Abstract
The Arabidopsis endonuclease RNase E (RNE) is localized in the chloroplast and is involved in processing of plastid ribonucleic acids (RNAs). By expression of a tandem affinity purification-tagged version of the plastid RNE in the Arabidopsis rne mutant background in combination with mass spectrometry, we identified the novel vascular plant-specific and co-regulated interaction partner of RNE, designated RHON1. RHON1 is essential for photoautotrophic growth and together with RNE forms a distinct ∼800 kDa complex. Additionally, RHON1 is part of various smaller RNA-containing complexes. RIP-chip and other association studies revealed that a helix-extended-helix-structured Rho-N motif at the C-terminus of RHON1 binds to and supports processing of specific plastid RNAs. In all respects, such as plastid RNA precursor accumulation, protein pattern, increased number and decreased size of chloroplasts and defective chloroplast development, the phenotype of rhon1 knockout mutants resembles that of rne lines. This strongly suggests that RHON1 supports RNE functions presumably by conferring sequence specificity to the endonuclease.
INTRODUCTION
The chloroplast is the product of an endosymbiotic event by which a cyanobacterium was ingested by a eukaryotic cell. Subsequently, most genes were lost from the chloroplast genome. The residual genes are embedded in the regulatory network of the chloroplast and are predominantly controlled by nuclear factors (1). Chloroplast biogenesis requires adaption of transcription rates, processing events and regulation of ribonucleic acid (RNA) stability (2,3). An important characteristic of chloroplast gene regulation is the predominance of posttranscriptional control, which is exerted at both gene-specific and global levels (4,5). Chloroplasts have retained some part of the general eubacterial RNA degradation system, such as masking of RNAs by polyadenylation (6,7). However, unlike in bacteria, nearly if not all polycistronic transcripts are processed by endo- and exonucleases, splicing activities and editing events (2,7,8). In several cases, processing within intergenic regions is required for subsequent translation of the 5′-processed product (9) and regulation of stability of individual messenger RNAs (mRNAs) at both ends (2,3,10).
In eubacteria, the endonuclease RNase E (RNE) is a well-studied enzyme and plays a major role in posttranscriptional regulation of gene expression by mediating processing of RNA and initiating RNA degradation (11). It is part of a high-molecular-weight (HMW) multiprotein complex, the so-called degradosome, with PNPase, Rhl B and the glycolytic enzyme enolase as major components in Escherichia coli (12). The N-terminal, catalytic part of the large multidomain protein RNE is essential for cell viability in E.coli. Mutations or deletions in this region lead to a reduced rate of RNA decay (13) and accumulation of partly degraded fragments with increased lifetimes (14). The C-terminal, non-catalytic domain serves as degradosome scaffold and helps targeting RNE to RNA but is not essential in E. coli, as the catalytic activity of the N-terminus is only little affected when part or all the C-terminus is deleted (15). RNE homologs can be found in cyanobacterial and land plant genomes, but it remains uncertain whether they originate from the highly similar RNE or the shorter form RNase G consisting mainly of the catalytic domain (7,16). Plant RNE differs from most bacterial forms mainly by the acquisition of a large N-terminal extension similar to the RNE of Streptomyces coelicolor (16,17), an insertion in the RNA-binding S1 motif of the catalytic domain and the lack of the C-terminal degradosome scaffold, which is replaced by a shorter chloroplast-specific region (Figure 1A). The lack of the degradosome scaffold of the plastid RNE along with the fact that the plastid PNPase forms a homo-multimer not associated with other proteins (18,19) and the suggestion that RNE forms homo-oligomers in vitro (17) led to the assumption that a degradosome homolog is not present in chloroplasts.
Figure 1.
Expression, purification and activity of Arabidopsis RNE and modified forms. (A) Schematic view of E. coli and Arabidopsis thaliana RNE protein, as well as the recombinant form (WT, aa 284-996), a mutated form identical to the final version of RNE-2 (lacking aa 688–693) and an S1-modified RNE (S1m, lacking aa 422–532). Numbers of aa are indicated according to the full-length sequence. Domains are coloured as follows: transit peptide, dark green; N-terminal extension, light grey; RNase H, dark grey; S1 domain, blue; S1 addition in plants, light green; 5′ sensor, yellow; DNase I-like, red; Zn link, black; small domain, purple; C-terminal extension, light grey and E. coli degradosome scaffold, rose. (B) The proteins were expressed in E. coli, purified by affinity and ion-exchange chromatography and analysed by SDS-PAGE and immunoblotting using His6-specific antibodies. (C) The endonucleolytic activity of Arabidopsis RNE (WT) and its recombinant form S1m was measured. 5′ mono- (P) or 5′-triphosphorylated (PPP), 32 P-UTP-labelled tRNATYRSu3 precursor RNA (Su3) was incubated with WT or S1m. Aliquots were removed at 5 and 15 min and purified RNA analysed by PAGE and autoradiography. (D) 5′-monophosphorylated, 32 P-UTP-labelled tRNATYRSu3 was incubated with Arabidopsis RNE (WT) or RNE-2 for 5, 15 and 45 min, following RNA isolation, denaturing gel electrophoresis and autoradiography.
The Arabidopsis nuclear genome encodes one RNE protein (At2g04270), which has been characterized in several recent studies, supporting its endonucleolytic function within the chloroplast stroma (17,20). It was proposed that the rne mutation causes mainly defective processing of the rpl22 mRNA coding for a ribosomal protein (21). However, the nature and necessity of interaction partners and an involvement of factors conferring binding specificity to RNA targets still remained unknown.
In this study, we analysed two different alleles of RNE, which accumulate unprocessed plastid precursor transcripts. We aimed to address the question whether the chloroplast RNE is associated with other proteins and how RNA targets are recognized. To that goal, RNE tagging and co-immunoprecipitation (Co-IP) experiments in combination with mass spectrometry were performed. Several lines of evidence confirmed a direct interaction of RNE with the previously uncharacterized RHON1 protein (At1g06190) and the localization of both proteins in the same HMW complex. The C-terminal domain of RHON1 binds to single-stranded (ss) RNA ensuring efficient processing of plastid transcripts by RNE. RHON1 is essentially required for photoautotrophic growth and seemed to repress the chloroplast division rate and to be an important key player in plastid gene expression.
MATERIALS AND METHODS
Plant material and growth conditions
Wild-type (WT) and mutant seedlings were grown for 3 weeks either on sucrose supplemented medium or on soil (22). Both rne-1 and rne-2 mutant lines were in Columbia, rhon1 in Wassilewskija background. The initially characterized rne-1 was obtained from SALK (20,21). The rne-2 mutant originated from a collection of ethyl methyl sulfonate (EMS)-induced mutations (23). The (transfer) T-DNA-tagged mutant line rhon1 was derived from the Feldman T-DNA collection (24).
Map-based cloning of rne-2 and rhon1
Mapping populations were generated by crossing wild-type plants (accession Landsberg erecta) with heterozygous mutant plants as pollen donor and examined using polymerase chain reaction (PCR)-based polymorphic markers.
Complementation of rne-1 and rhon1 mutants
The complementary deoxyribonucleic acid (cDNA) of RNE (17) and a reverse transcriptase (RT)-PCR-generated RHON1 cDNA were cloned in frame into the TAP-tag vector (K. Meierhoff, unpublished data) using the Gateway technology and stbl2-competent cells (both Invitrogen). Heterozygous mutant plants were transformed using the floral dip method (25).
Bimolecular fluorescence complementation and chlorophyll autofluorescence
The cDNAs of RNE and RHON1 were cloned in frame into split-YFP vectors (26) using Gateway technology (Invitrogen) and the same entry vector as for the TAP vector. Vectors were either transformed alone or co-transformed into Arabidopsis protoplasts (27). YFP fluorescence and chloroplast autofluorescence were visualized 48 h after incubation using a fluorescence microscope in ApoTome mode (Axio Imager, Zeiss) and identical exposure time for all samples. Analysis of chlorophyll autofluorescence of WT and mutants took place in the same manner (27).
Fluorometric and spectroscopic analyses
Chlorophyll a fluorescence analyses and P700 redox measurements were performed as described (28).
Protein analysis and antibody production
For isolation of soluble proteins, wild-type and mutant leaves were homogenized in isolation medium (0.3 M sorbitol, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid HEPES/KOH, 10 mM NaHCO3, 5 mM ethylene glycol-bis(2-aminoethylether)-N,N,N ′,N ′-tetraacetic acid (EGTA), 5 mM ethylenediaminetetraacetic acid, 5 mM MgCl2, pH 8.0) using a Warring blender after centrifugation at 1500g for 8 min with slow deceleration. Chloroplasts were lysed in radioimmunoprecipitation assay buffer (RIPA) buffer containing 1% NP-40, and after centrifugation at 18 000g, membrane proteins and soluble proteins were separated. In vivo labelling of plastid proteins with [35S]-Met/Cys was performed as described (29). Immunoblotting and antibodies used were described previously (3, 22, 28). For production of antibodies against RNE, the recombinant protein (17) was injected into rabbits at different intervals (Pineda-Antikörper-Service). The proteins RNETAP and RHON1TAP were identified using a monoclonal anti-HA antibody (Anti-HA-Peroxidase high affinity, Roche).
Co-IP of epitope-tagged proteins
Soluble chloroplast extracts of ∼30 g of rneTAP or rhon1TAP plants were prepared as described earlier and loaded onto a Strep-Tactin column (IBA) following the manufacturer’s instructions. The combined eluates were concentrated using Amicon Ultra filtration devices (Millipore) and either separated on Clear-Native (CN) and sodium dodecyl sulphate (SDS) gels for immunoblot analyses or used for identification of interaction partners by mass spectrometry (Mass Spectrometry Unit, Department Biology I, LMU Munich). Co-IPs for RIP-chip analyses were performed in tandem using first the Strep-Tactin column and then an HA-matrix-assisted (Roche) procedure.
CN polyacrylamide gel
For two-dimensional (2D) analysis of the HMW complex, Co-IP-purified and concentrated complexes were either treated with 1% RNase One (Promega) or loaded directly onto native 5–12% acrylamide gradient gels with a reduced Coomassie content (0.002%) when compared with Blue Native gels (30). After electrophoresis, lanes of the CN gels were denatured with SDS and β-mercaptoethanol and run on the second dimension in SDS-polyacrylamide gel (PAGE) with 10% acrylamide.
RIP-chip array design, hybridization and slot-blot analyses
Labelling of RHON1TAP-co-purified RNA and its hybridization on the Arabidopsis chloroplast microarray were carried out as described previously (31). Control experiments were performed using wild-type extracts. Fluorescence data and probes can be found in Supplementary Table S3. For slot-blot hybridizations, RNA was isolated as for RIP-chip analysis and 1/12 of the flow through and 1/6 of the pellet were spotted onto nylon membranes and hybridized with the probes indicated.
Generation of recombinant GST-Rho-N-Strep-fusion protein for RNA-binding studies
The last 120 nucleotides of RHON1 encoding the Rho-N motif were cloned into the pET41b+ vector (Novagen) and expressed in E. coli cells (Bl21pLysS, Novagen) using standard conditions. Precipitated cells were lysed by sonification in Strep-washing buffer supplemented with 10% v/v Sarkosyl 150 min after induction of expression. After 1:5 dilution with Strep-washing buffer supplemented with 2% Triton-X-100 and 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), cell extracts were incubated for 1 h in Strep-Tactin-Macroprep and subsequently treated with Strep-washing buffer. The glutathione S-transferase (GST)-Rho-N-Strep-fusion protein was eluted with Strep-elution buffer supplemented with 30% glycerol. Buffers and Macroprep were obtained from IBA.
Electrophoretic mobility shift assays
RNA-radiolabelling and gel shift assays were performed as described (3,32) using primers listed in Supplementary Table S1. RNAs were incubated with increasing protein concentrations (0, 80, 400 and 800 nM) in a buffer containing 40 mM Tris, pH 7.5, 100 mM NaCl, 0.1 mg/ml bovine serum albumin (BSA), 4 mM dithiothreitol (DTT) and 0.5 mg/ml Heparin at 25°C for 10 min.
In vitro activity assays of RNE protein
For in vitro studies, a truncated RNE protein comprising amino acids (aa) 284–996 and recombinant forms obtained by site-directed mutagenesis were expressed in E. coli, purified as described (17) and detected with His6-specific antibodies. Recombinant RNE-2 proteins were lacking the same six amino acids (688–693) as rne-2 mutants. An S1-modified RNE was constructed missing aa 422–532. Numbers of aa are indicated according to the full-length sequence. Cleavage activity assays with recombinant proteins were performed as described (17).
Isolation of total RNA, gel-blot analysis and RT-PCR
RNA extraction, labelling of DNA probes and gel blot analyses were performed as described (22). RT-PCRs were performed using the One-Step RT-PCR kit (Roche).
RESULTS
High-resolution mapping of HCF2 identified the endoribonuclease RNE
The non-photosynthetic high-chlorophyll-fluorescence (hcf) mutant hcf2 was originally identified in a screen of EMS-mutagenized Arabidopsis plants (23). We assigned the mutation in our mapping population using 1536 meiotic chromosomes to bacterial artificial chromosome T23O15 between two adjacent markers with 3 and 10 recombination events (Supplementary Figure S1A). Sequencing of genes located in between revealed a mutation (rne-2) in the RNE gene At2g04270 with a single G-to-A transition of the last nucleotide of the seventh intron (Supplementary Figure S1B). Sequencing of RT-PCR products uncovered an alternative splice site 18 nucleotides farther downstream within exon eight (Supplementary Figure S1B and C) resulting in a deletion of six amino acid residues within the RNase H fold of rne-2. RNE is absent in the rne-1 knockout line, whereas in rne-2, the protein lacking the six aa is expressed at normal levels (Supplementary Figure S1D). The protein runs at ∼140 kDa, although the predicted size of the mature protein lacking the transit peptide for chloroplast import is around 104 kDa. This anomaly in migration has also been reported for RNE in E. coli (33) and was mentioned in previous reports about plastid RNE (17,20).
RNE-2 and an S1 domain-modified RNE have no endonucleolytic activity in vitro
In E. coli, RNE plays an important role in processing of tRNA transcripts (34,35) and cleaves the tRNATYRSu3 precursor (Su3) with high efficiency (36). An over-expressed, truncated form of the chloroplast RNE protein lacking the first N-terminal 283 aa was reported previously to be enzymatically active in cleavage of Su3, producing the mature tRNA and a 5′-leader sequence (17). Moreover, similar to E. coli, Arabidopsis RNE exhibits an increased cleavage activity when the 5′-end of the substrate RNA is mono- (P) but not triphosphorylated (PPP) (17,37,38).
The endonucleolytic activity of the previously characterized Arabidopsis protein, comprising aa 284–996, was now compared with a modified RNE protein S1m, lacking the plant-specific insertion of domain S1 (aa 422–532), making the protein similar to the E. coli RNE (Figure 1A). In addition, the endonucleolytic activity of the corresponding version of RNE-2 lacking the six aa 688–693 was analysed (Figure 1A). All three forms were expressed in bacteria and isolated to high purity. Preparations of the recombinant enzymes were examined by immunoblot analysis using His6-specific antibodies (Figure 1B).
Only a very small portion of 5′-PPP-Su3 and 5′-P-Su3 was cleaved by S1m over an incubation period of 15 minutes (Figure 1C). In contrast, the wild-type form was able to cleave ∼50% of 5′-PPP-Su3 tRNA compared with almost complete cleavage of 5′-P-Su3. This indicates an indispensable role of the additional plant-specific motif within the S1 domain for RNE enzymatic activity. The recombinant RNE-2 was also unable to cleave 5′-P-Su3 transcripts (Figure 1D). This explains why the overall rne-2 phenotype is very similar to that of the rne-1 knockout line and demonstrates that the internal six aa in the RNase H fold are essential for activity.
Complementation and epitope tagging of rne-1
Homozygous rne seedlings developed pale cotyledons when grown on sucrose-supplemented medium and were only able to survive on soil when kept under optimal watering and light conditions albeit with severe growth retardations as reported recently (21) and confirmed in this study (Supplementary Figure S2A).
To further investigate the function of RNE and potential interactions, we complemented homozygous rne-1 mutants with the RNE full-length cDNA fused in frame to a TAP-tag, creating rneTAP lines. The TAP-tag consists of a tandem of three human influenza hemagglutinin (HA) epitopes followed by one StrepIII epitope, which are separated by a short linker sequence (Supplementary Figure S2B). The vector contains a weak promoter derived from the HCF173 gene to avoid over-expression of the tagged protein (39). This should allow identification of interaction partners using highly specific monoclonal antibodies raised against the epitopes of the tags in Co-IP experiments. Homozygosity of rneTAP plants was confirmed by PCR (Supplementary Figure S2C). No visible differences could be observed between epitope-tagged mutant lines and wild-type plants, suggesting that the TAP-tag does not impair the function of RNE (Supplementary Figure S2D). Immunoblot analyses of complemented rne lines revealed a signal at the expected size (RNE 140 kDa + TAP-tag = 147 kDa) in the chloroplast stroma using mono-specific HA antibodies and the StrepIII-tag allowed purification of RNE under native conditions (Supplementary Figure S2E).
Isolation of an RNE interacting protein in Arabidopsis
Stromal fractions were prepared from isolated chloroplasts and subjected to Co-IP using a Strep-tactin column. Co-IP eluates were shortly separated on a gel and subsequently in-gel digested with trypsin. The resulting peptides were analysed through microcapillary liquid chromatography tandem mass spectrometry MS/MS followed by protein database searches of the generated spectra. Three independent experiments identified several peptides of RNE together with a protein that could be assigned to the At1g06190 gene (Supplementary Table S2). Both proteins were completely absent when untagged wild-type plants or lines expressing other TAP-tagged proteins, such as HCF101 (40), PrfB3 (3) and PAC (41), were subjected to the same procedure (unpublished data), indicating the reliability of the TAP-tag purification and suggesting that the At1g06190 gene product is either directly or indirectly associated with RNE.
The chloroplast-targeted At1g06190 protein is found exclusively in vascular plants
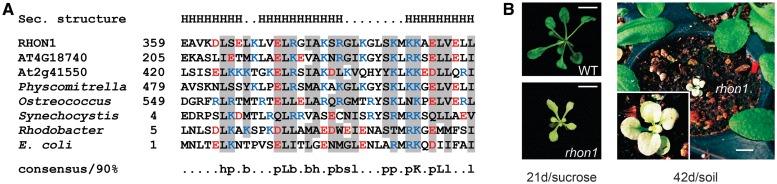
With the exception of Chlamydomonas reinhardtii, RNE homologs can be found in all photosynthetic lineages including green algae (17), whereas bioinformatical genome inspections revealed that At1g06190 holds homologies only among vascular plants. Domain analysis revealed that the protein contains a predicted transit peptide for transfer to the chloroplast (ChloroP) and two conserved regions of unknown function followed by a stretch consisting of highly negatively charged aa. The C-terminal domain is similar to the N-terminal part of the RNA-binding domain of bacterial transcription termination factor Rho (42,43), which has a helix-extended-helix structure and is called Rho-N (Figure 2A). Hence, we named the At1g06190 protein RHON1. Besides RHON1, the Arabidopsis genome encodes two other proteins with a putative Rho-N domain, predicted to be targeted to mitochondria (At4g18740) and cytoplasm (At2g41550) (Figure 2A).
Figure 2.
Multiple sequence alignment of Rho-N motifs and rhon1 phenotype. (A) The alignment shows the Rho-N motifs of RHON1 and proteins of unknown function in Arabidopsis thaliana (At4g18740, At2g41550), Physcomitrella patens (XP_001761620) and Ostreococcus lucimarinus (XP_001416615), as well as N-terminal sequences of Rho factors from Synechocystis, Rhodobacter and E. coli, starting with the aa indicated. The secondary structure shown above the alignment and the consensus sequence shown below were described in Aravind and Koonin (43). Negatively charged aa are coloured in red and positively charged in blue. H, helix; h, hydrophobic; p, polar; b, big; s, small and l, aliphatic. (B) The pale phenotype of a rhon1 mutant plant when compared with wild type grown on sucrose-supplemented MS media for 21 days under constant light is shown (left). A 6-week-old rhon1 mutant grown on soil displays an albinotic phenotype and dies after 7 weeks (right). Scale bars indicate the size of 1 cm.
The previously described rice protein OsBP-73 (Os03g0183100) was assumed to bind DNA (44) and exhibits 35% sequence similarity to the N-terminal part of RHON1. An in vitro interaction with the promoter of the nuclear waxy gene in rice suggested localization of OsBP-73 in the nucleus where it functions as transcription factor. However, localization of OsBP-73 was not addressed experimentally. Several subcellular localization algorithms predicted that RHON1 and OsBP-73 are present in the chloroplast (ChloroP, TargetP, PCLR). This is consistent with the identification of RHON1 peptides in recent chloroplast proteomics studies (45,46).
Molecular mapping, complementation and phenotype of rhon1 mutants
An Arabidopsis T-DNA mutant of the RHON1 locus affected in chloroplast RNA metabolism, and also exhibiting an hcf phenotype was previously identified in our group. Mutants of rhon1 were smaller and showed a pale-green phenotype when grown on sucrose-supplemented MS medium. When grown on soil, some rhon1 mutants could survive up to 7 weeks exhibiting an albinotic phenotype (Figure 2B). Segregation analysis using 2448 F2 progenies and 638 individual F3 plants derived from backcrossed mutant plants revealed that the mutation is either caused by or very closely located to one insertional event because the kanamycin resistance conferred by the T-DNA strictly co-segregated with the mutant phenotype. It was impossible to identify the T-DNA flanking sequences by inverse PCR because of multiple truncated T-DNA insertions in this locus as revealed by genomic Southern analysis. Therefore, we fine mapped the mutation on the upper part of chromosome 1 (Figure 3A) and localized the T-DNA insertions at position +481 relative to the start codon of RHON1 (Figure 3A and B). Using RT-PCR, we generated a full-length RHON1 cDNA encoding a protein of 401 aa.
Figure 3.
Mapping, phenotype and complementation of rhon1. (A) Molecular markers (H-M) used for fine mapping of rhon1 on chromosome 1 and the corresponding numbers of recombinant events are shown. Sequencing revealed that the T-DNA was inserted into the At1g06190 gene (RHON1). (B) Exons (grey boxes) and introns (black lines) show the structure of the genomic RHON1 gene. The T-DNA insertion at position +481 is marked. Arrows indicate the positions of primers used for PCR analysis. (C) RHON1 contains a putative chloroplast transit peptide (TP), two highly conserved regions (red) and the C-terminal motif Rho-N (yellow). For complementation of the mutant, a construct of the full-length protein fused to an HA-Strep TAP-tag was used. (D) WT and complemented rhon1TAP plants were grown on soil for 4 weeks in 12/12 hours light/dark conditions. Scale bars indicate the size of 1 cm. (E) PCR analysis demonstrates orientation of the T-DNA insertion in exon 2 of rhon1 and homozygosity of independent complemented lines. (F) Chlorophyll fluorescence was imaged from protoplasts isolated from WT, rne-1 and rhon1. Scale bars indicate the size of 10 µm.
All attempts to express the full-length RHON1 in E. coli cells or to immunize rabbits against synthetic RHON1 peptides to get functional antibodies failed. Thus, we complemented the rhon1 mutant with the cDNA fused to the same TAP-tag as for RNE (Figure 3C and D). Genotyping confirmed homozygosity of three independent lines (Figure 3E). Complemented rhon1TAP lines could not be distinguished phenotypically from the wild type (Figure 3D) indicating that the rhon1 mutant phenotype is solely caused by the multiple T-DNA insertions in the RHON1 gene.
The phenotype of rhon1 mainly resembles that of rne, but in contrast to rne, homozygous rhon1 mutants grown on soil are lethal after a few weeks and were therefore grown only on sucrose-supplemented medium. Chlorophyll a fluorescence imaging of isolated leaf protoplasts revealed a dramatic decrease in chloroplast size accompanied with an increase in chloroplast number in both rne and rhon1 mutants when compared with the wild type (Figure 3F).
RHON1 precipitates RNE
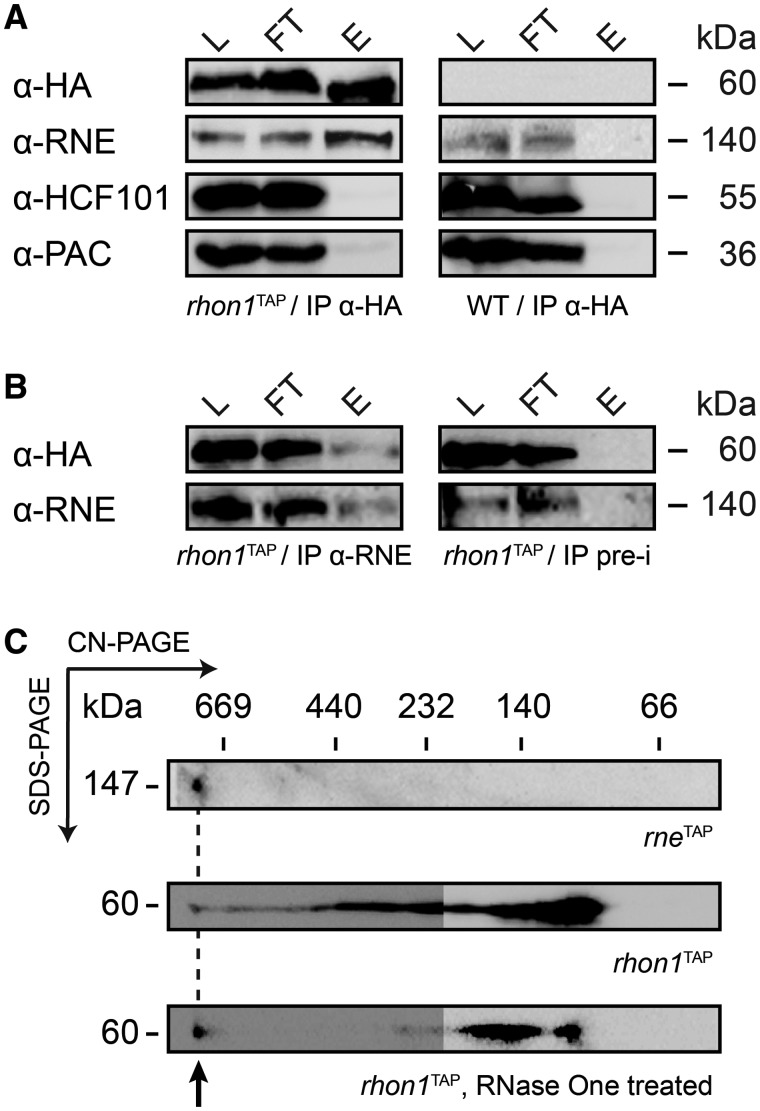
To confirm the RNE–RHON1 association by an independent experiment, we investigated Co-IPs of rhon1TAP lines. Using the HA antibody, we could observe a distinct RHON1 signal in lysate, flow through and eluate of rhon1TAP but not of untagged wild-type lines. Notably, the tagged RHON1 protein shows the same anomaly of migration in SDS-gels such as RNE, running at 60 kDa, which is ∼13 kDa higher than expected, probably due to the high amount of negatively charged aa (Supplementary Figure S3). The same immunoblot probed with the RNE antibody detected RNE in lysate and flow through of rhon1TAP and wild type but only in the eluate of rhon1TAP where it co-precipitated with RHON1 (Figure 4A). Control experiments using antibodies raised against HCF101 or PAC, which are involved in plastid Fe/S cluster biogenesis (40) and RNA metabolism (3,41), respectively, did not give any notable signal in rhon1TAP or wild-type eluates, suggesting that rhon1TAP precipitated specifically RNE. This data confirms that both proteins are stably associated in the same protein complex. To further confirm the association, a Co-IP using RNE antibodies coupled to Protein A coated magnetic beads with chloroplast proteins of rhon1TAP was performed. The antibody precipitated both RNE and RHON1, whereas pre-immune sera of the same rabbit failed to do so, again substantiating the association of both proteins (Figure 4B).
Figure 4.
RNE associates with RHON1 in a HMW complex. (A) Soluble protein extracts from rhon1TAP or WT were immunoprecipitated using an HA matrix. Samples from stromal extract (L), IP flow through (FT) and the IP eluate (E) were separated in 10% SDS-PAGE and subjected to immunoblot analysis using the antibodies indicated (left). Protein sizes are given in kDa (right). (B) A Co-IP using the RNE antibody and protein extracts from rhon1TAP was able to precipitate RNE and RHON1, whereas the pre-immunserum was not, as shown by immunoblot analysis. pre-i, pre-immunserum. (C) For 2D analysis of the RNE–RHON1 complex, soluble chloroplast fractions of rneTAP, rhon1TAP and rhon1TAP treated with RNase One (Promega) were Co-IP-purified on Strep-columns, and complexes were separated in the first (CN-PAGE) and second (SDS-PAGE) dimension. Blotted gels were immunodecorated with an HA antibody and sizes of complexes estimated using a HMW marker set (GE). The left parts of rhon1TAP immunoblots were overexposed. The HMW complex of ∼800 kDa (arrow) containing both RNE and RHON1 did not disappear on RNase One treatment (not shown for rneTAP).
Pleiotropic effects on photosynthetic performances and proteins in rne and rhon1
The hcf-phenotypes of pale rne and rhon1 mutant plants indicate defects in the photosynthetic apparatus. To investigate the deficiencies, the activities of photosystems (PS) I and II were determined using spectroscopic methods. The oxidized state and the weak signal of PSI demonstrate that electron transport towards PSI is limited and that PSI levels are reduced in the mutants, respectively (Supplementary Figure S4A). The maximum quantum yield of PSII (Fv/Fm), a standard parameter of PSII integrity, was reduced to ∼50% and 40% in rne and rhon1, respectively (Supplementary Figure S4B). The epitope-tagged complemented lines behaved comparably to wild type. As these data pointed out severe pleiotropic effects on photosynthetic capacity, we determined the content of photosynthetic proteins by immunoblot analyses. Levels of most PSI and PSII proteins and the plastid ATP synthase were reduced to equal amounts of less than 25% in both mutants, whereas amounts of the cytochrome b6f complex were more affected in rhon1 than in rne-1 (Supplementary Figure S4C). To check the molecular cause for reduced protein levels in rhon1, translation of plastid-encoded thylakoid proteins was estimated (47). It seemed that less radioactive label was incorporated into the PSII proteins D1, D2, CP47 and some smaller unknown proteins indicating translational deficiencies in rhon1 (Supplementary Figure S4D). Taken together, the pleiotropic phenotype of rhon1 is comparable with but to a certain extent more severe than that of rne-1 displaying an elevated importance and/or additional functions of RHON1 in comparison with RNE.
RNE and RHON1 are located in the same complex
As we confirmed that RHON1 always precipitates with RNE, we intended to elucidate the nature of the RHON1–RNE protein complex. The biochemical characterization of the plastid RNE turned out to be similarly difficult as reported for the bacterial enzyme, which is extremely sensitive to proteolysis and tends to form larger assemblies when over-expressed in E. coli cells (48). Because even freezing resulted in a severe loss of plastid RNE protein, biochemical analysis had to be conducted strictly with fresh preparations. This high rate of RNE protein degradation made classical separation methods such as size exclusion chromatography unfeasible. Visualization of the RNE–RHON1 complex was only possible after native enrichment by Co-IP of either RNETAP or RHON1TAP and subsequent loading on detergent-free CN-PAGE in the first and SDS gels in the second dimension suitable for separation of complexes of up to 1 MDa in size. Several independent 2D-PAGE experiments revealed the presence of RNETAP and RHON1TAP in a complex of ∼800 kDa running more than 1 cm into the separating gel (Figure 4C). Based on the highly variable degradosomes found in bacteria and mitochondria, the confirmation of a conserved function of RNE in RNA processing (Figure 1) and identical processing defects in rhon1 and rne (see later), this HMW assembly could hint to the presence of a degradosome-like complex in the chloroplast. Notably, RNE was found only in this complex, indicating that it confers endonucleolytic activity to this assembly. Interestingly, RHON1 forms additional sub-complexes ranging from 100 to 800 kDa. On RNase One treatment, sub-complexes >150 kDa disappeared almost completely and gave rise to a prominent and distinct RHON1TAP-containing complex of 150 kDa (Figure 4C) This indicates interactions of RHON1 with RNAs of a broad molecular weight range. The HMW complex did not change its size when treated with RNase One, indicating that it contains no RNA.
Direct interaction of RNE and RHON1 along with RHON1 dimerization
To test whether RNE directly interacts with RHON1 or whether they are connected to each other by additional proteins, we performed bimolecular fluorescence complementation (BiFC) analysis using transient expression of both proteins in Arabidopsis protoplasts (Figure 5). Co-expression of YFPC-tagged RNE and YFPN-tagged RHON1 in Arabidopsis protoplasts resulted in significant fluorescence (Figure 5A) suggesting a direct interaction of both proteins in the chloroplast, whereas the constructs expressed alone did not give any YFP signal (Figure 5B). Moreover, co-expression of RHON1-YFPN and RHON1-YFPC indicated that RHON1 has the ability to form homodimers or homomultimers (Figure 5C), which could be part of the RNA-binding RHON1 sub-complexes. No fluorescence was found in protoplasts with expression of RHON1-YFPC alone or co-expression of empty YFPN and YFPC vectors (Figure 5D). Consistent with a direct physical association, a specific reduction of RNE levels to ∼25% was observed in rhon1 mutants when compared with the wild type. This indicates that the proteins are also functionally connected to each other (Figure 6A).
Figure 5.
BiFC detection of RNE and RHON1 protein interactions and RHON1 dimerization. Interaction of proteins is depicted in green and was visualized by split YFP using fluorescence microscopy. Chlorophyll autofluorescence is shown in red. Merged pictures that appear orange confirm that protein interaction takes place inside the chloroplast. (A) RHON1-YFPN and RNE-YFPC constructs were transiently expressed in Arabidopsis protoplasts by co-transformation and show a clear YFP signal. (B) RHON1-YFPN or RNE-YFPC transformed alone do not show any signal. (C) Co-transformation of RHON1-YFPN and RHON1-YFPC shows a strong signal indicative for dimerization and/or multimerization of the protein. (D) RHON1-YFPC transformed alone, and co-transformed empty YFPN and YFPC vectors show no signal. Scale bars indicate the size of 10 µm.
Figure 6.
RNE levels in rhon1 and the phenotype of rne/rne rhon1/rhon1 double knockouts. (A) Comparison of RNE levels in wild type, rne and rhon1 mutants. Levels of the protein PAC were used as a loading control. (B) Double knockouts were generated by crossing heterozygous rne-1 and rhon1 mutants and selfing of the selected double heterozygous F1. The average Fv/Fm rate and standard deviation were determined from 10 independent plants. Representative mutant plants are shown above each column, whereas wild-type and double heterozygous plants are only shown in parts to maintain the same scale.
Double knockouts of rne-1 and rhon1 display an additive phenotype
The nature of both rne-1 and rhon1 mutations was strictly recessive. Fv/Fm levels were more reduced (0.07 ± 0.07), and the pale phenotype seemed to be more intense in the homozygous rne-1/rne-1 rhon1/rhon1 double knockout mutants than in the individual knockouts indicating an additive effect of the mutations on photosynthetic parameters and chloroplast development (Figure 6B). Therefore, we assume that RHON1 also has other functions in addition to supporting RNE performance.
RHON1 associates with 16 S and 23 S ribosomal RNA
As an initial survey experiment to identify the plastid RNA targets of RHON1 complexes, we performed RIP-chip assays. Co-immunoprecipitated RNAs from TAP-tagged lines and wild type as negative control were isolated from flow through and eluates using both HA- and Strep-tags and subsequently labelled with red- or green-fluorescing dyes, respectively. These RNAs were combined and hybridized to a tiling microarray of the Arabidopsis chloroplast genome (31). Replicate experiments revealed that the RNAs most strongly enriched in Co-IPs using rhon1TAP were the ribosomal 16 S and 23 S RNAs (Figure 7A, Supplementary Table S3). Slot-blot hybridization of immunoprecipitated RNAs from flow through and eluate verified the enrichment of 16 S and 23 S ribosomal RNA (rRNA) using full-length gene probes (Figure 7B), whereas 4.5 S, 5 S, ycf1 and rbcL RNAs were not bound efficiently. This suggests that RHON1-containing complexes associate with the highly abundant 16 S and 23 S rRNAs. Consistently, expression of these rRNAs is also severely affected in the mutants (see later). However, it remains uncertain whether RHON1 complexes are also associated with lower abundant mRNAs. Notably, RIP-chip analysis using the tagged RNE did not detect any association with RNAs in favour of the idea that the HMW complex contains no RNA.
Figure 7.
Association of RHON1 with rRNA transcripts. (A) Stroma of rhon1TAP or WT (control) was subjected to tandem IP with Strep-Tactin column and HA matrix. For RIP-chip analysis, co-purified RNAs after the second purification step were labelled with Cy5 and RNAs of the flow through from the first purification step were labelled with Cy3. The enrichment ratios (FCy5:FCy3) were normalized between duplicate assays. The median normalized values for replicate spots from the RHON1TAP purifications were divided by those from WT data and plotted according to fragment number on the Arabidopsis chloroplast genome tiling microarray. Fragments are numbered according to chromosomal position. The data used to generate this figure are provided in Supplementary Table S3. (B) RIP-chip data were validated by slot-blot hybridization; 1/12 of the RNA purified from the IP flow throughs of the first purification steps (FT1) and 1/6 of the RNA from the IP eluates of the second purification step (E2) were applied to slot-blots and hybridized with the indicated probes. (C) The RNA-binding capacity of RHON1 was analysed by gel-shift assays using radiolabelled short RNAs of the 16 S and 23 S rRNAs as indicated. Increasing concentrations of 80, 400 and 800 nm (black triangles) of the purified GST-Rho-N-Strep protein consisting of the last 40 aa of RHON1, harbouring the Rho-N domain, were used for binding experiments.
The C-terminal Rho-N domain of RHON1 is responsible for RNA binding
As the resolution of our RIP-chip and slot-blot analysis is limited to several hundred nucleotides, electrophoretic mobility shift assays were performed suitable for narrowing down the target sequences. Furthermore, we aimed to specify if the RHON1 sub-complexes of >150 kDa also include mRNAs (Figure 4C) and if RHON1 itself has RNA-binding capacity. Moreover, we addressed the question which motif of RHON1 is responsible for binding. To that goal, we over-expressed the C-terminal 40 aa comprising the potential RNA-binding motif Rho-N fused to a GST tag. We randomly selected regions of the 16 S and 23 S rRNAs and the intergenic region of 23 S-4.5 S for the association assays, each ∼70–100 nucleotides in length. It seemed that the Rho-N motif binds to all investigated RNAs, although with quite different stringencies (Figure 7C). Most efficient binding was observed for the region 23 S-1 and the 3′-intergenic region of 23 S-4.5 S harbouring the processing site. This result confirms binding of RHON1 to certain regions of the 23 S rRNA as revealed by RIP-chip data and provides evidence that especially RNAs which are not efficiently processed in both rne and rhon1 mutants are recognized by RHON1 (Figures 8 and 9). Only weak binding was found for probes of 16 S-1, 16 S-2, 23 S-4 and the 5′-intergenic region of the 23 S-4.5 S (Figure 7C). As rRNAs are often highly structured, the low-binding capacity of RHON1 to certain fragments could result from preferential binding to ss RNA. To study the nucleic acid-binding properties of RHON1 to ss and double-stranded (ds) RNAs, we used a small RNA fragment of the 23 S-4.5 S intergenic region in a gel mobility shift assay (Supplementary Figure S5A). RHON1 bound the ssRNA fragment with a similarly low stringency as before, whereas the dsRNA fragment was not bound at all, indicating that RHON1 preferentially binds ss RNA. However, binding to this fragment was still weak and is consequently rather caused by low sequence specificity than by a dsRNA structure. Interestingly, RHON1 was not capable of binding to a ss 31-mer of the maize chloroplast petA 5′ UTR, which is commonly used for such analyses (32) (Supplementary Figure S5B). Thus, RHON1 represents a novel RNA-binding protein recognizing specific regions of diverse RNAs by the Rho-N domain.
Figure 8.
RNA gel blot analyses of rRNAs in wild type, rne and rhon1. The rDNA gene cluster and all processing products (black lines) of rRNAs are shown down to scale. RNA gel blot analyses were performed using probes rrn16S, rrn23S, rrn4.5 S and rrn5S as indicated in grey boxes. A 2.7 kb transcript (dotted line and asterisks) is detected only in rhon1 and rne mutant lines. Equal loading was confirmed using methylene blue (M.B.) staining. The transcript lengths are indicated in kb.
Figure 9.
Processing of mRNAs and mRNA-binding studies. (A) Transcript levels of rbcL and petA are shown in WT Columbia (Col-0), rne-1, rne-2, WT Wassilewskija (Was) and rhon1; 10 μg leaf RNA of 3-week-old plants, grown on sucrose supplemented MS media, were loaded. (B) The RNA-binding capacity of the C-terminal 40 aa of RHON1 (Rho-N) motif to a radiolabelled RNA of the rbcL-accD intergenic region comprising the processing site was analysed by gel-shift assays. (C) Levels of petB transcripts in rne-1 and rhon1 were compared with those in wild-type accession Columbia. (D) The RNA-binding capacity of the Rho-N motif was examined using radioactive labelled RNAs of the petB 5′ and 3′ UTR.
Accumulation and processing of chloroplast rRNAs are affected in rne and rhon1
Binding of RHON1 to ribosomal 16 S and 23 S rRNAs suggested a crucial involvement in processing and/or stability of rRNAs. To visualize the integrity of plastid rRNAs, we stained a blot of denatured wild type, rne and rhon1 RNAs with methylene blue revealing a marked reduction of both 16 S and 23 S rRNAs in rhon1 but not in rne-1 (Figure 8). This again substantiates additional functions of RHON1, which are presumably not associated with RNE. We investigated the processing pattern of the rRNA transcription unit by performing RNA gel blot analysis. Accumulation of the 3.2 kb 23S–4.5 S dicistronic precursor transcript was observed in rne and even more in rhon1 mutants indicating loss of efficient endonucleolytic processing. This is accompanied by reduced levels of processed transcripts in rhon1 and to some extent in rne, such as the 0.5 kb, 1.1 kb and 1.8 kb RNAs originating from the 23 S rRNA. Hybridization with the 4.5 S probe revealed an additional band of 2.7 kb in both mutants, which is not present in the wild type or other rRNA processing mutants (49–52). As this band could not be detected with the probe of the 23 S 5′ region, it still comprises sequences of the 4.5S rRNA but lacks 0.5 kb of the 5′ region (Figure 8C). The 5S probe could not detect any HMW band although levels of the monocistronic 5 S rRNA were reduced in rhon1, supporting that the 2.7 kb band does not comprise the 5 S rRNA.
Processing defects of chloroplast mRNAs in rhon1 and rne mutants
In accordance with the identified functional and physical association of RHON1 and RNE, a recent chloroplast genome-wide transcriptomic approach revealed close clustering of the rne-2 and rhon1 (formerly named crp102 and crp135, respectively) mutant transcriptomes within the group of mutants exerting quite similar global functions in plastid gene expression (5).
A comparison of the macroarray data with RNA gel-blot analysis has not only confirmed alterations in the mutants’ RNA transcript levels but also revealed dramatic changes in the pattern of chloroplast transcripts in both mutants when compared with the wild type. A prominent processing defect was observed for the rbcL-accD intergenic region (Figure 9A). Amounts of precursor transcripts of 8.5 and 5.4 kb of the rbcL transcription unit, comprising sequences of the downstream located genes accD, psaI, ycf4, cemA and petA, accumulate in both mutants (Figure 9A, Supplementary Figure S5B). The inefficiently processed rbcL-accD intercistronic region harbours a processing site (53), which is also recognized by RHON1 as revealed by gel-shift analysis (Figure 9B). In the same way, processing of petB as part of the psbB transcription unit (3) was altered in both mutants (Figure 9C). Again, RHON1 binds to the 5′ and 3′ UTR of the monocistronic petB transcript (Figure 9D), which shows reduced levels in both rne and rhon1. Therefore, RNE and RHON1 are not only required for efficient cleavage of rRNAs but also for processing and presumably stabilization of mRNAs. Notably, most of the processed transcripts are present in both mutants, indicating that processing basically takes place although with reduced efficiency. This finding is indicative for the presence of additional, redundant endonucleases, such as RNase J (54). These endonucleases could also be supported by RHON1, thus partially replacing the catalytic function of RNE. Interestingly, all examined sites that are bound by RHON1 are also inefficiently processed. Further studies will have to prove that these sites are indeed identical with the RNE cleavage sites and elucidate the function of the RNE-containing HMW complex in this context. From our data, we assume that RHON1 facilitates efficient cleavage by RNE through determining sequence specificity and delivering target transcripts to the RNE–HMW complex.
DISCUSSION
Divergence of evolution, structure and function of RNE
RNE is a well-studied and highly conserved eubacterial enzyme that functions in mRNA decay and posttranscriptional gene expression. The Arabidopsis thaliana RNE homolog is localized in the chloroplast and shares in vitro biochemical similarities with the eubacterial protein (Figure 1). In this work, we present a new allele of the chloroplast RNE, rne-2 (Supplementary Figure S1). The EMS-induced mutation caused a deletion of six aa within the RNase H domain, leading to a significant loss of RNase activity, although the RNE-2 protein is expressed at normal levels (Figure 1D; Supplementary Figure S1D). This is similar to E. coli where the replacement of a single aa in the RNase H domain can severely affect the catalytic activity of the enzyme (55). Despite all similarities, RNE diverged in many respects in plants. The highly conserved RNA-binding S1 domain (55) is interrupted in plant RNEs by an insertion of more than 100 aa (Figure 1A). An over-expressed protein with a deletion of the S1 addition, making the protein similar to its E. coli counterpart in this region, is significantly less active as an endonuclease, indicating that this domain is also important for catalysis (Figure 1C). Despite the various processing defects in rne mutants, the presence of processed transcripts albeit at reduced amounts indicates that loss of RNE in Arabidopsis is partially compensated either by RNase J or by other RNases rendering the mutant viable. This is in contrast to the essential function of RNE in E. coli, which could be attributed to the lack of RNase J and, therefore, the inability to compensate for the lack of RNE. Apparently, the presence of RNE in Arabidopsis is neither essential nor a prerequisite for processing and degradation of plastid RNA. This clearly shows that RNE is not the main chloroplast RNA processing enzyme.
RNE forms a HMW complex together with RHON1
In E. coli, RNE forms the degradosome complex together with RhlB, enolase and PNPase, whereas degradosomes of other organisms are highly divergent due to the fact that the C-terminal degradosome assembly site highly varies in length and sequence. For example, the Bacillus subtilis degradosome consists of the endonuclease RNase Y, the two RNase J forms (J1 and J2), the PNPase, a DEAD-box helicase and two glycolytic enzymes (56). Interestingly, all genomes of Gram-positive bacteria such as B. subtilis lack RNE, whereas Gram-negative bacteria such as E. coli lack RNase J (57). The purple α-proteobacterium Rhodobacter capsulatus forms a degradosome-like complex consisting of RNE acting in concert with two DEAD-box helicases and the termination factor Rho, which is an RNA helicase as well (58). Notably, this degradosome-like complex is lacking PNPase. Cyanobacteria possess, similar to land plants, both RNase J and RNE (19,59,60). Components of the mitochondrial degradosome in yeast and Trypanosoma brucei comprise an RNase II-related exonuclease and a helicase, both not related to components of the bacterial degradosome (61,62). Plant RNE has acquired a novel, large N-terminal extension but is lacking the large C-terminal degradosome scaffold typical for bacterial RNE enzymes, which is now replaced by a plant-specific 130 aa extension (Figure 1A). Both terminal regions are of so far unknown function but could serve as scaffold for associated proteins.
Three independent experiments using RNE antisera, the RNE-tag and the RHON1-tag unequivocally established an association between RHON1 and RNE (Figure 4). Furthermore, BiFC analyses proved a direct physical contact between both proteins, indicating that they not only represent components of the same complex but also bind directly to each other (Figure 5A). Notably, only enrichment from at least 30 g starting leaf material by Co-IP experiments using the TAP-tags in the complemented rne and rhon1 mutant lines finally succeeded to characterize the HMW complex in vivo. The purified and enriched complex was still below the limit of detection in silver gels and could only be substantiated immunologically and by mass spectrometry. RNE was neither found in other complexes nor as a monomer indicating that the protein is present only in this HMW complex. This finding together with the proven enzymatic activity of RNE (Figure 1) provides indirect evidence that this complex exerts endonucleolytic activity similar to the bacterial degradosome. Several more factors, such as ribosomal proteins, helicases and other ribonucleases, were identified in RNE co-precipitates but still need to be reliably confirmed as true interaction partners. According to the presence in a homo-multimer, the plastid PNPase was never found to precipitate neither with RNE nor with RHON1. Association of RNE with RHON1 is in accord with co-sedimentation of both proteins with nucleoids (63,64).
RHON1 is a novel RNA-binding protein and supports RNE function
The vascular plant-specific RHON1 protein plays an indispensable role in plastid gene expression as revealed by seedling lethality and the rhon1 phenotype. Our data provide clear evidence for the presence of RNE and RHON1 as direct interaction partners in the same HMW complex, a tight co-regulation of protein expression and a close functional relationship between the two proteins (Figure 6A). This is also reflected by tight clustering of plastid transcriptomes of both mutants (5).
Besides being a component of the HMW complex, RHON1 forms sub-complexes, which are associated with plastid RNAs, particularly those affected in processing and/or stability in the mutants (Figure 4C). These RHON1 sub-complexes presumably contain RHON1 homodimers or multimers, as BiFC analysis revealed a strong dimerization of RHON1 proteins (Figure 5C). It is, therefore, likely that RHON1 multimers are also attached to the RNE-complex. No stable association of RNAs with the HMW complex could be detected, demonstrating that transcripts are only transiently bound to this complex in the course of endonucleolytic cleavage. Again, it is tempting to speculate that RHON1 targets transcripts to this complex to contribute to efficient catalytic activity of RNE similar to one of the degradosome functions in E. coli. This function of RHON1 is supported by the fact that RNA processing by RNE still occurs in rhon1 although less efficient. RHON1 target RNAs, such as the rbcL-accD and the 23 S-4.5 S intercistronic regions, both harbouring processing sites, could therefore coincide with the RNE-assisted cleavage sites. The additive effect in the corresponding double mutant, the stronger phenotype of rhon1 when compared with rne and the presence of a major fraction of RHON1 in smaller RNA-containing sub-complexes hints to additional roles of RHON1, e.g. targeting transcripts also to other RNases, such as RNase J, to support their endonucleolytic and/or exonucleolytic function.
RIP-chip and slot-blot analysis have shown that sub-complexes of RHON1 associate with RNA. As revealed by electrophoretic mobility shift assays, the motif responsible for RNA binding is the short C-terminal Rho-N domain of RHON1 (Figure 7C). Similar to related nucleic acid-binding domains, such as the SAP motif (43), the Rho-N domain has been recruited by quite diverse proteins of plants including mosses and some green algae and has often been transferred to their C-terminus (Figure 2). The Rho-N domain of RHON1 binds to specific regions of plastid ss RNAs with different strength (Figures 7C and 9B and D, Supplementary Figure S5) attesting a certain sequence specificity, which may be supported further by the remaining part of RHON1 in vivo.
RHON1 is an essential key player in plastid gene expression
Association of RHON1 to 16 S and 23 S RNA revealed an important role in facilitating rRNA maturation at least in part through RNE. This is evidenced by a severe loss of 16 S rRNA in rhon1 and accumulation of the 3.2 kb dicistronic 23 S-4.5 S precursor and a transcript of 2.7 kb, which appears in both mutants. The 2.7 kb transcript originates from an incorrectly processed 23 S-4.5 S precursor that is lacking the 23 S 5′-end of 0.5 kb (Figure 8). Processing of one of the two hidden breaks generates this 0.5 kb fragment on incorporation into ribosomes and usually depends on prior 23 S-4.5 S intercistronic cleavage (65). Therefore, it is likely that initial assembly of ribosomes takes place, but loss of 4.5 S cleavage in the mutants hinders subsequent assembly steps leading to translational deficiencies.
Several pleiotropic mutants exhibit defects in maturation of rRNAs (49–52, 66–68). However, it is still unclear whether all these deficiencies are primary or secondary effects (69) as the molecular mechanisms of rRNA maturation are rather unclear and numerous proteins were assumed to be involved (7). For example, defects of the endonucleases CSP41a and b in processing of 23 S RNA (70) were recently proposed to represent secondary effects (71). In none of the previously described mutants, the 2.7 kb 23S-4.5 S transcript was detectable. This indicates a primary effect of RNE and RHON1 on rRNA processing.
Another intriguing observation is the high number of chloroplasts in rne and rhon1 mutants, accompanied by a smaller size when compared with wild type. This suggests a faster plastid multiplication rate caused by diminished repression of division (Figure 3F). This finding could not be observed in mutants with similar defects in chloroplast development or RNA metabolism, e.g. apo1 (29, 72), prfB3 (3) or other mutants generally affected in plastid gene expression (5) (unpublished data). E. coli degradosomes form cytoskeleton-like structures together with MinD, a repressor of cell division (73). In analogy to rhon1, minD mutants in E. coli divide more frequently resulting in minicells (74). Interestingly, MinD was shown to directly associate with bacterial RNE in a yeast-two-hybrid screen suggesting that the bacterial cytoskeleton is involved in compartmentalization of protein activities such as RNA processing and degradation (73). Accordingly, chloroplasts possess highly organized filamentous scaffolds composed of FtsZ named plastoskeleton that are most likely involved in plastid division (75). Similar to E. coli, positioning of the plastid-division apparatus requires the plastid-targeted form of MinD, AtMinD1 (76). A functional interaction between RNE and AtMinD1 is possibly conserved in the chloroplast, causing plastid division defects when RNE or RHON1 are lacking. Alternatively, defective processing of plastid transcripts functioning in chloroplast division could account for elevated chloroplast division rates. Candidates could be ycf1 and ycf2 genes with still unknown function. To our knowledge, RHON1 and RNE are the first proteins linking plastid RNA metabolism to chloroplast division.
The discovery of the essential role of RHON1 in RNA processing and the evolution of a plant-specific RNE-containing HMW complex lead to the conclusion that the plastid RNA processing/degradation machinery is more sophisticated than previously thought and that a degradosome-like complex is presumably existing in the chloroplast. We anticipate that future research on this complex, its compartmentalization and association partners will shed new light on chloroplast gene expression, division and development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–6 and Supplementary Reference [77].
FUNDING
The Binational Science Foundation [2009253 to G.S.]; the Binational Agricultural Research & Development Fund [4152-8C to G.S.]; Deutsche Forschungsgemeinschaft [SCHM1698/4 to C.S.L. and SFB-TR1 B2 to J.M.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Elli Gerick for excellent technical assistance. They thank Karin Meierhoff and Randy Dinkins for providing the TAP-vector and hcf2 seeds, respectively. They thank Kathrin Wittkowski for experimental help, Ben Luisi for help in the analysis of the plant RNE S1 domain structure and Dario Leister for reading of the manuscript.
REFERENCES
- 1.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu. Rev. Plant. Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 2.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoppel R, Lezhneva L, Schwenkert S, Torabi S, Felder S, Meierhoff K, Westhoff P, Meurer J. Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell. 2011;23:2680–2695. doi: 10.1105/tpc.111.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monde RA, Schuster G, Stern DB. Processing and degradation of chloroplast mRNA. Biochimie. 2000;82:573–582. doi: 10.1016/s0300-9084(00)00606-4. [DOI] [PubMed] [Google Scholar]
- 5.Cho WK, Geimer S, Meurer J. Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana. DNA Res. 2009;16:31–44. doi: 10.1093/dnares/dsn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 7.Stoppel R, Meurer J. The cutting crew - ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 2012;63:1663–1673. doi: 10.1093/jxb/err401. [DOI] [PubMed] [Google Scholar]
- 8.Bollenbach TJ, Schuster G, Portnoy V, Stern DB. Processing, degradation and polyadenylation of chloroplast transcripts. Topics Curr. Genet. 2008;19:175–211. [Google Scholar]
- 9.Bollenbach TJ, Schuster G, Stern DB. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas-Suarez M, Borner T, Barkan A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012;40:3092–3105. doi: 10.1093/nar/gkr1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 12.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 13.Mudd EA, Krisch HM, Higgins CF. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol. Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 14.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 15.Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, Cohen SN. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 2003;48:349–360. doi: 10.1046/j.1365-2958.2003.03435.x. [DOI] [PubMed] [Google Scholar]
- 17.Schein A, Sheffy-Levin S, Glaser F, Schuster G. The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA. 2008;14:1057–1068. doi: 10.1261/rna.907608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage RE, Shabanowitz J, Hunt DF, Schuster G, Gruissem W. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- 19.Rott R, Zipor G, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- 20.Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung WI, Day A. A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J. Exp. Bot. 2008;59:2597–2610. doi: 10.1093/jxb/ern126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter M, Piepenburg K, Schottler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 2010;64:851–863. doi: 10.1111/j.1365-313X.2010.04377.x. [DOI] [PubMed] [Google Scholar]
- 22.Meurer J, Lezhneva L, Amann K, Godel M, Bezhani S, Sherameti I, Oelmuller R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinkins RD, Bandaranayake H, Green BR, Griffiths AJ. A nuclear photosynthetic electron transport mutant of Arabidopsis thaliana with altered expression of the chloroplast petA gene. Curr. Genet. 1994;25:282–288. doi: 10.1007/BF00357174. [DOI] [PubMed] [Google Scholar]
- 24.Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- 25.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 26.Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 27.Dovzhenko A, Dal Bosco C, Meurer J, Koop HU. Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma. 2003;222:107–111. doi: 10.1007/s00709-003-0011-9. [DOI] [PubMed] [Google Scholar]
- 28.Meurer J, Meierhoff K, Westhoff P. Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta. 1996;198:385–396. doi: 10.1007/BF00620055. [DOI] [PubMed] [Google Scholar]
- 29.Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J. ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell. 2004;16:3084–3097. doi: 10.1105/tpc.104.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwenkert S, Umate P, Dal Bosco C, Volz S, Mlcochova L, Zoryan M, Eichacker LA, Ohad I, Herrmann RG, Meurer J. PsbI affects the stability, function, and phosphorylation patterns of photosystem II assemblies in tobacco. J. Biol. Chem. 2006;281:34227–34238. doi: 10.1074/jbc.M604888200. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casaregola S, Jacq A, Laoudj D, McGurk G, Margarson S, Tempete M, Norris V, Holland IB. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soderbom F, Svard SG, Kirsebom LA. RNase E cleavage in the 5′ leader of a tRNA precursor. J. Mol. Biol. 2005;352:22–27. doi: 10.1016/j.jmb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Jiang X, Diwa A, Belasco JG. Regions of RNase E important for 5′-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 2000;182:2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 39.Schult K, Meierhoff K, Paradies S, Toller T, Wolff P, Westhoff P. The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell. 2007;19:1329–1346. doi: 10.1105/tpc.106.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwenkert S, Netz DJ, Frazzon J, Pierik AJ, Bill E, Gross J, Lill R, Meurer J. Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 2010;425:207–214. doi: 10.1042/BJ20091290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meurer J, Grevelding C, Westhoff P, Reiss B. The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 1998;258:342–351. doi: 10.1007/s004380050740. [DOI] [PubMed] [Google Scholar]
- 42.Allison TJ, Wood TC, Briercheck DM, Rastinejad F, Richardson JP, Rule GS. Crystal structure of the RNA-binding domain from transcription termination factor rho. Nat. Struct. Biol. 1998;5:352–356. doi: 10.1038/nsb0598-352. [DOI] [PubMed] [Google Scholar]
- 43.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Tang WH, Hong MM, Wang ZY. OsBP-73, a rice gene, encodes a novel DNA-binding protein with a SAP-like domain and its genetic interference by double-stranded RNA inhibits rice growth. Plant Mol. Biol. 2003;52:579–590. doi: 10.1023/a:1024854101965. [DOI] [PubMed] [Google Scholar]
- 45.Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lezhneva L, Meurer J. The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 2004;38:740–753. doi: 10.1111/j.1365-313X.2004.02081.x. [DOI] [PubMed] [Google Scholar]
- 48.Coburn GA, Mackie GA. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 49.Bellaoui M, Keddie JS, Gruissem W. DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Mol. Biol. 2003;53:531–543. doi: 10.1023/B:PLAN.0000019061.79773.06. [DOI] [PubMed] [Google Scholar]
- 50.Bisanz C, Begot L, Carol P, Perez P, Bligny M, Pesey H, Gallois JL, Lerbs-Mache S, Mache R. The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 2003;51:651–663. doi: 10.1023/a:1022557825768. [DOI] [PubMed] [Google Scholar]
- 51.Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D. RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimura K, Ashida H, Ogawa T, Yokota A. A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J. 2010;63:766–777. doi: 10.1111/j.1365-313X.2010.04276.x. [DOI] [PubMed] [Google Scholar]
- 53.Stern DB, Gruissem W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- 54.Sharwood RE, Hotto AM, Bollenbach TJ, Stern DB. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA. 2011;17:230–243. doi: 10.1261/rna.2336611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 56.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stulke J. RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J. Bacteriol. 2011;193:5431–5441. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Condon C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010;7:316–321. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- 58.Jager S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, Klug G. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 2001;29:4581–4588. doi: 10.1093/nar/29.22.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaberdin VR, Miczak A, Jakobsen JS, Lin-Chao S, McDowall KJ, von Gabain A. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl Acad. Sci. USA. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mattiacio JL, Read LK. Evidence for a degradosome-like complex in the mitochondria of Trypanosoma brucei. FEBS Lett. 2009;583:2333–2338. doi: 10.1016/j.febslet.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daoud R, Forget L, Lang BF. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2012;40:1728–1736. doi: 10.1093/nar/gkr941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phinney BS, Thelen JJ. Proteomic characterization of a triton-insoluble fraction from chloroplasts defines a novel group of proteins associated with macromolecular structures. J. Proteome Res. 2005;4:497–506. doi: 10.1021/pr049791k. [DOI] [PubMed] [Google Scholar]
- 64.Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–189. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leaver CJ. Molecular integrity of chloroplast ribosomal ribonucleic acid. Biochem. J. 1973;135:237–240. doi: 10.1042/bj1350237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Y, Li C, Wang H, Chen H, Berg H, Xia Y. AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. Plant J. 2011;67:13–25. doi: 10.1111/j.1365-313X.2011.04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Komatsu T, Kawaide H, Saito C, Yamagami A, Shimada S, Nakazawa M, Matsui M, Nakano A, Tsujimoto M, Natsume M, et al. The chloroplast protein BPG2 functions in brassinosteroid-mediated post-transcriptional accumulation of chloroplast rRNA. Plant J. 2010;61:409–422. doi: 10.1111/j.1365-313X.2009.04077.x. [DOI] [PubMed] [Google Scholar]
- 68.Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beligni MV, Mayfield SP. Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism. Plant Mol. Biol. 2008;67:389–401. doi: 10.1007/s11103-008-9328-2. [DOI] [PubMed] [Google Scholar]
- 71.Qi Y, Armbruster U, Schmitz-Linneweber C, Delannoy E, de Longevialle AF, Ruhle T, Small I, Jahns P, Leister D. Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J. Exp. Bot. 2012;63:1251–1270. doi: 10.1093/jxb/err347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watkins KP, Rojas M, Friso G, van Wijk KJ, Meurer J, Barkan A. APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell. 2011;23:1082–1092. doi: 10.1105/tpc.111.084335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taghbalout A, Rothfield L. RNaseE and the other constituents of the RNA degradosome are components of the bacterial cytoskeleton. Proc. Natl. Acad. Sci. USA. 2007;104:1667–1672. doi: 10.1073/pnas.0610491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 75.Kiessling J, Kruse S, Rensing SA, Harter K, Decker EL, Reski R. Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J. Cell. Biol. 2000;151:945–950. doi: 10.1083/jcb.151.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 2000;10:507–516. doi: 10.1016/s0960-9822(00)00466-8. [DOI] [PubMed] [Google Scholar]
- 77.Meurer J, Berger A, Westhoff P. A nuclear mutant of Arabidopsis with impaired stability on distinct transcripts of the plastid psbB, psbD/C, ndhH, and ndhC operons. Plant Cell. 1996;8:1193–1207. doi: 10.1105/tpc.8.7.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.