Abstract
Mutations in the gene of human RNase T2 are associated with white matter disease of the human brain. Although brain abnormalities (bilateral temporal lobe cysts and multifocal white matter lesions) and clinical symptoms (psychomotor impairments, spasticity and epilepsy) are well characterized, the pathomechanism of RNase T2 deficiency remains unclear. RNase T2 is the only member of the Rh/T2/S family of acidic hydrolases in humans. In recent years, new functions such as tumor suppressing properties of RNase T2 have been reported that are independent of its catalytic activity. We determined the X-ray structure of human RNase T2 at 1.6 Å resolution. The α+β core fold shows high similarity to those of known T2 RNase structures from plants, while, in contrast, the external loop regions show distinct structural differences. The catalytic features of RNase T2 in presence of bivalent cations were analyzed and the structural consequences of known clinical mutations were investigated. Our data provide further insight into the function of human RNase T2 and may prove useful in understanding its mode of action independent of its enzymatic activity.
INTRODUCTION
RNase T2 is the only member of the Rh/T2/S family of acidic hydrolases in humans. We recently found that mutations in the RNase T2 gene are associated with white matter disease of human brain that manifests in early infancy (1). Affected patients are diagnosed with cystic leukoencephalopathy and show psychomotor retardation, spasticity and epilepsy. Comparable brain abnormalities were also described for a transgenic zebrafish model deficient in RNase T2 activity (2). Although the pathomechanism of RNase T2-deficient cystic leukoencephalopathy is still unclear, associated brain magnetic resonance imaging patterns of affected patients are very similar to those of children suffering from an intrauterine infection with cytomegalovirus (CMV). To counteract cellular antiviral mechanisms, CMV inactivates the endoribonuclease RNase L, which is regulated up on viral infections and stops viral protein synthesis by messenger RNA (mRNA) and ribosomal RNA (rRNA) degradation (3). RNase T2 therefore might play a similar role as RNase L in cellular immune response processes, and studies on parasitic worm eggs support this hypothesis: the RNase T2 homologous omega-1, secreted by eggs of the blood fluke Schistosoma mansoni, triggers a host T helper 2 (Th2) cell-polarized immune response followed by worm egg excretion to start new cycles of parasite infection (4,5).
T2 endoribonucleases can also be involved in phosphate scavenging processes. EcRNase 1 is present in the periplasm of Escherichia coli and recruits environmental RNA nucleotides (6). On phosphate starvation in tomato plants, ribonuclease LE is expressed at a higher rate and extracellular ribonuclease activity is increased (7,8). RNS2, a T2 endoribonuclease from Arabidopsis thaliana, is important for rRNA decay in plant vacuoles and endoplasmic reticulum-derived compartments (9).
RNase T2 proteins can modulate cellular functions independently of their catalytic function. Actibind, a T2 ribonuclease from fungus Aspergillus niger, showed an anticlonogenic, tumor suppressing effect even when its catalytic activity was inactivated (10,11). There is also evidence that human RNase T2 acts as a tumor suppressor. In ovarian cancer cell line Hey3Met2, RNase T2 showed in vivo tumor suppressor activity, and two point mutations that eliminate the catalytic activity of RNase T2 showed no effect on its anti-metastatic properties in Hey3Met2 cells in nude mice experiments (12). Control of ovarian tumor genesis by RNase T2 thereby occurs through modification of the cellular microenvironment and induction of immunocompetent cells of the monocyte/macrophage lineage (13).
Interestingly, while a broad range of biological functions has been reported for T2 ribonucleases, the known three-dimensional structures of these enzymes from bacteria, fungi and plants are highly conserved with a typical α+β core structure (14). The protein core consists of a 4–8 strand β-sheet and the exterior of the protein’s tertiary structure is formed by helices.
To our knowledge, no T2 ribonuclease structures from organisms higher than plants have been determined so far. We therefore conducted a structural analysis of human RNase T2 to reveal potential conformational features that differ from those of known structures and may help to explain RNase T2 functions in higher mammals and consequently its role in human neurological diseases.
Herein, we report the crystal structure of human RNase T2 at a resolution of 1.6 Å and compare its features to those of plant homologs; inhibition of RNase T2 by bivalent cations as well as functional consequences of clinical known mutations are also discussed.
MATERIALS AND METHODS
Cell culture medium was purchased from Invitrogen (New York, USA). Oligonucleotides for the introduction of the C-terminal tag were synthesized by MWG-Biotech (Munich, Germany). All reagents were purchased from Sigma-Aldrich (Seelze, Germany), unless otherwise indicated.
Expression of RNase T2
The amino acid sequence ‘HHHHH’ was introduced at the C terminus of the RNase T2 to facilitate purification. Transfection of the modified RNase T2 complementary DNA (cDNA) and selection of HEK 293 cells were performed as described previously (1,15). To produce RNase T2 with predominantly mannose-type glycosylation sites by α-mannosidase inhibition for later EndoH treatment, cells were grown with kifunensine (Biomol, Hamburg, Germany) (1 mg/l medium). RNase T2 was purified from the cell culture supernatant (100 ml total volume). The medium was cleared by centrifugation at 3000g and 4°C for 60 min and then filtered with a 0.2 -µm pore membrane. After adding 20 mM K2HPO4, pH 7.5, 0.5 M NaCl and 40 mM imidazole, the crude solution was loaded on a HisTrap HP column (GE Healthcare, Munich, Germany). Bound RNase T2 was eluted with an imidazole gradient and was typically released at 70–90 mM imidazole. For deglycosylation, the purified RNase T2 fractions were pooled and concentrated using a centrifugal filter (Millipore, Schwalbach, Germany).
Deglycosylation of RNase T2 and gel filtration
For cleavage of high mannose oligosaccharides in the N-linked glycoprotein, 88 μl of concentrated protein solution (10 µg/µl) from HisTrap were digested with 2 μl EndoH (New England Biolabs, Frankfurt, Germany) (C: 1000 u/ml) in 10 μl NEB G5 Buffer for 4 h at 37°C leaving one N-acetyl glucosamine residue at each of the N-glycosylation sites.
To remove remaining glycosylated protein after digestion in the sample, affinity chromatography with a GE Healthcare 1 ml ConA column (flow rate 0.1 ml/min, Buffer A: 20 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 1 mM MnCl2 and 1 mM CaCl2, Buffer B: 0.5 M Methyl-α-d-glucopyranoside, 20 mM Tris–HCl, pH 7.5 and 0.5 M NaCl) was applied. Again, the protein was concentrated to a volume of 700 µl. Finally, the protein was gel filtrated using a SuperDex 75 column (flow rate 0.2 ml/min, buffer: 200 mM acetate, pH 6.0 and 50 mM NaCl, injection volume 100 µl) and could be eluted as a single peak. It was concentrated as before and rebuffered in 20 mM acetate pH 6.0 and 20 mM NaCl. The final concentration was 9.2 mg/ml.
Mass spectrometry
Both glycosylated and deglycosylated RNase T2 were reduced with 50 mM dithiothreitol, alkylated with 100 mM 2-chloroacetamide incubated with trypsin (Promega GmbH, Mannheim, Germany) at an enzyme-to-substrate ratio of 1:20 overnight at 37°C and analyzed by electrospray ionization liquid chromatography tandem mass spectrometry (LC-MS/MS). The LC-MS/MS analysis was done on an LTQ Orbitrap Velos (Thermo Fisher Scientific, Germany). An extensive description of mass spectrometry analysis methods and results is provided in the Supplementary Material.
Crystallization
Hanging drop crystallization was set up on VDXm pre-greased plates (Hampton Research, Aliso Viejo, USA) (0.6 ml reservoir) with MD CrystalClene cover slips holding a 2 μl drop of a 1:1 reservoir/protein solution. A block-shaped crystal (diameter 50 μm) was grown from microcrystalline and viscous precipitate that had formed in a condition of 0.2 M NH4NO3 (p.a. Merck, Darmstadt, Germany), 15% polyethylene glycol 3350 (Hampton Research) and water after 9 weeks. For cryoprotection, the crystals were soaked in a mixture of glucose (99% purity, Merck)/reservoir solution 1:2 (wt/vol). They were mounted on MiTeGen MicroMounts and flash cooled by plunging into liquid nitrogen.
Data collection and processing
Datasets were measured at BESSY MX 14.1 beam line with a Rayonix MX-225 3 × 3 CCD detector. Data collection statistics for three native datasets are summarized in Table 1. Statistics for an additional long-wavelength dataset can be found in the Supplementary Material. The crystals belonged to the monoclinic space group P21, a typical space group for T2 RNases (16). The data were integrated with XDS (17), converted with XDS2SAD for absorption correction with SADABS (18). XPREP (19) was used for merging and data quality analysis.
Table 1.
Summary of the collection statistics
| Source | BESSY MX 14.1 | ||
|---|---|---|---|
| Unit cell dimensions | a = 31.322, b = 68.092, c = 47.984, β = 90.827° | ||
| Space group | P21 | ||
| Wavelength (Å) | 0.9184 | 0.9184 | 0.9810 |
| Oscillation range | 95° | 103.5° | 237° |
| Resolution range (Å) | 27.56–1.73 (1.83–1.73) | 28.31–1.74 (1.84–1.74) | 28.31–1.59 (1.69–1.59) |
| Number of observations | 40 334 (5303) | 43 694 (6281) | 75 521 (11 987) |
| Uniquea | 20 100 (2714) | 20 030 (2926) | 26 238 (4246) |
| Redundancya | 1.96 (1.73) | 2.13 (1.94) | 2.80 (2.55) |
| Completenessa (%) | 97.5 (88.6) | 97.7 (90.5) | 97.2 (90.3) |
| Mean I/σ (I) | 17.00 (4.11) | 20.47 (6.48) | 16.74 (5.04) |
| Rint (%) | 3.06 (19.54) | 2.63 (13.59) | 3.07 (19.10) |
| Rrim (%) | 4.15 (26.80) | 3.45 (17.83) | 3.75 (23.38) |
| Rpim (%) | 2.79 (18.25) | 2.21 (11.44) | 2.11 (13.31) |
The three datasets were merged for structure solution and refinement. Values in parentheses refer to outer shell.
aFriedel pairs merged.
Structure solution and refinement
For structure solution, the unmodified protein molecule of RNase LE (20) was used as search model. The correct PHASER (21) solution was identified and improved by density modification and subsequent poly-Ala tracing with SHELXE (22). In a SHELXE ‘free lunch’ map extended to 1.0 Å resolution, the initial model with 178 residues was built from the poly-Ala backbone using COOT (23). The structure was refined with REFMAC (24).
After checking with the TLS-MD server (25), TLS refinement (26) was applied, with only one domain consisting of the whole enzyme. Checks with WHATIF (27) as well as with MOLPROBITY (28) lead to a significant improvement of the model.
The REFMAC weighting scheme was optimized. Refinement statistics are given in Table 2. The sulfur positions were confirmed using the anomalous dataset and the program ANODE (29). Using the SSM web service (30), no significant intermolecular contact surface could be found, so the molecules appear to be biological monomers. Topological analysis of the protein was performed with PDBsum (31).
Table 2.
Summary of the refinement statistics
| Refinement statistics | |
|---|---|
| Resolution range | 27.56–1.59 |
| Number of reflections in working set | 25 064 |
| Completeness of the working set | 98.00 |
| Number of reflections in test set | 1347 |
| Solvent content (%) | 43.99 |
| Number of protein atoms | 1642 |
| Number of water molecules | 182 |
| Protein molecules per ASU | 1 |
| R (%) | 15.38 |
| Rfree (%) | 19.03 |
| Average B factors (Å2) | |
| Overall | 18.89 |
| Protein atoms | 18.27 |
| Water molecules | 21.93 |
| Observed r.m.s.d. from ideal geometrya | |
| Bond lengths (Å) | 0.021 |
| Bond angles (°) | 1.057 |
| Ramachandran plot, residues inb | |
| Favored regions | 97.37 |
| Allowed regions | 2.63 |
Values in parentheses refer to the outer shell.
aREFMAC output.
bCalculated with MOLPROBITY.
Zinc binding site calculation
Potential zinc binding sites were recognized by the FEATURE metal scanning server (32) using default options; hits above a Z-score threshold of 20.0 were assessed.
Computer graphics
Figures of human RNase T2 were produced with the program PyMOL (PYMOL molecule rendering; DeLano Scientific, Palo Alto, USA) (33).
RNase activity assay
RNase activity assays were performed as previously described (1). Enzymatic activity toward yeast RNA was measured in terms of the increase in absorbance at 260 nm due to soluble nucleotide production after digestion at 37°C of substrate at pH 4.6 or 6.0, respectively. Briefly, 1 µl of a stock solution containing 4.5 μg/µl of RNase T2 was added on ice to a reaction mix containing RNA at a concentration of 4 mg/ml buffer (0.2 M sodium acetate and 0.05 M NaCl, pH 4.6 or 6.0) in a total volume of 400 μl. Blank tubes were prepared in the same way, except that the RNase T2 was replaced by buffer solution. As a control, half of the reaction mixture was precipitated immediately. The remaining 200 μl of the reaction mixture was incubated for 30 min at 37°C and then 50 μl of stop mixture (0.75% (wt/vol) uranyl acetate in 25% (vol/vol) perchloric acid) was added. The reaction mixture was cooled on ice for 15 min and centrifuged at 14 000g for 15 min at 4°C to remove precipitated RNA. Fifty microliters of the supernatant fluid containing the free nucleotides were diluted 40-fold with distilled water, and absorbance determined at 260 nm. To analyze the effect of bivalent metal ions on RNase T2 activity, stock solutions of RNase T2 containing 2.25 µg/µl of enzyme and 5 or 50 mM of Zn2+, Cu2+, Co2+, Ni2+ or Mg2+ were prepared. After 2 h of incubation at 37°C, 2 µl of stock solution was applied in activity assays as described above, the activity was measured in comparison to equal amounts of untreated enzyme. To examine the effect of ethylenediaminetetraacetic acid (EDTA), 20 µl of the RNase T2 solution pretreated with 50 mM Zn2+ where mixed with 20 µl of 100 mM ETDA, 0.2 M sodium acetate and 0.05 M NaCl at pH 6.0. After incubation for 30 min at 37°C, 4 µl of this mixture was measured for activity as described above in comparison to equal amounts of untreated enzyme.
RESULTS
This is the first structure determination to be reported for human RNase T2.
Quality of the structure
The structure of human RNase T2 has been determined by X-ray diffraction from crystals in the monoclinic space group P21 at a resolution of 1.6 Å. Data collection and refinement statistics are shown in Tables 1 and 2. The structural refinement of human RNase T2 converged to an Rfree of 19.03% and an R-value of 15.39%. The overall geometry shows no abnormalities, with an all-atom clash score of 5.97 and 0.6% bad rotamers as given by MOLPROBITY (23). The structure had no outliers and 97.4% of all residues were in the favored regions of the Ramachandran plot.
Overall structure and fold
Human RNase T2 crystallizes, like almost all other T2 RNases, with one monomer in the asymmetric unit, which also comprises the biological active unit.
The structure shows a typical fold for members of the T2 RNase family with seven α-helices and eight β-strands constituting an α+β motif (34), as most of the central anti-parallel β-sheet is clearly separated in the sequence from the helical parts (Figures 1 and 2). The central β-sheet consists of six strands (β1–2, β4–7) and is flanked by a large helical region containing α-helices α1–4. The 15-residue long α-helix 5 runs across the central β-sheet and is joined by a three amino acid linker with α-helix 6; α7 flanks the central β-sheet opposite the region containing helices α1–α4. The two β-strands β3 and β8 form a small sheet at the bottom of the protein. Most of the annotated catalytic residues are located in a distorted helix region consisting of residues 110–121 (α3, α4). The catalytic site was annotated by comparison with homologous structures and comprises residues His 65, His 113, Glu 114, Lys 117 and His 118 (see ‘Discussion’ section).
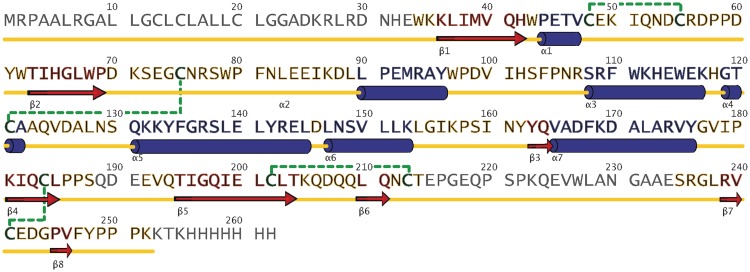
Figure 1.
The sequence of human RNase T2. The first 24 residues correspond to a signal peptide, which is cleaved. β-strands are marked with red arrows and α-helices with dark blue barrels. Gray residues are not visible in the electron density; the other colors refer to Figure 2.
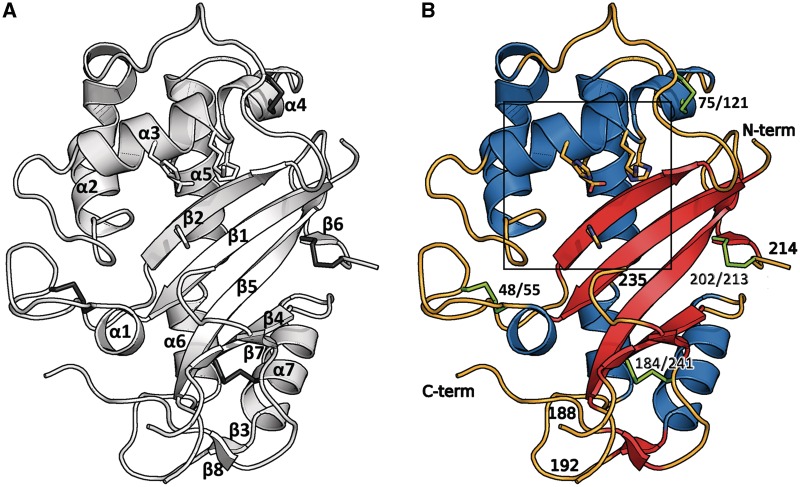
Figure 2.
The structure shows a typical α+β core fold. The catalytic residues are shown as sticks. (A) Topology of human RNase T2: the structure shows the typical fold of this family with seven α-helices and eight β-strands compromising two anti-parallel sheets. (B) Residues 188–192 and 214–235 and the terminal ends are disordered and could not be resolved in the electron density. Disulfide bridges are shown in green. The box refers to Figure 6.
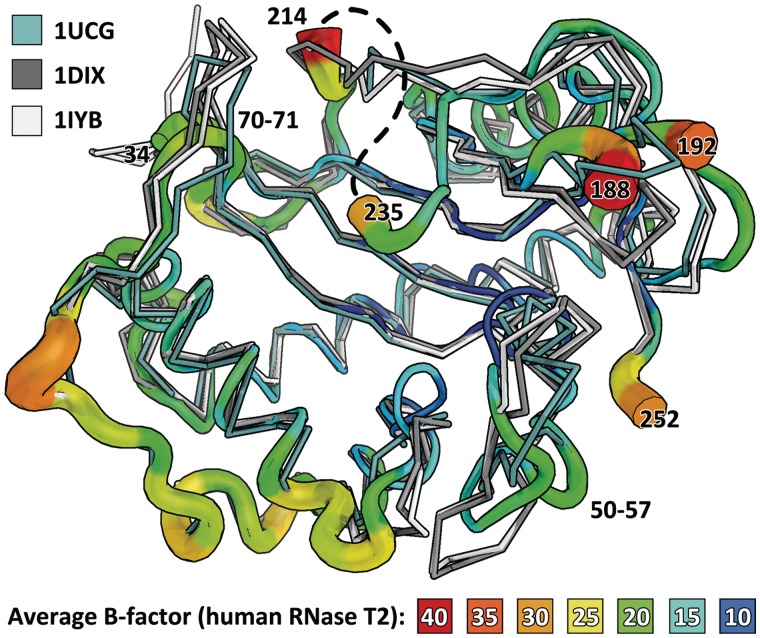
Human RNase T2 has four disulfide bridges, connecting cysteine residues 48/55, 75/121, 184/241 and 202/213, which can also be found in the nearest structural relatives (Table 3). No model could be built for residues 188–192 and 214–235 as well as the terminal ends as they were disordered.
Table 3.
Structural alignment statistics, as given by SSM Tool (30)
| Name | PDB | Residues | Sequence identity (%) | r.m.s.d. (Å) | Color as in Figure 5 |
|---|---|---|---|---|---|
| RNase LE | 1DIX | 208 | 33 | 1.50 | Gray |
| RNase MC1a | 1UCG | 190 | 28 | 1.61 | Pale cyan |
| RNase MC1 | 1BK7 | 190 | 29 | 1.59 | Not shown |
| RNase NW, chain B | 1IYB | 208 | 31 | 1.66 | Light gray |
aFor the figure, the mutant N71T was chosen as the structure shows a slightly lower r.m.s.d. with our protein than the wild type. All r.m.s.d. values are in comparison to RNase T2, but only for the compared sequence part.
Mass spectrometry of the wild-type protein reveals three glycosylation sites at asparagine residues 76, 106 and 212. Two bound N-acetyl glucosamine residues left over from the EndoH deglycosylation are clearly visible in the electron density at Asn 76 and Asn 212. Asn 106 does not have corresponding electron density in its vicinity, which might be because of disorder or carbohydrate cleavage.
In silico analysis of potential zinc binding sites
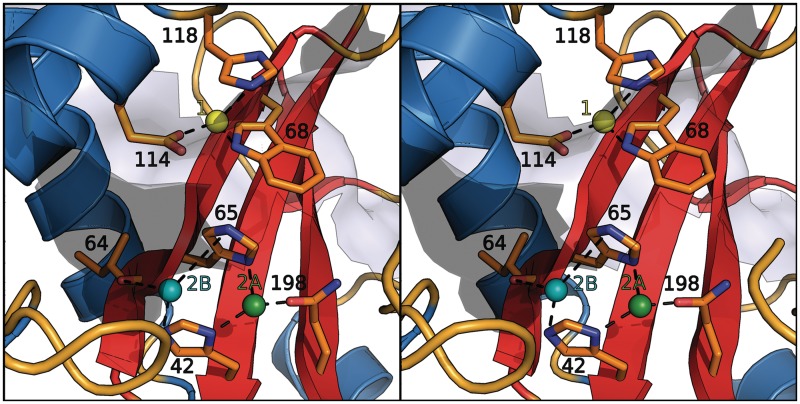
Furthermore, investigation of the structure revealed a well-established water position directly behind the catalytic site between residues His 65, Tyr 42 and Gln 198 that showed some properties of a zinc binding site. For RNase T2 from Aspergillus oryzae, zinc and copper ions were reported to be potent inhibitors of catalytic hydrolase activity (35,36). We therefore performed an in silico prediction of potential zinc binding sites in human RNase T2 and analyzed the effect of bivalent metal ions on RNase T2 catalysis in activity assays. A number of potential zinc binding positions were predicted with the FEATURE (32) web server. The Z-score threshold of the potential zinc binding sites was between 20.0 and 27.7 and clustered in three sites (Figure 3):
-
(1)
eight hits between the residues Trp 68, His 118 and Glu 114;
-
(2A)
four hits between the residues His 65, Glu 198 and His 42, forming a 2-histidine-1-carboxylate motif;
-
(2B)
two hits between His 42, His 65 and the carbonyl group of Ile 64.
Figure 3.
The potential zinc binding sites predicted by the FEATURE web server (32). Site 1 is colored in yellow, 2A in green and 2B in cyan; the involved residues are labeled. While the dashed lines refer to potential bonds, the residues will most likely change conformation to accommodate the metal coordination.
Site 1 corresponds approximately to the binding site of phosphate in the structure of Actibind. Leeuw et al. (37) also suggested that this site might be important for the catalyzed reaction.
RNase T2 activity is inhibited by zinc and copper ions but is independent of glycosylation
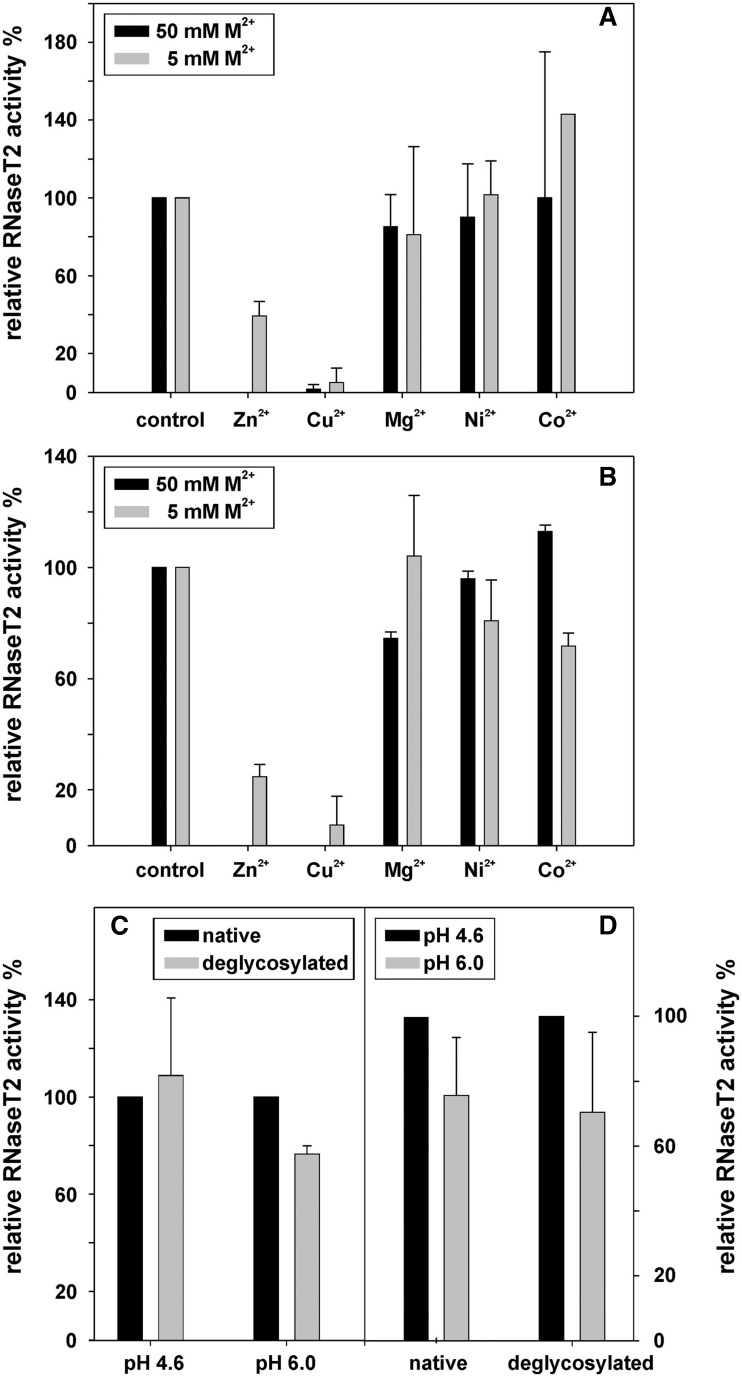
Catalytic activity of soluble RNase T2 from Aspergillus oryzae has been reported to be reduced by zinc and copper ions (35,36). For human RNase T2 at pH 6.0, we find that the residual activity in the presence of 5 mM zinc sulfate is only 40% of that of the untreated protein (Figure 4A); with Cu2+ ions, the activity was even reduced to 6%. No significant inhibitory effects were measured with equimolar solutions of nickel, cobalt or calcium. If treated with 50 mM bivalent metal ions, no residual enzymatic activity was detectable for zinc and copper, while for cobalt, nickel and magnesium no pronounced reduction in activity was measured. The inhibitory effects of zinc were reversible and could be eliminated by incubation with 100 mM EDTA at 37°C for 30 min (this was not measured for copper).
Figure 4.
The activity of RNase T2 was measured under different conditions. All values are means of two independent experiments and uncertainty ranges represent standard deviations. (A) At pH 6.0, the effects of several bivalent cations on RNase T2 activity were determined at 5 and 50 mM ion concentration. The activity bars are normalized relative to an untreated control. (B) The effects of several bivalent cations on RNase T2 activity were determined at 5 and 50 mM ion concentrations at pH 4.6. At this pH, RNase T2 showed an activity around 4.5 U/µg, with one unit defined as the amount of enzyme that produces a change of 1 OD260 in 30 min at 37°C. All activities are normalized relative to an untreated control. (C) The activity of native RNase T2 in comparison to that of deglycosylated RNase T2 at pH 4.6 and 6.0. The activity of the native protein was normalized as 100%. (D) The pH dependency of enzymatic activity was measured for native and for deglycosylated RNase T2, with the activity at pH 4.6 normalized to 100%.
Since T2 RNases were reported to show their highest activity under more acidic conditions, these experiments were repeated at pH 4.6. Indeed, at pH 4.6, the enzymatic activity was 23% higher than at pH 6.0 for the native protein (Figure 4D). Although no significant inhibitory effect of cobalt, nickel or magnesium was detectable at 50 mM concentration, equimolar amounts of zinc and copper eliminated enzymatic activity completely (Figure 4B). The inhibitory effects of zinc and copper at a concentration of 5 mM were found to be similar as to those at pH 6.0.
As RNase T2 was crystallized after deglycosylation with EndoH, the influence of glycosylation on the enzymatic activity was tested. However, no significant differences could be observed when the in vitro activity of untreated RNase T2 was compared with that of EndoH digested protein (Figure 4C).
Comparison with other structures and main-chain displacement
Structural alignment with PDBeFold (SSM) (30) was used to identify structural relatives of RNase T2 in the PDB. Six structures related to human RNase T2 derived from three different RNases were investigated: RNase LE from tomato Solanum lycopersicum (20), Ribonuclease MC from bitter gourd Momordica charantia (38,39) and Ribonuclease NW from Nicotiana glutinosa (40). A secondary structure matching superposition of all three proteins with human RNase T2 revealed that the core fold is strictly conserved, while the outer loops show conformational differences. Loop 50-57 bends inwards when compared with the other T2 RNases. Human RNase T2 has a beta turn at residues 69–72, while the structural relatives contain a more extended loop at this position.
In the structures RNase LE, NW and MC, as well as in the recently published structure of Actibind (PDB 3D3Z (37)) and even in the more distant structural relative RNase 1 from E. coli (PDB code 2Z70 (6)), the β-sheet 4 is overlaid by a loop region. In human RNase T2, there seems to be a longer, partly disordered (residues 215–234) loop with a different conformation. The primary sequences of the structural relatives differ significantly after cysteine 213 and only residues 240–251 align to the C-terminal region of the other proteins. The displacement of the protein main chain is illustrated as B factor putty in Figure 5; the width and color of the ribbon correspond to the average B factor of the residues. The structural alignment is shown in this figure and summarized in Table 3.
Figure 5.
Overlay of the B factor putty representation of human RNase T2 and three other members of its family: RNase MC1 (1UCG) in pale cyan, RNase Le (1DIX) in gray and RNase NW (1IYB) in light gray. The average displacement, given by the putty, is tightly related to the differences between the structures, suggesting that the rigid part mainly conveys the catalytic function while outer loops adapt. The structures from plant T2 RNases have a prolonged loop between residues 70 and 71. Also, the disordered loop 214–235 (approximate position is painted as a dashed line) seems not to have an equivalent in the other structures, and possibly involves an insertion. Loop 50-57 of RNase T2 bends inwards when compared with the other T2 RNases. Apart from these differences in the outer regions, the core fold is highly conserved and rigid, as can be seen from its low average B factor.
DISCUSSION
Catalysis
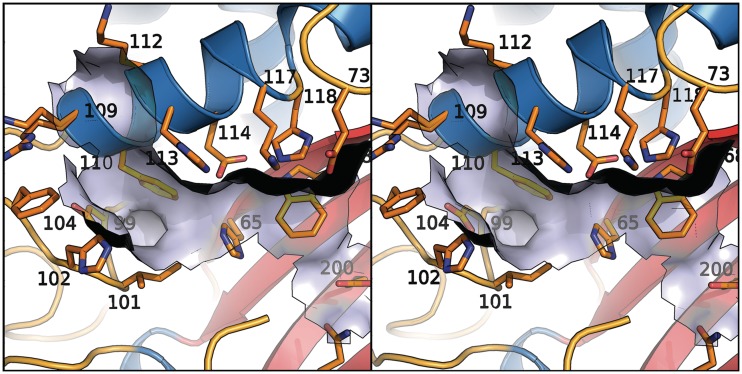
RNA cleavage by T2 RNases is mediated by histidine residues in the conserved active site motifs CASI and CASII (14). Mutations in these histidine residues lead to inactivation of the enzyme both in vivo and in vitro (16), and zinc has been shown to inhibit the cleavage in vitro. Substrate binding involves hydrophobic interactions and hydrogen bonding to the phosphate group (16). The substrate binding sites have been identified by co-crystallization with mono- and di-nucleotides (20,41). In human RNase T2, CASI spans residues 62–69, the aligned CASII spans residues 110–121. The latter motif almost forms a perfect α-helix, but the carbonyl groups of residues 116 and 117 do not make proper hydrogen bonds and His 118 has slightly deviant phi and psi angles. This results in a distortion in the helix, possibly allowing for a higher energy state and better mobility of the catalytic residues 117 and 118. Due to the highly conserved geometrical catalytic environment of RNase T2, the enzymatic reaction mechanism is proposed to be analogous to the one postulated by Kurihara et al. (42). A comparison with the active site of RNase LE (20) as well as the reaction mechanism is given in Supplementary Figures S1 and S2. The RNA binding surface with relevant residues is modeled in Figure 6.
Figure 6.
Surface of the assumed RNA binding pocket shown with numbered residues. Residue 109 is disordered.
Role of zinc
On visual inspection, we predicted a zinc binding site for RNase T2 in the region of His 65 which was supported by in silico analysis of RNase T2 with the FEATURE web server. The program also found a potential zinc binding region in the catalytic cleft of RNase T2, though the enzyme does not require divalent ions as cofactor. Using RNase activity assays, we showed that zinc and also copper inhibit human RNase T2 even at 5 mM concentration. A comparable inhibition by the same metal ions was already reported for RNase T2 from fungus Aspergillus oryzae. Despite this, it is uncertain how many zinc or copper ions are involved and whether they bind to distinct regions of human RNase T2. Furthermore, crystallization experiments in the presence of zinc or copper would be needed to clarify this question, but each of the predicted zinc binding regions would readily explain the observed reversible inhibition of the enzyme on treatment with EDTA. It remains unclear whether zinc binding is of physiological relevance. Coordination in Site 2B might also affect the main-chain conformation and hence, influence the shape of the protein surface with potential consequences for protein–protein interactions.
Clinical relevant mutations
A number of inherited mutations in the gene encoding for RNase T2 are known to be associated with white matter disease of human brain (1). Analyzing the role of these mutations using the X-ray structure of RNase T2, almost each one has severe consequences for the protein’s conformation (Table 4). The point mutation Cys184Arg destroys the disulfide bridge Cys 184–Cys 241 which might result in destabilization of the loop in which Cys 241 sits and therefore the lower part of the major β-sheet. The remaining unlinked SH group of Cys 241 might promote precipitation and instability. Thus, our structural data underline the disease causing effect of the Cys184Arg mutation of the affected patient.
Table 4.
Known mutations in RNase T2 with clinical relevance. (1)
| Mutation type | Affected amino acids | Comment |
|---|---|---|
| Missense | Cys184Arg | Disulfide bridge 184–241 missing, destabilizing and sensitizing for oxidation |
| Deletion | Met 1–Glu 92 | Secretion signal peptide deleted; deletion of central part of α+β motif containing β1, β2, α1 and α2 |
| Deletion | Ala 17–Leu 21 | Secretion signal peptide deleted |
| Deletion | Asp 30–Trp 86 | Deletion of central part of α+β motif containing β1, β2, α1 and α2 |
| Deletion | Asp 30–Trp 111 | Deletion of central part of α+β motif containing β1, β2, α1 and α2 |
| Deletion | Pro 69–Trp 111 | Deletion of central part of α+β motif containing β2 and α2 |
| Deletion | Val 165–Gln 189 | Deletion of central part of α+β motif containing β4 and α7 |
| Stop | Asp 88–XX–Stop | Rest of protein is not produced |
| Stop | Asp 88–XXX–Stop | Rest of protein is not produced |
The other known patient mutations within the RNase T2 gene influence mRNA splicing and result in a variety of large internal deletions that all affect the α+β core motif.
Structural differences to other members of the T2 family/TRAF2
Comparison between human RNase T2 and plant T2 RNases shows that the α+β core motif is highly conserved with no pronounced deviations between the main-chain atom positions. However, conformational differences were found in parts of the outer loops. Some of these regions could be important for protein–protein interactions of RNase T2. It was recently shown that RNase T2 co-localizes with processing (P-) bodies which increase upon metabolic stress and that cells lacking RNase T2 show a reduced number of P-bodies (43). It has still to be seen whether or which outer loop regions of RNase T2 are involved in P-body formation. Minor main-chain deviations in surface regions could be due to crystallization conditions, crystal packing or similar effects, but the region between Cys213 and Val240 clearly differs between human RNase T2 and the compared structures. Since no model could be built for that part of the peptide chain, this region is assumed to comprise a highly disordered loop. This loop contains 21 or 22 amino acid residues more than the corresponding ordered regions in RNase LE, Ribonuclease MC and Ribonuclease NW. Analysis of the RNase T2 amino acid sequence between Cys213 and Val240 using the ELM tool for the identification of eukaryotic linear motifs for functional sites in proteins in silico (44) revealed a potential tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2) binding motif, PKQE, at Positions 222–225. TRAF2 plays an important role in cellular signaling for anti-inflammatory and apoptotic response (45). TRAF2 binds as an associated factor to the cytosolic domain of TNFα receptor and modulates TNFα action on the MAP kinase and NFκB pathway.
There is evidence that human RNase T2 acts as a tumor suppressor and might be involved in immune response processes (4,5,10–13). In contrast to RNase A homolog Onconase from frog Rana pipiens, whose antitumoral activity depends on its ribonucleolytic properties, the tumor suppressive features of RNase T2 seem not to be linked to its catalytic function (46). A TRAF2 binding site—particularly in a protein region that clearly differs from that of the related plant enzymes—could be a starting point to explain further functions of RNase T2 independent of its enzymatic activity. However, further experiments are needed to verify the predicted TRAF2 site and to evaluate its physiological relevance.
ACCESSION NUMBERS
PDB-ID: 3T0O.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1 and 2 and Supplementary Methods and Results.
FUNDING
German Bundesministerium für Bildung und Forschung (BMBF) [LEUKONET]. Funding for open access charge: BMBF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christian Grosse for his support in crystallization and data collection.
REFERENCES
- 1.Henneke M, Diekmann S, Ohlenbusch A, Kaiser J, Engelbrecht V, Kohlschutter A, Krätzner R, Madruga-Garrido M, Mayer M, Opitz L, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat. Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- 2.Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gärtner J, Alia A, et al. RnaseT2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for Rnase T2 in degrading ribosomal RNA. Proc. Natl Acad. Sci. USA. 2011;108:1099–1103. doi: 10.1073/pnas.1009811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Child SJ, Jarrahian S, Harper VM, Geballe AP. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J. Virol. 2002;76:4912–4918. doi: 10.1128/JVI.76.10.4912-4918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everts B, Perona-Wright G, Smits HH, Hokke CH, van der Ham AJ, Fitzsimmons CM, Doenhoff MJ, van der Bosch J, Mohrs K, Haas H, et al. Omega-1, a glycoprotein secreted by Schistosoma mansoni eggs, drives Th2 responses. J. Exp. Med. 2009;206:1673–1680. doi: 10.1084/jem.20082460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinfelder S, Andersen JF, Cannons JL, Feng CG, Joshi M, Dwyer D, Caspar P, Schwartzberg PL, Sher A, Jankovic D. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1) J. Exp. Med. 2009;206:1681–1690. doi: 10.1084/jem.20082462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez SM, Panjikar S, Van Belle K, Wyns L, Messens J, Loris R. Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli. Protein Sci. 2008;17:681–690. doi: 10.1110/ps.073420708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loffler A, Abel S, Jost W, Beintema JJ, Glund K. Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiol. 1992;98:1472–1478. doi: 10.1104/pp.98.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurnberger T, Abel S, Jost W, Glund K. Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol. 1990;92:970–976. doi: 10.1104/pp.92.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl Acad. Sci. USA. 2011;108:1093–1098. doi: 10.1073/pnas.1009809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roiz L, Smirnoff P, Bar-Eli M, Schwartz B, Shoseyov O. ACTIBIND, an actin-binding fungal T2-RNase with antiangiogenic and anticarcinogenic characteristics. Cancer. 2006;106:2295–2308. doi: 10.1002/cncr.21878. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz B, Shoseyov O, Melnikova VO, McCarty M, Leslie M, Roiz L, Smirnoff P, Hu GF, Lev D, Bar-Eli M. ACTIBIND, a T2 RNase, competes with angiogenin and inhibits human melanoma growth, angiogenesis, and metastasis. Cancer Res. 2007;67:5258–5266. doi: 10.1158/0008-5472.CAN-07-0129. [DOI] [PubMed] [Google Scholar]
- 12.Acquati F, Possati L, Ferrante L, Campomenosi P, Talevi S, Bardelli S, Margiotta C, Russo A, Bortoletto E, Rocchetti R, et al. Tumor and metastasis suppression by the human RNASET2 gene. Int. J. Oncol. 2005;26:1159–1168. [PubMed] [Google Scholar]
- 13.Acquati F, Monti L, Lualdi M, Fabbri M, Sacco MG, Gribaldo L, Taramelli R. Molecular signature induced by RNASET2, a tumor antagonizing gene, in ovarian cancer cells. Oncotarget. 2011;2:477–484. doi: 10.18632/oncotarget.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luhtala N, Parker R. T2 Family ribonucleases: ancient enzymes with diverse roles. Trends Biochem. Sci. 2010;35:253–259. doi: 10.1016/j.tibs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal A, Kraetzner R, Gruene T, Grapp M, Schreiber K, Gronborg M, Urlaub H, Becker S, Asif AR, Gärtner J, et al. Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis. J. Biol. Chem. 2009;284:3976–3984. doi: 10.1074/jbc.M806947200. [DOI] [PubMed] [Google Scholar]
- 16.Deshpande RA, Shankar V. Ribonucleases from T2 family. Crit. Rev. Microbiol. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheldrick GM. Madison, WI: Bruker; 2009. SADABS—Bruker AXS area detector scaling and absorption correction, version. 2009/2001. [Google Scholar]
- 19.Sheldrick GM. Madison, WI: Bruker; 2011. XPREP—Bruker data preparation and reciprocal space exploration, version 2011/2011. [Google Scholar]
- 20.Tanaka N, Arai J, Inokuchi N, Koyama T, Ohgi K, Irie M, Nakamura KT. Crystal structure of a plant ribonuclease, RNase LE. J. Mol. Biol. 2000;298:859–873. doi: 10.1006/jmbi.2000.3707. [DOI] [PubMed] [Google Scholar]
- 21.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheldrick GM. Experimental phasing with SHELXC/D/E:combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 26.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G. Homology modelling, model and software evaluation: three related resources. Bioinformatics. 1998;14:523–528. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 28.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta. Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorn A, Sheldrick GM. ANODE: ANOmalous and heavy-atom DEnsity calculation. J. Appl. Crystallogr. 2011;44:1285–1287. doi: 10.1107/S0021889811041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert JC, Altman RB. Robust recognition of zinc binding sites in proteins. Protein Sci. 2008;17:54–65. doi: 10.1110/ps.073138508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mura C, McCrimmon CM, Vertrees J, Sawaya MR. An introduction to biomolecular graphics. PLoS Comput. Biol. 2010;6:e1000918. doi: 10.1371/journal.pcbi.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levitt M, Chothia C. Structural patterns in globular proteins. Nature. 1976;261:552–558. doi: 10.1038/261552a0. [DOI] [PubMed] [Google Scholar]
- 35.Reddy LG, Shankar V. Preparation and properties of RNase T2 immobilized on concanavalin A-sepharose. Appl. Biochem. Biotechnol. 1989;22:237–246. doi: 10.1007/BF02921759. [DOI] [PubMed] [Google Scholar]
- 36.Uchida T. Purification and properties of RNase T2. J. Biochem. 1966;60:115–132. doi: 10.1093/oxfordjournals.jbchem.a128410. [DOI] [PubMed] [Google Scholar]
- 37.de Leeuw M, Gonzalez A, Lanir A, Roiz L, Smirnoff P, Schwartz B, Shoseyov O, Almog O. The 1.8 A crystal structure of ACTIBIND suggests a mode of action for T2 ribonucleases as antitumorigenic agents. J. Med. Chem. 2012;55:1013–1020. doi: 10.1021/jm1015507. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa A, Tanaka I, Sakai R, Nakashima T, Funatsu G, Kimura M. Crystal structure of a ribonuclease from the seeds of bitter gourd (Momordica charantia) at 1.75 A resolution. Biochim. Biophys. Acta. 1999;1433:253–260. doi: 10.1016/s0167-4838(99)00126-0. [DOI] [PubMed] [Google Scholar]
- 39.Numata T, Suzuki A, Kakuta Y, Kimura K, Yao M, Tanaka I, Yoshida Y, Ueda T, Kimura M. Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5′-GMP: structural basis for alterations in substrate specificity. Biochemistry. 2003;42:5270–5278. doi: 10.1021/bi034103g. [DOI] [PubMed] [Google Scholar]
- 40.Kawano S, Kakuta Y, Kimura M. Guanine binding site of the Nicotiana glutinosa ribonuclease NW revealed by X-ray crystallography. Biochemistry. 2002;41:15195–15202. doi: 10.1021/bi026247l. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki A, Yao M, Tanaka I, Numata T, Kikukawa S, Yamasaki N, Kimura M. Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2′-UMP or 3′-UMP, reveal structural basis for uridine specificity. Biochem. Biophys. Res. Commun. 2000;275:572–576. doi: 10.1006/bbrc.2000.3318. [DOI] [PubMed] [Google Scholar]
- 42.Kurihara H, Nonaka T, Mitsui Y, Ohgi K, Irie M, Nakamura KT. The crystal structure of ribonuclease Rh from Rhizopus niveus at 2.0 A resolution. J. Mol. Biol. 1996;255:310–320. doi: 10.1006/jmbi.1996.0025. [DOI] [PubMed] [Google Scholar]
- 43.Vidalino L, Monti L, Haase A, Moro A, Acquati F, Taramelli R, Macchi P. Intracellular trafficking of RNASET2, a novel component of P-bodies. Biol. Cell. 2012;104:13–21. doi: 10.1111/boc.201100092. [DOI] [PubMed] [Google Scholar]
- 44.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. ELM—the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wajant H, Scheurich P. TNFR1-induced activation of the classical NF-kappaB pathway. FEBS J. 2011;278:862–876. doi: 10.1111/j.1742-4658.2011.08015.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee JE, Bae E, Bingman CA, Phillips GN, Jr, Raines RT. Structural basis for catalysis by onconase. J. Mol. Biol. 2008;375:165–177. doi: 10.1016/j.jmb.2007.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.