Abstract
Streptomyces davawensis is the only organism known to synthesize the antibiotic roseoflavin, a riboflavin (vitamin B2) analog. Roseoflavin is converted to roseoflavin mononucleotide (RoFMN) and roseoflavin adenine dinucleotide in the cytoplasm of target cells. (Ribo-)Flavin mononucleotide (FMN) riboswitches are genetic elements, which in many bacteria control genes responsible for the biosynthesis and transport of riboflavin. Streptomyces davawensis is roseoflavin resistant, and the closely related bacterium Streptomyces coelicolor is roseoflavin sensitive. The two bacteria served as models to investigate roseoflavin resistance of S. davawensis and to analyze the mode of action of roseoflavin in S. coelicolor. Our experiments demonstrate that the ribB FMN riboswitch of S. davawensis (in contrast to the corresponding riboswitch of S. coelicolor) is able to discriminate between the two very similar flavins FMN and RoFMN and shows opposite responses to the latter ligands.
INTRODUCTION
Riboswitches represent gene regulatory systems that consist of a metabolite-responsive aptamer and an overlapping expression platform and are predominantly located in the 5′-untranslated regions (UTRs) of messenger ribonucleic acids (mRNAs) (1). Flavin mononucleotide (FMN) is one of the biologically active forms of riboflavin. In bacteria, the synthesis of FMN from riboflavin and adenosine triphosphate is catalyzed by flavokinase (RibC or RibF) (2). FMN controls riboflavin biosynthesis in the model bacterium Bacillus subtilis by binding the aptamer portion of the ribG FMN riboswitch present in the 5′-UTR of the ribGBAH mRNA causing transcription termination (3,4). In the Gram-positive bacteria, Streptomyces davawensis and Streptomyces coelicolor two gene loci responsible for riboflavin biosynthesis (ribBMAH and ribG) have been identified. The corresponding gene products RibBAH and RibG are similar to the B. subtilis enzymes RibGBAH and catalyze the synthesis of riboflavin from guanosine-5′-triphosphate (GTP) and two molecules of ribulose-5′-phosphate (5). The gene ribM within the ribBMAH gene cluster of S. davawensis and S. coelicolor encodes a riboflavin permease, which is also able to import roseoflavin (6). Sequence analysis of S. coelicolor and S. davawensis revealed that expression of the ribBMAH genes may be regulated by an FMN riboswitch (ribB FMN riboswitch) (5). FMN binding by the ribB FMN riboswitch aptamer portion was predicted to prevent initiation of translation of the ribBMAH-mRNA by a mechanism involving sequestration of the ribB ribosomal binding site (7) rather than termination of transcription as observed for the ribG FMN riboswitch of B. subtilis (1,3).
The antibiotic roseoflavin (Supplementary Figure S1) is synthesized by S. davawensis (8) and was found to bind to FMN riboswitches and to affect gene expression (9–11). Additional support for flavins (or flavin analogs) targeting FMN riboswitches came from structural models for the FMN riboswitch of the Gram-negative bacterium Fusobacterium nucleatum bound to riboflavin, FMN and roseoflavin (12).
Roseoflavin is the only natural antimetabolite known to affect riboswitches and thus aroused our interest. Understanding the mechanism of roseoflavin resistance of the producer organism, S. davawensis was the immediate objective for initiating this study. Beyond that, the results show the unique ability of the ribB FMN riboswitch of S. davawensis to enhance or reduce gene expression depending on the flavin ligand.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Streptomyces davawensis (JCM strain 768) was aerobically grown at 37°C and pH 7.2 in a nutrient broth (YS) containing yeast extract (2 g l−1) and soluble potato starch (10 g l−1). Streptomyces coelicolor was aerobically grown at 30°C in YS. Escherichia coli was aerobically cultivated on lysogeny broth (LB) (13).
Isolation of total deoxyribonucleic acid and other molecular biology/cloning techniques
For the isolation of total deoxyribonucleic acid (DNA) from Streptomycetes, a modified protocol of the ‘Kirby Mix Procedure’ was used (14). Other molecular biology techniques were carried out according to standard protocols (13).
Chemicals
Riboflavin, FMN and flavin adenine dinucleotide (FAD) were purchased from Sigma (Munich, Germany). Roseoflavin was obtained from Chemos (Regenstauf, Germany). The riboflavin analog 8-demethyl-8-amino riboflavin (AF) was prepared synthetically by Sandro Ghisla (University of Constance, Germany) and was a gift from Peter Macheroux (Graz University of Technology, Austria).
Analysis of flavins
Flavins were analyzed by high performance liquid chromatography (HPLC) coupled to a mass spectrometer (Agilent Technologies, Waldbronn, Germany) as described earlier (15).
Enzymatic synthesis of flavins
Roseoflavin mononucleotide (RoFMN), roseoflavin adenine dinucleotide (RoFAD) and 8-demethyl-8-amino riboflavin mononucleotide (AFMN) were enzymatically synthesized and purified as described earlier (15).
Construction of plasmids used for in vitro transcription/translation
The plasmids for generating transcriptional and translational fusions of FMN riboswitches to the firefly luciferase reporter gene (luc) were constructed using pT7luc (Promega, Mannheim, Germany) and a modified version (pT7lucmod) (Supplementary Figure S2). The DNA fragments coding for the different FMN riboswitches were generated by polymerase chain reaction (PCR) using genomic DNA as a template and modifying oligonucleotides (Supplementary Table S3).
In vitro transcription/translation assays
The coupled transcription/translation (TK/TL) assay was performed using the E. coli T7 S30 Extract System for Circular DNA Kit (Promega, Mannheim, Germany). Luciferase activity was determined using a microtiter plate reader (Tecan Genios Pro microplate reader, Tecan, Mainz, Germany). T50 values were estimated by fitting the plot of the luciferase activity percentage versus the flavin concentration using the SigmaPlot 9 software (Systat Software, Erkrath, Germany). The data fit to a first-order exponential decay equation.
Protein concentration determination
Protein concentration was estimated by the method of Bradford (16).
Measuring riboflavin synthase in Streptomycetes
Spore suspensions (108 spores) from S. davawensis and S. coelicolor were used to inoculate 1 l flasks containing 100 ml YS broth, and cultures were grown overnight (14 h). The mycelia were harvested by centrifugation (4000g), washed three times and suspended in 100 ml minimal medium. Following a medium change, the cells were grown for another 20, 30 or 40 h and again harvested by centrifugation. The mycelia were washed five times with 100 mM potassium phosphate pH 7.4, suspended in the same buffer and disrupted using a French press (2000 bar, 10°C). The resulting cell extract was tested for riboflavin synthase activity: assay mixtures containing 100 mM potassium phosphate (pH 7.4), 10 mM ethylenediaminetetraacetic acid, 7.5 mM 2-mercaptoethanol and 0.6 mM 6,7-dimethyl-8-ribityllumazine were pre-warmed (5 min, 37°C), and the reaction was started by the addition of cell extracts. Riboflavin formation was monitored by HPLC.
Selection and analysis of spontaneous roseoflavin-resistant S. coelicolor mutants
Streptomyces coelicolor spores (108) were spread on mannitol/soya flour (MS) agar containing 200 µM roseoflavin, and the plates were incubated at 30°C for 4 days. The largest (apparently roseoflavin resistant) colonies were isolated and used as an inoculum for 5 ml cultures in YS broth. The isolated strains were aerobically grown for 16 h at 30°C, and cells were harvested by centrifugation. Genomic DNA was extracted from the different isolates using the ‘Genomic DNA Extraction Kit’ (Fermentas, St. Leon-Rot, Germany) and was used as a template for subsequent PCR reactions. The FMN riboswitch region including the rib promoter was amplified by PCR. The resulting PCR products were subjected to DNA sequencing.
In-line probing assay
In-line probing assays were performed as described previously (17).
Fluorescence measurements
The intensity of fluorescence emission was measured at 535 nm with an excitation at 485 nm. Assays were performed on the basis of the intrinsic fluorescence of FMN and RoFMN, which is quenched on the specific interaction of the flavins with the riboswitch aptamers. RoFMN fluorescence was ∼20 times weaker when compared with FMN. Each experiment was performed three times at 25°C using a Tecan Genios Pro microplate reader. FMN riboswitch RNA molecules (containing both the aptamer and the expression platform) were in vitro synthesized by T7 RNA polymerase (Promega, Mannheim) using the plasmids generating translational fusion as templates.
RESULTS
Streptomyces davawensis is roseoflavin resistant
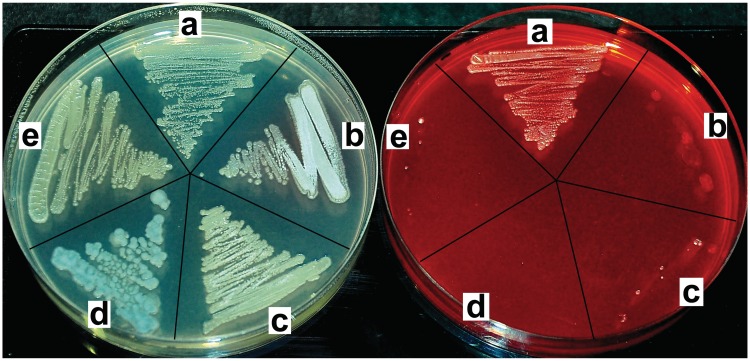
It was reported earlier that roseoflavin inhibits growth of the Gram-positive bacteria Staphylococcus aureus (MIC from 0.25 to 6.25 µg ml−1), Bacillus subtilis (MIC 1.56 µg ml−1), Bacillus cereus and Micrococcus luteus (Sarcina lutea) (8). In this work, S. davawensis and some closely related species, Streptomyces lividans, Streptomyces coelicolor, Streptomyces albus and Streptomyces avermitilis were tested with respect to roseoflavin sensitivity. Spores of S. lividans, S. coelicolor, S. albus and S. avermitilis were not able to produce colonies on a solid growth medium containing 200 µM roseoflavin, and we concluded that these bacteria are roseoflavin sensitive (Figure 1). The three latter species all contain a gene homologous to S. davawensis ribM, which was shown to be responsible for riboflavin and roseoflavin uptake (5,6). The S. lividans genomic data have not been published yet, and thus, the presence of a ribM homologous gene could not be verified. In contrast to the other Streptomycetes, spores of S. davawensis were able to germinate and produce colonies in the presence of 200 µM roseoflavin showing that S. davawensis is roseoflavin resistant. Notably, RibC homologous enzymes are present in all five Streptomycetes, and it is thus very likely that in these organisms imported flavins (and flavin analogs) are phosphorylated and adenylylated.
Figure 1.
Streptomyces davawensis is roseoflavin resistant. Streptomyces davawensis (a) and the closely related species Streptomyces lividans (b), S. coelicolor (c), Streptomyces albus (d) and Streptomyces avermitilis (e) were tested with respect to roseoflavin sensitivity. To each of the sectors of the YS plates, ∼50 000 spores of the different Streptomyces species were applied and incubated aerobically for 60 h at 30°C. Only the YS plate to the right contained the antibiotic roseoflavin (200 µM).
RoFMN and RoFAD are present in the cytoplasm of S. coelicolor and S. davawensis
Streptomyces coelicolor and S. davawensis were cultured in the presence of sub-lethal concentrations of roseoflavin (50 µM). Exponentially growing cells were harvested, thoroughly washed and disrupted. Subsequently, the corresponding cell-free extracts were analyzed by liquid chromatography (LC-MS) for their flavin content, which was normalized to the total protein content of the cells. Analysis of S. coelicolor cell-free extracts revealed the presence of both RoFMN (1.2 ± 0.3 µM) and RoFAD (0.1 ± 0.05 µM). Similar concentrations of RoFMN/RoFAD were detected in the corresponding cell-free extracts of S. davawensis (RoFMN, 1.4 ± 0.2 µM; RoFAD, 0.2 ± 0.06 µM). Streptomyces davawensis naturally produces roseoflavin in the stationary growth phase. Correspondingly, it was of interest whether cell-free extracts of stationary cultures not pulsed with roseoflavin-contained RoFMN and RoFAD as well. Indeed, cell free-extracts from these cells were found to contain 0.9 µM RoFMN (±0.1 µM) and 0.1 µM RoFAD (±0.03 µM). Free flavins (riboflavin and roseoflavin) were detected in both bacteria in trace amounts only (<0.01 µM). Thus, S. davawensis is roseoflavin resistant but is not a riboflavin overproducer.
The addition of roseoflavin reduces riboflavin synthase activity in S. coelicolor but not in S. davawensis
Expression of the ribBMAH genes was monitored in S. coelicolor and in S. davawensis cultures by measuring the activity of riboflavin synthase (RibB), the product of the first gene of the cluster catalyzing the last step in riboflavin biosynthesis. RibB activity was measured at different time points (20, 30 and 40 h) following a change of the culture medium and the addition of flavins (Table 1). It was found that on addition of riboflavin to cultures of S. coelicolor, RibB activity decreased up to 47.7% when compared with the control level. When roseoflavin was added to the cultures, a stronger decrease of RibB activity was detected (up to 20.7% after 20 h). After 40 h of growth in the presence of roseoflavin, no RibB activity was detected in cells of S. coelicolor. In S. davawensis, the addition of riboflavin reduced RibB activity up to 23.5%. The addition of roseoflavin, however, had no such effect (RibB activity remained at >90%) showing that expression of the ribBMAH genes in S. davawensis was only slightly reduced by this antibiotic. The data (Table 1) also revealed that the specific RibB activity in S. davawensis (20 h: 0.485 µM mg−1 h−1) was quite similar to what was found in S. coelicolor at that time point (20 h: 0.319 µM mg−1 h−1). However, in S. davawensis, RibB activity strongly increased at 30 h of growth and at 40 h was almost 60 times higher when compared with S. coelicolor. Notably, this did not lead to an increased riboflavin level in the cytoplasm of S. davawensis.
Table 1.
Monitoring ribBMAH gene expression by measuring the activity of riboflavin synthase (RibB)
| MM 20 h (µM mg−1 h−1) | MM 30 h (µM mg−1 h−1) | MM 40 h (µM mg−1 h−1) | YS 40 h (µM mg−1 h−1) | |
|---|---|---|---|---|
| S. davawensis | ||||
| Control | 0.485 ± 0.040 | 1.862 ± 0.151 | 5.140 ± 0.502 | 0.451 ± 0.050 |
| RF 50 µM | 0.225 ± 0.030 | 0.614 ± 0.060 | 1.208 ± 0.100 | 0.226 ± 0.018 |
| RoF 50 µM | 0.474 ± 0.050 | 1.781 ± 0.070 | 4.575 ± 0.313 | 1.168 ± 0.235 |
| S. coelicolor | ||||
| Control | 0.319 ± 0.015 | 0.254 ± 0.040 | 0.088 ± 0.005 | |
| RF 50 µM | 0.176 ± 0.014 | 0.129 ± 0.011 | 0.042 ± 0.003 | |
| RoF50 µM | 0.066 ± 0.002 | 0.053 ± 0.004 | 0 |
| MM 20 h (%) | MM 30 h (%) | MM 40 h (%) | YS 40 h (%) | |
|---|---|---|---|---|
| S. davawensis | ||||
| Control | 100 ± 8.2 | 100 ± 8.1 | 100 ± 9.7 | 100 ± 11.0 |
| RF 50 µM | 46.4 ± 13.3 | 33.0 ± 9.7 | 23.5 ± 8.3 | 50.1 ± 8.0 |
| RoF 50 µM | 97.7 ± 10.5 | 95.6 ± 3.9 | 90.0 ± 6.8 | 259.8 ± 20.1 |
| S. coelicolor | ||||
| Control | 100 ± 4.7 | 100 ± 15.7 | 100 ± 5.7 | |
| RF 50 µM | 55.1 ± 7.9 | 50.8 ± 8.5 | 47.7 ± 7.1 | |
| RoF 50 µM | 20.7 ± 3.0 | 20.8 ± 7.5 | 0 |
RibB activity was determined in cell extracts of S. coelicolor and S. davawenis cultures. Cells were pregrown, washed, resuspended in fresh culture medium containing the indicated concentration of flavins and grown further. RibB activity was measured at different time points (20, 30 and 40 h) following the change of the culture medium and addition of flavins (riboflavin, RF; roseoflavin, RoF). For culturing the cells, either a minimal medium (MM) or a complex medium (YS) was used. The activities (µMol h−1) are presented per mg of total protein or normalized to 100% (control level). Mean values of three independent experiments are shown.
When grown on minimal medium, S. davawensis produced comparably small concentrations of roseoflavin (1 µM). However, when grown in YS culture medium containing small concentrations (4 µM) of the precursor riboflavin, considerably more roseoflavin (19 µM) was produced by S. davawensis. We, therefore, performed similar experiments as described earlier with S. davawensis growing in YS. A strongly increased RibB activity (to 259.8%) was detected on addition of roseoflavin to the cultures. In contrast, the addition of riboflavin reduced RibB activity to 50.1%. An increase of RibB activity in S. davawensis on stimulation with roseoflavin is perfectly in line with the fact that riboflavin is the precursor of roseoflavin (18) and thus is needed in larger concentrations when roseoflavin synthesis is triggered in the stationary growth phase.
The ribB FMN riboswitches of S. davawensis and S. coelicolor regulate at the translational level
To be able to quantify the activity of FMN riboswitches from different bacteria, an in vitro transcription/translation assay was established based on the plasmid pT7luc. Between the bacteriophage T7 promoter and the luciferase reporter gene, FMN riboswitches from B. subtilis, S. coelicolor and S. davawensis were placed. Two different plasmid constructs were generated for each FMN riboswitch to be tested producing transcriptional as well as translational fusions (Supplementary Figure S2). The plasmids were used as templates in a T7 RNA polymerase based in vitro transcription/translation assay. Before analyzing the yet uncharacterized Streptomyces riboswitches, the assay was validated using the well-characterized ribG FMN riboswitch from B. subtilis. The presence of FMN in the reaction reduced luciferase gene expression regardless of whether the plasmid generating a transcriptional fusion (reduction to 40%; ±2%) or a translational fusion (reduction to 42%; ±3%) to the reporter gene luc was used. Apparently, the ribG FMN riboswitch from B. subtilis acted at the transcriptional level as was reported earlier (3, 4). In contrast, luc expression was not reduced when the FMN riboswitches from S. coelicolor and S. davawensis were tested in the in vitro transcription/translation assay using the plasmids generating transcriptional fusions. However, FMN reduced luciferase gene expression using the plasmids for translational fusions: An identical reduction to 40% (±2%) was observed for both Streptomycetes riboswitches. The latter result shows that the FMN riboswitches of S. davawensis and S. coelicolor indeed regulate at the translational level as was hypothesized earlier (7). Similar results were obtained by in vivo experiments in B. subtilis using transcriptional lacZ fusions (data not shown).
RoFMN triggers the B. subtilis ribG FMN riboswitch to repress gene expression
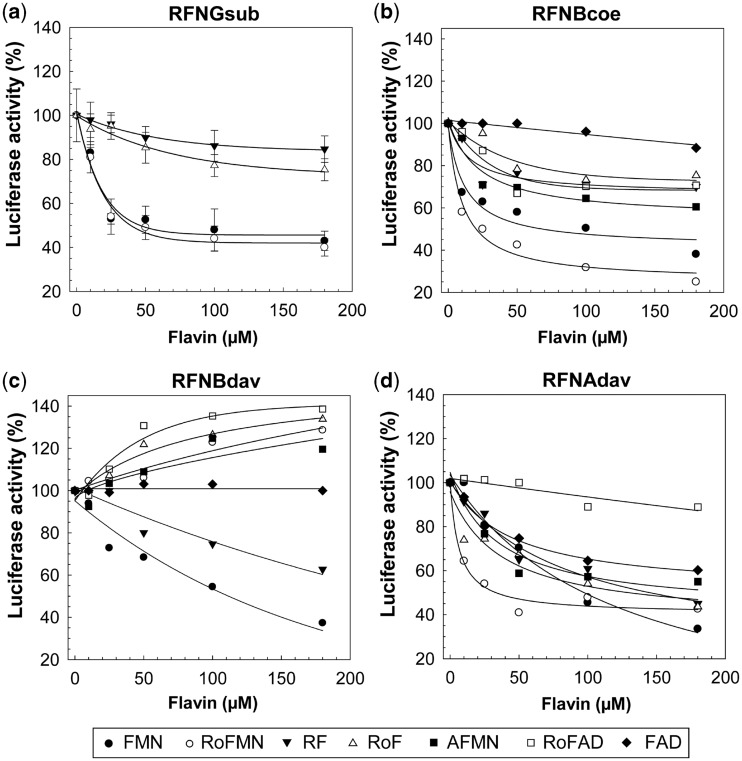
It was reported earlier that the bifunctional flavokinase/FAD synthetase of B. subtilis (RibC) accepts roseoflavin as a substrate and produces RoFMN and RoFAD (19). Moreover, our present data show that the latter cofactor analogs are present in roseoflavin-treated cells. Hence, it seemed feasible that RoFMN (and not roseoflavin) was the true effector of FMN riboswitches. Indeed, the addition of increasing concentrations of RoFMN (0–180 µM) led to reduced Luc activity (by up to 60%) when the B. subtilis ribG FMN riboswitch was tested using in vitro transcription/translation (Figure 2a). Notably, a very similar effect was found when FMN was added. The addition of riboflavin and roseoflavin had a much smaller effect.
Figure 2.
Flavins and their effect on FMN riboswitches. (a) A reporter plasmid producing a transcriptional fusion of the ribG FMN riboswitch of Bacillus subtilis, and the reporter gene luc (coding for firefly luciferase) was used as a DNA template for an in vitro transcription/translation assay driven by RNA polymerase of bacteriophage T7 (Supplementary Figure S2a). The addition of FMN resulted in less activity of Luc indicating that less luc transcript was produced. RoFMN had a very similar effect. The addition of riboflavin or roseoflavin also resulted in reduced luciferase activity; however, the effect was less pronounced. (b) and (c) Reporter plasmids producing translational luc fusions (Supplementary Figure S2b) of the ribB FMN riboswitches of S. coelicolor (b, RFNBcoe) and S. davawensis (c, RFNBdav) were used as templates for in vitro transcription/translation assays as in (a). Most importantly, in contrast to RFNBdav (c), RFNBcoe (b) was negatively affected by RoFMN. A genome-wide screen for FMN riboswitches in S. davawensis identified a second (ribA) FMN riboswitch (Supplementary Figure S3) controlling a gene cluster with unknown function. The activity of the ribA FMN riboswitch (RFNAdav, d) was tested as described earlier. RFNAdav (in contrast to RFNBdav) was found to be negatively affected by RoFMN. Mean values of three independent experiments are shown. For clarity, standard deviations (SDs) are shown in (a) only.
The S. coelicolor and S. davawensis ribB FMN riboswitches respond differently to the flavin analogs RoFMN and AFMN
According to the results up to this point, our study suggested that formation of RoFMN targeting FMN riboswitches was responsible for the antibiotic effect of roseoflavin. As S. davawensis, in contrast to S. coelicolor, was found to be roseoflavin resistant, we hypothesized that the ribB FMN riboswitches of the two organisms may react differently with respect to treatment with RoFMN. To test this, the ribB FMN riboswitches of S. coelicolor and S. davawensis were characterized using in vitro transcription/translation assays. For the S. coelicolor FMN riboswitch, both FMN and RoFMN significantly reduced the amount of luciferase activity in vitro (Figure 2b). FMN induced a reduction by up to 62% (±7%), and for RoFMN, the reduction was up to 75% (±6%). Similar experiments (Figure 2c) were carried out with the S. davawensis riboswitch, and it was found that luciferase activity was strongly reduced on addition of FMN (by up to 63%, ±7%) to the assay. The extent of reduction was similar to what was found for the S. coelicolor riboswitch. Most strikingly, however, the addition of RoFMN to the assay containing the S. davawensis riboswitch resulted in an increase of luciferase activity by 28% (±9%).
The amount of flavin needed for a 50% reduction (T50) of luciferase activity in the transcription/translation assays is a measure for the apparent ligand affinity of the FMN riboswitch aptamers and was estimated from the data in Figure 2. T50 of the B. subtilis aptamer was 46 µM (±6 µM) for FMN and 40 µM (±4 µM) for RoFMN. T50 of the S. coelicolor aptamer was 50 µM (±8 µM) for FMN and 20 µM for RoFMN (±5 µM). T50 of the S. davawensis aptamer was 102 µM (±6 µM) for FMN. For RoFMN, an activation was observed, and thus, T50 could not be calculated.
The FMN riboswitches from S. coelicolor and S. davawensis were also tested with respect to modulation by other flavins. 8-Demethyl-8-amino-riboflavin (AF, the 5′-phosphorylated form is AFMN) is the direct precursor of roseoflavin (20) and was found to exhibit antibiotic activity as well (M. Mack, unpublished data). In summary, the S. coelicolor FMN riboswitch was found to be negatively affected by all flavins tested (roseoflavin, riboflavin, RoFMN, FMN, RoFAD and AFMN) except for FAD (Figure 2b). Except for FMN, only riboflavin was able to decrease the luciferase activity level when testing the S. davawensis riboswitch (Figure 2c). RoFAD, RoFMN, roseoflavin and AFMN stimulated luciferase gene expression by up to 38% (180 µM RoFAD).
Analysis of a second FMN riboswitch present in the genome of S. davawensis
A genome-wide screen in S. davawensis identified a second sequence element (‘ribA FMN riboswitch’) matching the profile of FMN riboswitch aptamers (Supplementary Figure S3). The ribA FMN riboswitch is present in the 5′-UTR of a gene cluster (comprising seven genes) with unknown function. Interestingly, the product of the first putative gene within this cluster is similar (up to 54% on the amino acid level) to 3,4-dihydroxy-2-butanone 4-phosphate synthases (EC 4.1.99.12) from other microorganisms. The latter enzymes catalyze an initial step in riboflavin biosynthesis, the formation of 3,4-dihydroxy-2-butanone 4-phosphate from ribulose-5-phosphate (21).
The ribA FMN riboswitch was analyzed using in vitro transcription/translation and, in contrast to the S. davawensis ribB FMN riboswitch, was found to be negatively affected by both RoFMN (T50 = 25 ± 4 µM) and FMN (T50 = 94 ± 5 µM) (Figure 2d). Besides RoFAD, all the other flavins tested negatively affected expression of the luciferase gene under control of this FMN riboswitch. Only the translational fusion of the ribA FMN riboswitch produced a decrease in luciferase activity on FMN addition, strongly suggesting that it also operates at the translational level.
The nucleotide A61 of the S. davawensis ribB riboswitch is responsible for discrimination between FMN and RoFMN
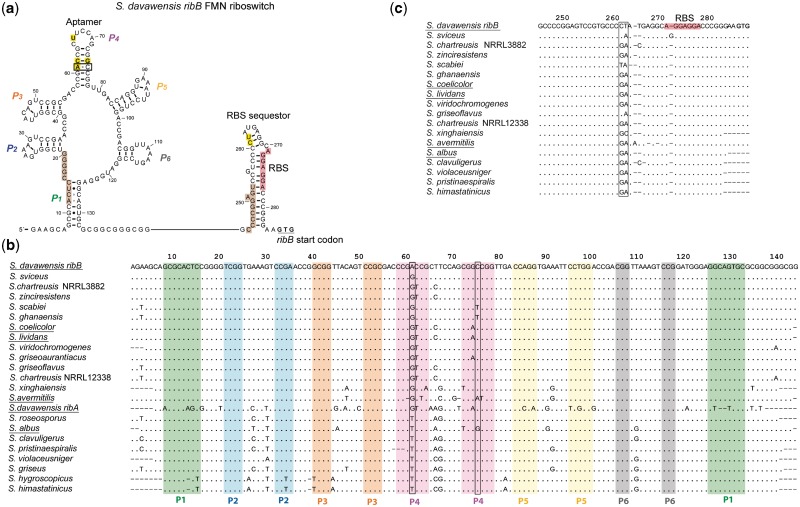
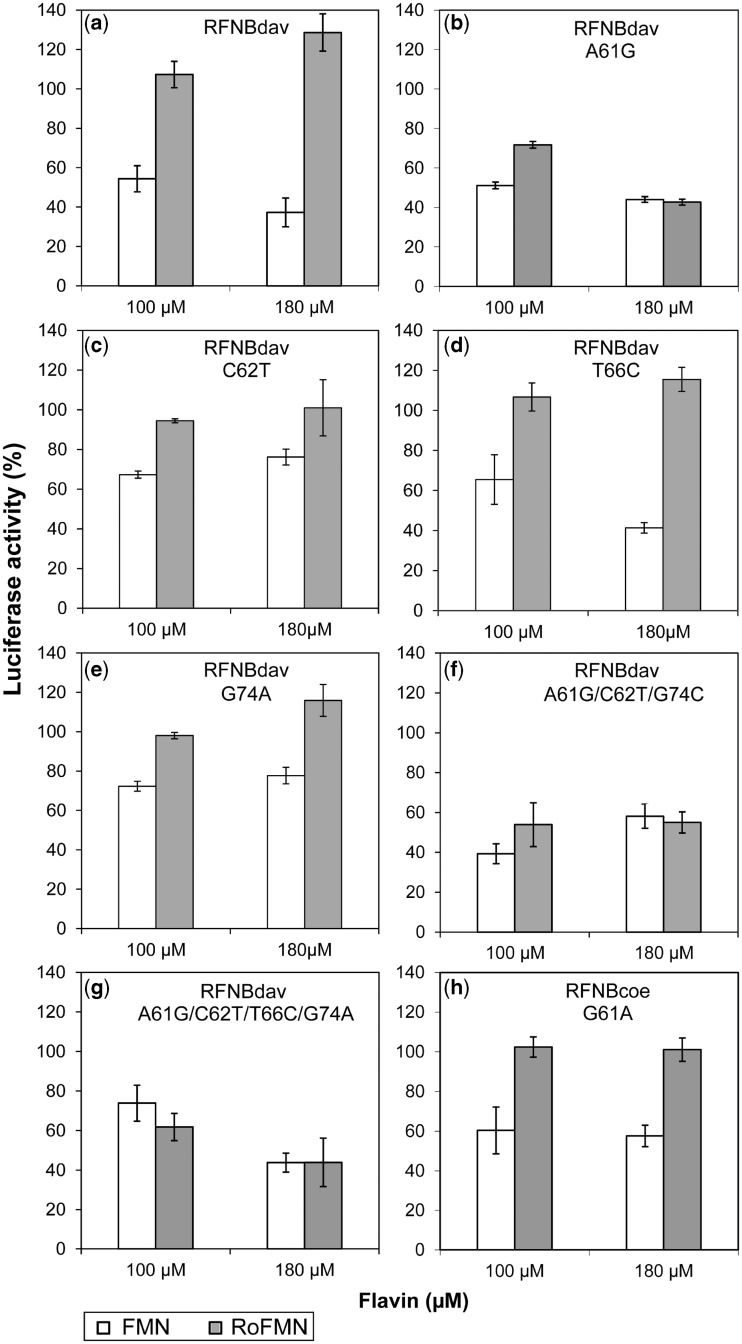
The ribB FMN riboswitch aptamers of S. coelicolor and S. davawensis differ at four nucleotide positions only (61, 62, 66 and 74; Figure 3a and Supplementary Figure S3). Sequence analysis of ribB FMN riboswitch aptamers of roseoflavin-sensitive Streptomyces species tested in this work (S. coelicolor, S. lividans, S. albus and S. avermitilis) revealed that roseoflavin-resistant S. davawensis is the only species to contain an A at position 61 (Figure 3b). Positions 62, 66 and 74 appeared not to be key residues with respect to roseoflavin/RoFMN resistance as S. coelicolor, S. lividans, S. avermitilis and/or S. albus FMN riboswitch aptamers share identical nucleotides with the S. davawensis ribB riboswitch. Moreover, none of the 23 Streptomyces FMN riboswitches listed in the RFAM database (22) revealed the nucleotide pair 61AC75 (Figure 3a). Variants of the riboswitch aptamers of S. coelicolor and S. davawensis were generated by site-directed mutagenesis and tested using in vitro transcription/translation. The single-nucleotide exchange A61G produced a mutant S. davawensis ribB FMN riboswitch (RFNBdavA61G), which apparently was negatively affected by both FMN and RoFMN (Figure 4b). In contrast, the wild-type riboswitch was negatively affected by FMN but found to be more active in the presence of RoFMN (Figures 4a and 2c). The single-nucleotide exchanges C62T (Figure 4c) and G74A (Figure 4e) produced mutant S. davawensis FMN riboswitches, which were less sensitive with respect to both FMN and RoFMN indicating that destabilization of the P4 stem (Figure 3a) affects aptamer/riboswitch function. The exchange T66C responded like the wild type to FMN and RoFMN (Figure 4d). The mutant ribB FMN riboswitches A61G/C62T/G74A (Figure 4f) and A61G/C62T/T66C/G74A (Figure 4g; corresponds to the S. coelicolor aptamer) both were negatively affected by RoFMN. Most interestingly, the exchange G61A within the S. coelicolor ribB FMN riboswitch (removing Watson-Crick base pairing at 61GC75) produced a RoFMN-insensitive riboswitch (Figure 5h) still responding to FMN. Notably, S. davawensis is the only species to contain the nucleotides 261CT262 in the downstream expression platform (Figure 3c).
Figure 3.
Comparison of different Streptomyces FMN riboswitches. (a) Sequence, secondary structure and expected transcriptional intermediates of the 5′-UTR of the ribB mRNA from S. davawensis [ribosomal binding site (RBS), nucleotides in red]. The solid black line represents a highly variable RNA stretch. The key nucleotide pair (61AC75) with respect to RoFMN resistance is boxed. Nucleotides differing to the S. coelicolor aptamer are boxed yellow. Nucleotides marked brown are responsible for anti-sequestration. (b) Nucleotide sequences of aptamer portions of FMN riboswitches from different Streptomycetes. The aptamers are highly similar and only the nucleotides differing to the S. davawensis ribB FMN riboswitch (top sequence) are shown. The S. davawensis aptamer is the only sequence containing an A at position 61 (boxed). The other Streptomyces species show either a G or a T (U) at this position. A61 is not able to pair with C75 (boxed). (c) Nucleotides encompassing the ribB ribosomal binding sites (red) of different Streptomycete ribB FMN riboswitches are shown. Streptomyces davawensis is the only species to contain the nucleotides C and T at positions 261 and 262 (boxed).
Figure 4.
Nucleotide A61 of the S. davawensis ribB FMN riboswitch is responsible for the discrimination between FMN and RoFMN. Reporter plasmids producing translational luc-fusions of ribB FMN riboswitches from S. davawensis and S. coelicolor were used as templates for in vitro transcription/translation assays (Figure 2). Wild-type (a) and mutant FMN riboswitches of S. davawensis (b–g) and a mutant FMN riboswitch of S. coelicolor (h) were tested in the presence of FMN or RoFMN. The single-nucleotide exchange A61G (b) produced an S. davawensis FMN riboswitch which, in contrast to the wild-type riboswitch (a), was negatively affected by both FMN and RoFMN. The single-nucleotide exchanges C62T (c), T66C (d) and G74A (e) produced similar results as in (a). Most interestingly, the single mutation G61A within the S. coelicolor FMN riboswitch aptamer (removing Watson–Crick DNA base pairing at nucleotide 61G-75C) produced a RoFMN-insensitive FMN riboswitch (h). Mean values of three independent experiments are shown.
Figure 5.
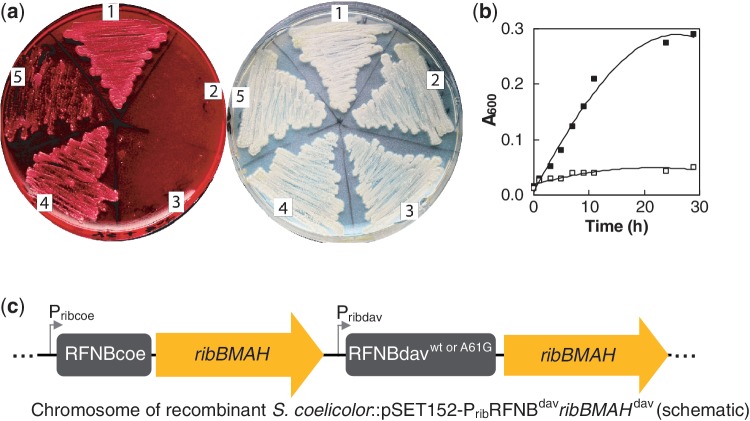
The S. davawensis FMN riboswitch confers roseoflavin resistance to S. coelicolor. Spores of different Streptomyces strains were applied to each of the sectors of the YS plates shown (1, wild-type S. davawensis; 2, wild-type S. coelicolor; 3, S. coelicolor::pSET152; 4, S. coelicolor::pSET152-PribRFNBdavribBMAHdav and 5, S. coelicolor:: pSET152-PribRFNBdavA61GribBMAHdav), and the plates were aerobically incubated at 30°C for 60 h. The recombinant S. coelicolor in sector 4 was generated by introducing the S. davawensis ribBMAH cluster into the chromosome of S. coelicolor by homologous recombination. The strain in sector 3 is a control containing empty pSET152 at Φ31. The recombinant S. coelicolor in sector 5 was generated by introducing the S. davawensis ribBMAH-genes (including the roseoflavin-sensitive A61G mutant FMN ribB riboswitch). (a) The plate to the left contained 200 µM roseoflavin, and the plate to the right did not contain any antibiotic. The spores in sector 5 of the left plate produced less mycelium when compared with the spores in sector 4. (b) Growth of the strains S. coelicolor::pSET152-PribRFNBdavribBMAHdav (solid squares) and S. coelicolor::pSET152-PribRFNBdavA61GribBMAHdav (open squares) was also monitored in a liquid culture (minimal medium). (c) Schematic drawing showing the chromosome of recombinant S. coelicolor strains containing two different gene clusters controlled by two different promoters (with very similar activity) and two different FMN ribB riboswitches (RFNBcoe, from S. coelicolor, and RFNBdav from S. davawensis).
The S. davawensis FMN riboswitch confers roseoflavin resistance to S. coelicolor
The following experiment was initiated to in vivo validate the in vitro results strongly suggesting that a single-nucleotide change conferred roseoflavin/RoFMN resistance to S. davawensis. The S. davawensis ribBMAH genes including the RoFMN insensitive (wild type) ribB FMN riboswitch and the corresponding promoter Prib were integrated into the chromosome of S. coelicolor by homologous recombination at the Φ31 site (Figure 5c) using pSET152 (23). The resulting strain S. coelicolor::pSET152-PribRFNBdavribBMAHdav was found to be roseoflavin resistant (Figure 5a). It has long been known that oversynthesis of riboflavin in the cytoplasm compensates for the toxic effect of roseoflavin and results in roseoflavin resistance (24). Therefore, it was important to exclude the possibility that increased activities of the products of the chromosomally integrated S. davawensis ribBMAH genes (possibly leading to increased riboflavin levels) were responsible for roseoflavin resistance. To exclude this possibility, the S. davawensis ribBMAH-genes including the roseoflavin-sensitive A61G mutant FMN riboswitch were chromosomally integrated in S. coelicolor. The corresponding recombinant strain S. coelicolor::pSET152-PribRFNBdavA61GribBMAHdav was grown on a solid medium in the presence of roseoflavin, and only sparse growth was observed (Figure 5a). Similarly, when grown in liquid medium, only little growth was observed (Figure 5b). The recombinant S. davawensis strains were further characterized by measuring cellular riboflavin synthase (RibB) activity (Supplementary Table S1). On addition of riboflavin to the culture medium of the roseoflavin-resistant strain S. coelicolor::pSET152-PribRFNBdavribBMAHdav, RibB activity decreased to 42%, whereas roseoflavin addition only caused a small reduction of RibB activity (91%). This experiment showed that S. coelicolor::pSET152-PribRFNBdavribBMAHdav essentially responded like S. davawensis to treatment with roseoflavin. In case of the roseoflavin-sensitive S. coelicolor::pSET152-PribRFNBdavA61GribBMAHdav strain, RibB activity decreased to 58% on addition of riboflavin to the culture medium. An even stronger decrease of RibB activity was found when roseoflavin was added to the cultures (47%), showing that the mutation A61G in the riboswitch leads to a roseoflavin response similar to what was found for S. coelicolor wild type. Notably, the total specific RibB activities were very similar in all S. coelicolor strains irrespective whether the S. davawensis ribBMAH genes (including the rib promoter and the FMN riboswitches) were present or not. Furthermore, the rib promoters from S. coelicolor and S. davawensis in vivo had an almost identical activity (data not shown).
A spontaneous roseoflavin-resistant mutant of S. coelicolor carries the point mutation G61A within the ribB FMN riboswitch
Spores of wild-type S. coelicolor were plated on a growth medium supplemented with roseoflavin and spontaneous roseoflavin-resistant cells (producing colonies) were detected with a frequency of 1 × 10−8. The regions upstream of the ribBMAH gene clusters (comprising the promoter Prib and the ribB FMN riboswitch) of 20 independent roseoflavin-resistant strains were amplified by PCR and analyzed by DNA sequencing. One of 20 isolated roseoflavin-resistant S. coelicolor strains showed the mutation G61A in the FMN riboswitch (Supplementary Figure S3). The remaining 19 strains did not show mutations in the corresponding 500 bp PCR fragment. The reason for roseoflavin resistance of the latter strains is most likely due to a mutation in ribC (25,26).
The FMN riboswitch aptamers of S. coelicolor and S. davawensis bind FMN and RoFMN with high affinity
An in-line probing assay (17) was used to test ribB FMN riboswitch aptamers from S. coelicolor and S. davawensis for FMN and RoFMN binding. We found that the apparent affinities of the S. coelicolor and S. davawensis ribB FMN riboswitch aptamers were exceedingly high for FMN and also for RoFMN. Indeed, the KD values were so small (KD ≪ 5 nM) that they could not reliably be measured with in-line probing. To overcome this, a single mutation was placed in the P1 stem (Supplementary Figure S3), as it is known that similar mutations in other riboswitches weaken affinities but do not alter ligand specificities (27). The KD estimates using in-line probing are summarized in the Supplementary Table S2 and revealed that all the FMN riboswitch aptamers tested had a similar affinity for FMN and RoFMN. At first sight, this result with respect to the S. davawensis ribB FMN riboswitch aptamer was surprising given the expectation that the S. davawensis FMN riboswitch, being not negatively affected by RoFMN in in vitro transcription/translation experiments, would contain a metabolite-sensing aptamer with a much higher KD value for RoFMN when compared with FMN. The data earlier, however, suggest that the S. davawensis aptamer does not selectively discriminate against RoFMN under equilibrium conditions. The control of gene expression mediated by metabolite-sensing riboswitches is predicted to be either thermodynamically or kinetically driven (27,28). For example, it has been shown that the B. subtilis ribG FMN riboswitch does not reach thermodynamic equilibrium with FMN in a time frame that is relevant for gene control. Rather, the latter riboswitch is kinetically driven, and therefore, the rate constant for ligand association is more meaningful than the KD value when evaluating the concentration of ligand needed to trigger gene control. Our working hypothesis was that the S. davawensis ribB riboswitch functions as a kinetically driven riboswitch as well and thus differently responds to FMN/RoFMN on a seconds or minutes timescale (28). As in-line probing assays extended for 40 h, this kinetic discrimination effect in the case of the S. davawensis ribB FMN riboswitch (i.e. a slower rate of association of RoFMN) may have been masked by a slower off rate of RoFMN when compared with FMN.
The ribB FMN riboswitches of S. davawensis and S. coelicolor operate differently
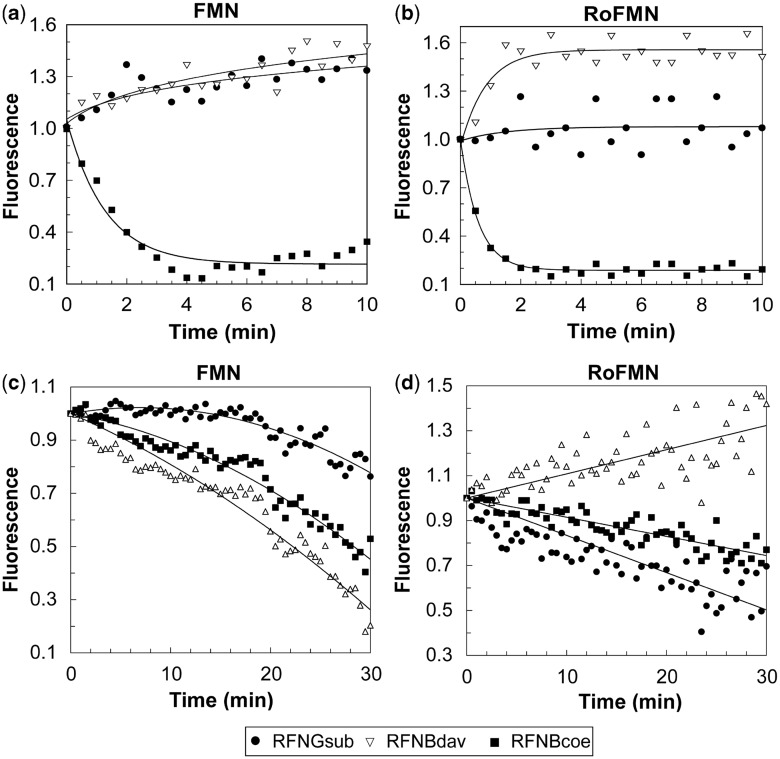
Fluorescence measurement assays were conducted to study the basis of the ligand–riboswitch interaction for the ribB FMN riboswitches of S. coelicolor and S. davawensis. The kinetically driven ribG FMN riboswitch from B. subtilis was used as a control. FMN riboswitch RNAs (aptamer portions in combination with the expression platforms) in vitro were synthesized and were incubated in the presence of either FMN (1 µM) or RoFMN (1 µM). As expected (28), the B. subtilis riboswitch RNA did not induce fluorescence quenching due to ligand (FMN or RoFMN) binding (Figure 6a and b). In contrast, the presence of the S. coelicolor riboswitch RNA led to significant fluorescence quenching of FMN and RoFMN indicating strong binding of both ligands (Figure 6a and b). The latter result suggested that the S. coelicolor riboswitch RNA (in contrast to the B. subtilis riboswitch RNA) is able to remain receptive for a considerably longer time, which would allow it to function thermodynamically. Very similar results as for the B. subtilis ribG FMN riboswitch were found for the S. davawensis ribB FMN riboswitch, which apparently also formed an RNA unable to bind either FMN or RoFMN when it was complete (Figure 6a and b). We, therefore, tentatively concluded that the S. davawensis FMN riboswitch is kinetically driven.
Figure 6.
Monitoring riboswitch-FMN/RoFMN complex formation by real-time fluorometry. Each image shows the change of fluorescence with time. The measurements were carried out to study ligand binding of the ribB FMN riboswitches from S. coelicolor (RFNBcoe) and S. davawensis (RFNBdav). The kinetically driven ribG FMN riboswitch from B. subtilis (RFNGbs) was used as a control. Riboswitch RNAs (aptamer portion in combination with the expression platform) in vitro were synthesized by T7 RNA polymerase and were incubated in the presence of 1 µM FMN or 1 µM RoFMN. Fluorescence quenching due to FMN (a) or RoFMN (b) binding was determined. The change of fluorescence was also measured during the in vitro transcription process in the presence of 1 µM FMN (c) or 1 µM RoFMN (d). The observed fluorescence for each experiment was corrected to yield a plot where the initial signal was 1 (corresponding to the fluorescence of the free flavin).
In the subsequent experiment, fluorescence quenching induced by FMN or RoFMN was monitored during the formation of different riboswitch RNAs in the time course of transcription. FMN (1 µM) or RoFMN (1 µM) were added to an in vitro transcription assay, and fluorescence was measured for 30 min (Figure 6c and d). The nascent riboswitch RNAs of B. subtilis and S. coelicolor induced fluorescence quenching in the presence of FMN and RoFMN indicating that both ligands are able to bind to nascent RNA molecules. However, in contrast to the completed RNA (Figure 6a and b), the S. davawensis ribB riboswitch RNA as a nascent transcript was able to bind FMN but unable to bind RoFMN (Figure 6c and d). This finding was perfectly in line with the results generated by the in vitro transcription/translation assays, which revealed that RoFMN, in contrast to FMN, did not negatively affect S. davawensis riboswitch function.
DISCUSSION
Streptomyces davawensis is roseoflavin resistant to concentrations exceeding the level synthesized by S. davawensis under laboratory conditions. Moreover, RoFMN and RoFAD indeed are present in the cytoplasm of S. davawensis and also of S. coelicolor cells treated with roseoflavin. Our in vitro transcription/translation data suggest that in S. davawensis and in S. coelicolor, expression of the rib genes is controlled by the concentration of FMN present in the cytoplasm and that RoFMN (and not roseoflavin) triggers roseoflavin-sensitive FMN riboswitches. Interestingly, the ribB FMN riboswitches of S. davawensis and S. coelicolor were found to respond very differently with respect to the addition of RoFMN to in vitro transcription/translation assays: The S. coelicolor ribB FMN riboswitch was turned off by RoFMN. In contrast, the S. davawensis ribB FMN riboswitch provoked enhanced gene expression in the presence of RoFMN. Both riboswitches, however, were turned off by FMN. These in vitro results were strongly supported by our in vivo data, which showed that RibB activity was strongly reduced in S. coelicolor cells on treatment with roseoflavin but not in cells of S. davawensis. Furthermore, RoFMN was found to block the S. coelicolor FMN riboswitch more efficiently when compared with FMN (and also reduced RibB activity more strongly when compared with FMN), which explains why roseoflavin is able to inhibit growth and act as an antibiotic. Even if the supply with essential riboflavin/FMN/FAD would only be slightly reduced in the presence of roseoflavin, this may constitute a major disadvantage for competing cells in a natural habitat.
The critical residue responsible for the different responses of S. davawensis and of S. coelicolor to roseoflavin/RoFMN is nucleotide 61 present within the aptamer portion of the ribB FMN riboswitches. An A at nucleotide 61 (S. davawensis) accounts for roseoflavin resistance, a G or U at this position (S. coelicolor, B. subtilis and other Streptomyces species; Supplementary Figure S3) results in roseoflavin sensitivity. Notably, the RoFMN-sensitive ribA FMN riboswitch of S. davawensis also contains a G at this critical position. We propose that nucleotide A61 within the S. davawensis ribB riboswitch aptamer specifically slows down RoFMN binding. Consequently, the sequestering RNA structure leading to reduced translation is not able to form which explains why S. davawensis is not sensitive to roseoflavin. In contrast, FMN binding is considerably faster, which explains why the S. davawensis ribB riboswitch is still regulated by FMN. Moreover, we propose that the downstream expression platform of the S. davawensis ribB riboswitch plays an important role with respect to the discrimination between FMN and RoFMN. Our fluorescence measurements suggest that the S. davawensis riboswitch is kinetically driven and remains receptive for modulator binding only in the relatively short time frame between aptamer formation and synthesis of the anti-ribosomal binding site stem. In contrast, the S. coelicolor riboswitch RNA seems to remain receptive to modulator binding for a longer time and thus is able to switch from the ‘ON’ to the ‘OFF’ mode at any time depending on whether FMN or RoFMN is present.
It is more difficult to find an explanation for the significantly increased activity of the S. davawensis ribB FMN riboswitch in the presence of RoFMN. Our fluorescence measurements argue against RoFMN binding to the complete riboswitch. In contrast, in-line probing data suggest tight binding of RoFMN to the aptamer portion under equilibrium conditions. Structural experiments will have to be carried out to resolve this apparent contradiction and possibly will reveal that RoFMN leads to a conformational change promoting access of the ribosomes to their binding site within the 5′-UTR of the ribBMAH mRNA.
Notably, roseoflavin-sensitive cells contain multiple targets for roseoflavin, FMN riboswitches and also flavoenzymes (29), which makes flavin analogs even more attractive as basic structures for developing novel antibacterial compounds. The current knowledge with respect to roseoflavin activity and resistance is summarized in Supplementary Figure S4.
Our work on roseoflavin underscores the potential generality of targeting riboswitches with new antibacterial drugs. Roseoflavin is a unique chemical being the only known natural compound negatively affecting riboswitches. As riboswitches are widespread in bacteria, we expect that a large number of highly interesting natural anti-infectives synthesized by yet unknown organisms remain to be discovered.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–4.
FUNDING
The National Council for Scientific and Technological Development (CNPq, Brazil) in cooperation with the German Academic Exchange Service (DAAD) (to D.P.); the state of Baden-Württemberg (HBIGS) (to A.M.). Funding for open access charge: Mannheim University of Applied Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 2.Bacher A. Riboflavin kinase and FAD synthetase. In: Muller F, editor. Chemistry and Biochemistry of Flavoenzymes. Vol. 1. Boca Raton, FL: CRC press; 1991. pp. 349–370. [Google Scholar]
- 3.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grill S, Yamaguchi H, Wagner H, Zwahlen L, Kusch U, Mack M. Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch. Microbiol. 2007;188:377–387. doi: 10.1007/s00203-007-0258-1. [DOI] [PubMed] [Google Scholar]
- 6.Hemberger S, Pedrolli DB, Stolz J, Vogl C, Lehmann M, Mack M. RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains. BMC Biotechnol. 2011;11:119. doi: 10.1186/1472-6750-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otani S, Takatsu M, Nakano M, Kasai S, Miura R. Letter: roseoflavin, a new antimicrobial pigment from Streptomyces. J. Antibiot. 1974;27:88–89. [PubMed] [Google Scholar]
- 9.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansjo M, Johansson J. The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol. 2011;8:674–680. doi: 10.4161/rna.8.4.15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ott E, Stolz J, Lehmann M, Mack M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 2009;6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- 12.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 14.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. pp. 168–169. [Google Scholar]
- 15.Pedrolli DB, Nakanishi S, Barile M, Mansurova M, Carmona EC, Lux A, Gartner W, Mack M. The antibiotics roseoflavin and 8-demethyl-8-amino-riboflavin from Streptomyces davawensis are metabolized by human flavokinase and human FAD synthetase. Biochem. Pharmacol. 2011;82:1853–1859. doi: 10.1016/j.bcp.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 17.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 18.Matsui K, Juri N, Kubo Y, Kasai S. Formation of roseoflavin from guanine through riboflavin. J. Biochem. 1979;86:167–175. [PubMed] [Google Scholar]
- 19.Grill S, Busenbender S, Pfeiffer M, Kohler U, Mack M. The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin. J. Bacteriol. 2008;190:1546–1553. doi: 10.1128/JB.01586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowitsch F, Kuhm C, Kellner R, Kalinowski J, Pelzer S, Macheroux P, Mack M. A novel N,N-8-amino-8-demethyl-d-riboflavin Dimethyltransferase (RosA) catalyzing the two terminal steps of roseoflavin biosynthesis in Streptomyces davawensis. J. Biol. Chem. 2011;286:38275–38285. doi: 10.1074/jbc.M111.292300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer M, Bacher A. Biosynthesis of flavocoenzymes. Nat. Prod. Rep. 2005;22:324–350. doi: 10.1039/b210142b. [DOI] [PubMed] [Google Scholar]
- 22.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 24.Matsui K, Wang H, Hirota T, Matsukawa H, Kasai S, Shinagawa K, Otani S. Riboflavin production by roseoflavin-resistant strains of some bacteria. Agric. Biol. Chem. 1982;46:2003–2008. [Google Scholar]
- 25.Mack M, van Loon AP, Hohmann HP. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 1998;180:950–955. doi: 10.1128/jb.180.4.950-955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coquard D, Huecas M, Ott M, van Dijl JM, van Loon AP, Hohmann HP. Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol. Gen. Genet. 1997;254:81–84. doi: 10.1007/s004380050393. [DOI] [PubMed] [Google Scholar]
- 27.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 28.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 29.Macheroux P, Kappes B, Ealick SE. Flavogenomics—a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.