuclease, a protein known to play a very early role in the sensing and nucleolytic processing of DSBs and in the choice of HR and NHEJ pathways (17). To our surprise, we observed a marked elevation of MRE11 nuclear foci formation in the p21−/− cells. In the absence of MMC, a ∼3-fold increase (P < 0.001) in the percentage of nuclei exhibiting MRE11 foci was observed for the p21−/− cells, compared with both wild-type (p53+/+/p21+/+) and p53−/− cells (Figure 6A and B). A dose-dependent increase in MRE11 nuclear foci formation was observed for both the wild-type and the p53−/− cells following exposure to MMC. However, under the same conditions, no appreciable change in the percentage of p21−/− nuclei harboring MRE11 foci was detected (Figure 6A and B). Differences in the percentage of nuclei containing MRE11 foci could not be attributed to changes in MRE11 protein levels, as measured by immunoblotting analysis (Figure 6C). We also examined BRCA1 and CtIP nuclear foci formation in these cells. BRCA1 and CtIP also act early in the DNA damage response and, together with MRE11, are thought to regulate the extent of nucleolytic processing of DSBs (18–21). Similar to that observed for MRE11, albeit to a lesser degree, we observed increased basal levels of BRCA1 and CtIP nuclear foci formation in p21−/− cells (Supplementary Figure S6A and B). However, in contrast to that observed for MRE11, BRCA1 and CtIP, no differences in the percentage nuclei positive for 53BP1 foci formation were observed between p21+/+ and p21−/− cells, both in the absence or presence of MMC (Supplementary Figure S6C).
Figure 6.
Elevated MRE11 nuclease activity contributes to the increased chromosome instability of p21−/− cells. (A–C) HCT116 wild-type, p21−/− and p53−/− cells were incubated in the absence or presence of 10, 20 and 40 nM MMC for 16 h. (A and B) The number of nuclei displaying >5 discrete MRE11 foci were quantified using immunofluorescence microscopy and plotted. Representative images of MRE11 nuclear foci are shown in (A). At least 300 nuclei were scored per treatment per experiment, and this experiment was performed at least three times with similar results. (C) Whole-cell lysates were prepared and resolved proteins were immunoblotted with anti-MRE11 and anti-α-tubulin antibodies. (D–F) p21−/− cells were incubated in the absence or presence of 20 nM MMC and 5 and 10 µM mirin for 16 h. Metaphase spreads were prepared, and chromosome aberrations, including gaps and breaks (D), dicentric chromosomes (E) and complex aberrations (F), including radial formations, were scored. At least 80 metaphases were scored per treatment, and this experiment was repeated twice with similar findings. Error bars represent standard errors of the means. *P < 0.05; **P < 0.01; ***P < 0.001.
To determine whether increased MRE11 nuclear foci formation might contribute to the increased sensitivity of the p21−/− cells to the clastogenic effects of MMC, we incubated p21−/− cells in the absence and presence of both MMC and mirin, a chemical inhibitor of MRE11 nuclease activity (22). Treatment of p21−/− cells with mirin had no discernible effect on spontaneous chromosome instability (Figure 6D–F). However, co-treatment of p21−/− cells with MMC and mirin resulted in a pronounced decrease in susceptibility to the clastogenic effects of MMC (Figure 6D–F). For example, we observed a ∼2-fold reduction (P < 0.05) in gaps and breaks in cells co-treated with MMC and mirin, compared with cells treated with MMC alone (Figure 6D). In addition, under the same conditions, we observed a ∼8-fold reduction in dicentric chromosomes (P < 0.05 for 10 µM mirin) (Figure 6E). Under these conditions, we did not observe a significant difference in percent survival between cells incubated in the absence or presence of MMC and mirin (Supplementary Figure S7). Collectively, these results suggest that in the absence of p21, inappropriate MRE11 nuclease activity may promote error-prone DSB repair activity and increased chromosome instability.
Increased CDK-mediated BRCA2 S3291 phosphorylation in p21−/− cells
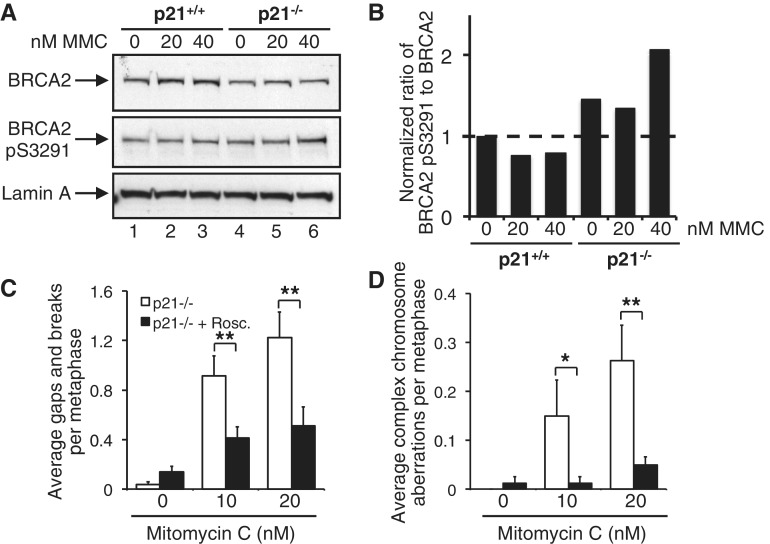
Previous studies have demonstrated that BRCA2 S3291 phosphorylation leads to the destabilization of RAD51 nucleoprotein filaments (23). Misregulation of this process has been shown to lead to inappropriate MRE11-dependent DNA replication fork instability (24). Therefore, we next examined the levels of both unmodified BRCA2 and BRCA2 phosphorylated on S3291, a CDK-dependent post-translational modification (10), in p21+/+ and p21−/− cells. Total BRCA2 levels were uniformly reduced in the absence of p21 (Figure 7A, lanes 4–6). In addition, we observed increased BRCA2 S3291 phosphorylation both in the absence and presence of MMC. For example, following exposure to 40 nM MMC, we detected a ∼2-fold increased BRCA2 pS3291:BRCA2 ratio in the absence of p21 (Figure 7A and B), suggesting that increased CDK-mediated BRCA2 phosphorylation might also contribute to the chromosome instability phenotype of p21−/− cells. To determine whether this was indeed true, we next incubated p21−/− cells in the absence and presence of both MMC and roscovitine, a cell-permeable reversible CDK inhibitor and again scored metaphase chromosome aberrations. Importantly, co-treatment of p21−/− cells with MMC and roscovitine led to a significant reduction in the frequencies of both chromosome gaps and breaks and complex chromosome aberrations. For example, we observed a ∼5-fold reduction (P < 0.01) in complex chromosome aberrations in p21−/− cells co-treated with 20 nM MMC and 3 µM roscovitine, compared with p21−/− cells treated with MMC alone (Figure 7D). Under these conditions, no appreciable differences in cell cycle stage distribution, mitotic index or percent survival were observed for p21−/− cells incubated in the absence or presence of MMC and roscovitine (Supplementary Figure S8A–C). Collectively, our results strongly suggest that increased CDK-mediated BRCA2 S3291 phosphorylation promotes chromosome instability in MMC-treated p21−/− cells.
Figure 7.
An increased CDK-mediated BRCA2 S3291 phosphorylation in p21−/− cells. (A and B) HCT116 p21+/+ and p21−/− cells were incubated in the absence or presence of 20 or 40 nM MMC for 16 h. Whole-cell lysates were prepared, and resolved proteins were immunoblotted with anti-BRCA2, anti-BRCA2 pS3291 and anti-lamin A antibodies. (B) Band intensities from (A) were quantified using ImageJ and the ratios of BRCA2 pS3291 to BRCA2 plotted. (C and D) p21−/− cells were incubated in the absence or presence of 10 and 20 nM MMC and 3 µM roscovitine (Rosc.) for 16 h. Metaphase spreads were prepared, and chromosome aberrations, including gaps and breaks (C) and complex chromosome aberrations (D), including radial formations, were scored. At least 80 metaphases were scored per treatment, and this experiment was repeated twice with similar findings. Error bars represent standard errors of the means. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
In this study, we have established that p21 plays a novel role in the promotion of error-free replication-coupled DSB repair. This role is mechanistically distinct from its well-known function in the G1-S-phase checkpoint. A colorectal carcinoma cell line harboring a homozygous p21 deletion, p21-null primary MEFs and cells depleted of p21 using siRNA, all display hypersensitivity to the clastogenic effects of the DNA crosslinking agent MMC. Importantly, hypersensitivity of the p21−/− cells to MMC was rescued by transient re-expression of p21. In addition, we show that p21−/− cells are hypersensitive to the clastogenic effects of the topoisomerase I and II poisons CPT and VP-16, respectively. All three DNA-damaging agents are potent inducers of DSBs, albeit via different mechanisms, discussed further below. In support of our findings, a recent report has described the physical accumulation of p21 at laser-irradiated sites and co-localizing with γH2AX (25). In addition to chromosome gaps and breaks, p21−/− cells demonstrate a markedly increased frequency of DNA damage-inducible chromatid-type aberrations, including dicentric chromosomes and complex chromatid interchanges. These aberrations most likely arise as a consequence of increased NHEJ. Consistent with this hypothesis, we observed elevated and persistent levels of phosphorylated DNA-PKCS S2056 in p21−/− cells, a marker of NHEJ (15), suggesting the inappropriate engagement of DNA-PKCS with DSB ends. In addition, we observed persistent DNA damage-inducible γH2AX nuclear foci formation in the absence of p21 strongly suggesting an outright defect in DSB repair. Furthermore, we have established an important role for p21 in replication-coupled HR: p21-deficient cells display reduced DNA damage-inducible SCEs and are defective for replication-dependent HR repair of a site-specific ICL, similar to that recently described for FA patient cells (9). Thus, we have established that, in addition to its role in the G1-S phase checkpoint, p21 is a negative regulator of error-prone NHEJ and a positive regulator of error-free HR. Similarly, an important role for p21 in the negative regulation of the error-prone translesion DNA synthesis pathway has been established (26).
Using several approaches, we have also firmly established that this newly discovered function for p21 in DSB repair is mechanistically distinct from its role in G1- to S-phase progression, nor is it an indirect consequence of altered cell cycle progression in general. First, we demonstrate that asynchronous p21−/− cells are markedly more sensitive to MMC than similarly derived asynchronous p53−/− cells. The G1-S-phase checkpoint is similarly abrogated in both cell lines (7), arguing that a defective G1-S-phase checkpoint is not the primary cause of the pronounced sensitivity of the p21−/− cells to DSB-inducing agents. Second, we show that cell cycle-synchronized p21−/− cells display markedly greater sensitivity to MMC than synchronized wild-type and p53−/− cells, arguing that this phenotype is not a general consequence of altered cell cycle progression. For the latter experiment, it is important to note that the cell cycle stage distributions and mitotic indices of MMC-treated p21−/− and p53−/− cells were very similar, indicating that differences in the rate of S and G2 progression do not underlie the observed phenotype. Instead, we demonstrate that p21−/− cells exhibit elevated levels of MRE11 nuclear foci formation, even in the absence of exposure to DNA-damaging agents. This phenotype was not observed for the p53−/− cells. We also demonstrate that treatment of p21−/− cells with mirin, a chemical inhibitor of MRE11 nuclease activity, rescues the MMC hypersensitivity of these cells, strongly suggesting that inappropriate MRE11 nuclease activity promotes chromosome instability in the absence of p21. Several recent studies have described important roles for MRE11 in NHEJ (27–29). However, the inappropriate use of NHEJ can lead to genome rearrangements and chromosome instability, consistent with our observations. MRE11 may act alone or in concert with other nucleases such as CtIP and EXO1 (30–32). Our findings of increased spontaneous CtIP nuclear foci formation in the p21−/− cells suggest a cooperative role for CtIP. However, the contribution of EXO1 to this phenotype remains to be determined.
Recently, it has been established that the BRCA2 protein prevents nascent DNA strand degradation on DNA replication fork stalling by stabilizing RAD51 nucleoprotein filaments on single-stranded DNA (ssDNA) (24,33). In the absence of BRCA2, or RAD51, inadvertent MRE11 nuclease activity promotes fork degradation and chromosome instability (24,33). BRCA2 is subjected to CDK-mediated phosphorylation on S3291 (10). S3291 phosphorylation blocks the binding of RAD51 oligomers with the carboxy-terminus of BRCA2, leading to the destabilization of RAD51 nucleoprotein filaments (10). On exposure to DNA-damaging agents CDK-mediated BRCA2 S3291 phosphorylation is decreased, resulting in stimulation of RAD51 binding to the carboxy-terminus of BRCA2, and increased HR activity (10). Our results suggest that p21 plays a key role in the coordination of this process via the inhibition of CDK activity (see Figure 8). In the absence of p21, most likely as a consequence of unrestricted CDK activity, both spontaneous and DNA damage-inducible BRCA2 S3291 phosphorylation are elevated. We hypothesize that this leads to the continuous disassembly of RAD51 nucleoprotein filaments, facilitating inappropriate MRE11 nuclease activity. Consistent with this hypothesis, we demonstrate that the MMC-hypersensitivity of p21−/− cells can be rescued by the inhibition of CDK activity or MRE11 nuclease activity. Although we detected an increase in spontaneous RAD51 nuclear foci formation in the absence of p21, we did not observe differences in the formation or disappearance of RAD51 nuclear foci between p21+/+ and p21−/− cells following exposure to MMC. RAD51 nucleoprotein loading onto ssDNA is mediated by the BRC repeats of BRCA2 and is thus unlikely to be affected by BRCA2 S3291 phosphorylation (34–36). Furthermore, standard epifluorescence microscopy methods may not be sufficiently sensitive to detect rapid RAD51 turnover. BRCA2 S3291 phosphorylation has also been suggested to lead to BRCA2 proteolysis (10,37), consistent with our observation of decreased overall levels of BRCA2 in the absence of p21. It is also important to note that although we did observe increased spontaneous γH2AX and DNA-PKCS pS2056 nuclear foci formation in the absence of p21, indicative of increased DSBs, we did not detect spontaneous metaphase chromosome aberrations in these cells. However, the presence of submicroscopic chromosome aberrations including small interstitial deletions or duplications, which would not be readily detectable using the conventional cytogenetic analyses used in this study, remains a distinct possibility.
Figure 8.
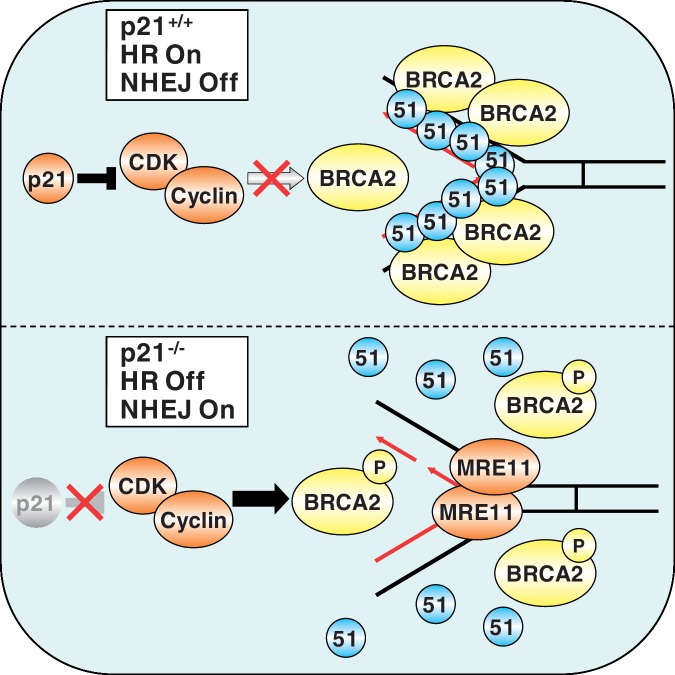
Proposed model for the role of p21 in replication-coupled DNA double-strand break repair. Following exposure to DNA-damaging agents that block DNA replication fork progression, p21 physically binds to, and inhibits, CDK, thereby preventing phosphorylation of BRCA2 on S3291. RAD51 nucleoprotein filaments assemble on ssDNA and are stabilized through interaction with the unphosphorylated carboxy-terminus of BRCA2, thereby promoting HR repair. In the absence of p21, unrestricted CDK activity leads to the constitutive phosphorylation of BRCA2 on S3291. S3291 phosphorylation precludes binding of RAD51 to the carboxy-terminus of BRCA2, resulting in destabilization of RAD51 nucleoprotein filaments. Deprotected ssDNA now becomes susceptible to opportunistic and inadvertent MRE11 nuclease activity, leading to increased error-prone NHEJ and chromosome instability.
Here, we also make the observation that p21 is required for HR between sister chromatids following exposure to MMC and CPT, but not VP-16, providing further mechanistic insight into p21’s role in the maintenance of chromosome stability. VP-16 forms a ternary complex with topoisomerase II and DNA and prevents re-ligation of the severed DNA duplex, thereby generating two-ended DSBs irrespective of cell cycle stage (38). In contrast, MMC-induced ICLs are converted to one-ended DSBs only on encounter with a DNA replication fork (12). Similarly, CPT forms a ternary complex with topoisomerase I and DNA and prevents re-ligation of the single-nicked DNA strand, again giving rise to a one-ended DSB on encounter with the replication fork. Our results suggest that p21 preferentially promotes one-ended DSB-induced sister chromatid recombination at stalled DNA replication forks, yet may be dispensable for two-ended DSB HR mechanisms such as synthesis-dependent strand annealing.
Taken together, our findings represent an important advancement in our understanding of the molecular mechanisms regulating the repair of DSBs, particularly upon DNA replication fork arrest. In a capacity mechanistically distinct from its role in the G1-S-phase checkpoint, p21-mediated inhibition of CDK activity facilitates the interaction between RAD51 and the carboxy-terminus of BRCA2, thereby promoting conservative HR DNA repair. In contrast, the absence of p21 renders stalled DNA replication forks susceptible to opportunistic and inappropriate MRE11-mediated nucleolytic attack, driving potentially error-prone repair pathways such as NHEJ, ultimately leading to increased genome instability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8 and Supplementary Methods.
FUNDING
Leukemia Research Foundation New Investigator grant (NGH); National Center for Research Resources [Rhode Island IDeA Network of Biomedical Research Excellence grant P20RR016457-09 to Z.A.S., PI/NGH, Faculty Development]; National Science Foundation EPSCoR [1004057]; National Institutes of Health/National Heart, Lung and Blood Institute [R21HL095991 (NGH) and R01HL101977 (NGH)]. Funding for open access charge: University of Rhode Island.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Howlett and Sun laboratories for helpful discussions. They also thank Bert Vogelstein, Tyler Jacks and Alexander J. Bishop for cells and the Charles Lee laboratory for SKY/M-FISH analysis.
REFERENCES
- 1.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haber JE. Partners and pathways repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 3.Boulton SJ. Cellular functions of the BRCA tumour-suppressor proteins. Biochem. Soc. Trans. 2006;34:633–645. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR, Wilson TE. SnapShot: nonhomologous DNA end joining (NHEJ) Cell. 2010;142:496–496 e491. doi: 10.1016/j.cell.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 5.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 7.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 8.Nakanishi K, Cavallo F, Brunet E, Jasin M. Homologous recombination assay for interstrand cross-link repair. Methods Mol. Biol. 2011;745:283–291. doi: 10.1007/978-1-61779-129-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat. Struct. Mol. Biol. 2011;18:500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 11.Rego MA, Harney JA, Mauro M, Shen M, Howlett NG. Regulation of the activation of the Fanconi anemia pathway by the p21 cyclin-dependent kinase inhibitor. Oncogene. 2012;31:366–375. doi: 10.1038/onc.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell Biol. 2006;26:7520–7528. doi: 10.1128/MCB.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 24.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike M, Yutoku Y, Koike A. Accumulation of p21 proteins at DNA damage sites independent of p53 and core NHEJ factors following irradiation. Biochem. Biophys. Res. Commun. 2011;412:39–43. doi: 10.1016/j.bbrc.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 26.Avkin S, Sevilya Z, Toube L, Geacintov N, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 29.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mimitou EP, Symington LS. DNA end resection—unraveling the tail. DNA Repair (Amst) 2011;10:344–348. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2011;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl. Acad. Sci. USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 37.Bertwistle D, Swift S, Marston NJ, Jackson LE, Crossland S, Crompton MR, Marshall CJ, Ashworth A. Nuclear location and cell cycle regulation of the BRCA2 protein. Cancer Res. 1997;57:5485–5488. [PubMed] [Google Scholar]
- 38.Yang L, Rowe TC, Liu LF. Identification of DNA topoisomerase II as an intracellular target of antitumor epipodophyllotoxins in simian virus 40-infected monkey cells. Cancer Res. 1985;45:5872–5876. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.