Abstract
Regulation of bacterial gene networks by small non-coding RNAs (sRNAs) requires base pairing with messenger RNA (mRNA) targets, which is facilitated by Hfq protein. Hfq is recruited to sRNAs and mRNAs through U-rich- and A-rich-binding sites, respectively, but their distance from the sRNA–mRNA complementary region varies widely among different genes. To determine whether distance and binding orientation affect Hfq’s chaperone function, we engineered ‘toy’ RNAs containing strong Hfq-binding sites at defined distances from the complementary target site. We show that RNA annealing is fastest when the distal face of Hfq binds an A-rich sequence immediately 3′ of the target. This recruitment advantage is lost when Hfq binds >20 nt away from the target, but is partially restored by secondary structure that shortens this distance. Although recruitment through Hfq’s distal face accelerates RNA annealing, tight binding of six Us to Hfq’s proximal face inhibits annealing. Finally, we show that ectopic A-rich motifs dramatically accelerate base pairing between DsrA sRNA and a minimal rpoS mRNA in the presence of Hfq, demonstrating that proximity and orientation predict the activity of Hfq on long RNAs.
INTRODUCTION
In bacteria, small non-coding RNAs (sRNAs) perform post-transcriptional regulation of diverse gene expression pathways such as iron metabolism, carbon source use and osmotic shock (1–3). sRNAs often act by base pairing with complementary sequences in target messenger RNAs (mRNAs), either down-regulating gene expression by masking the ribosome-binding site (RBS) or up-regulating expression by opening an inhibitory structure and unmasking the RBS (Figure 1). Thus, an important feature of sRNA regulation is how stably and rapidly the two complementary RNAs interact and base pair (4,5).
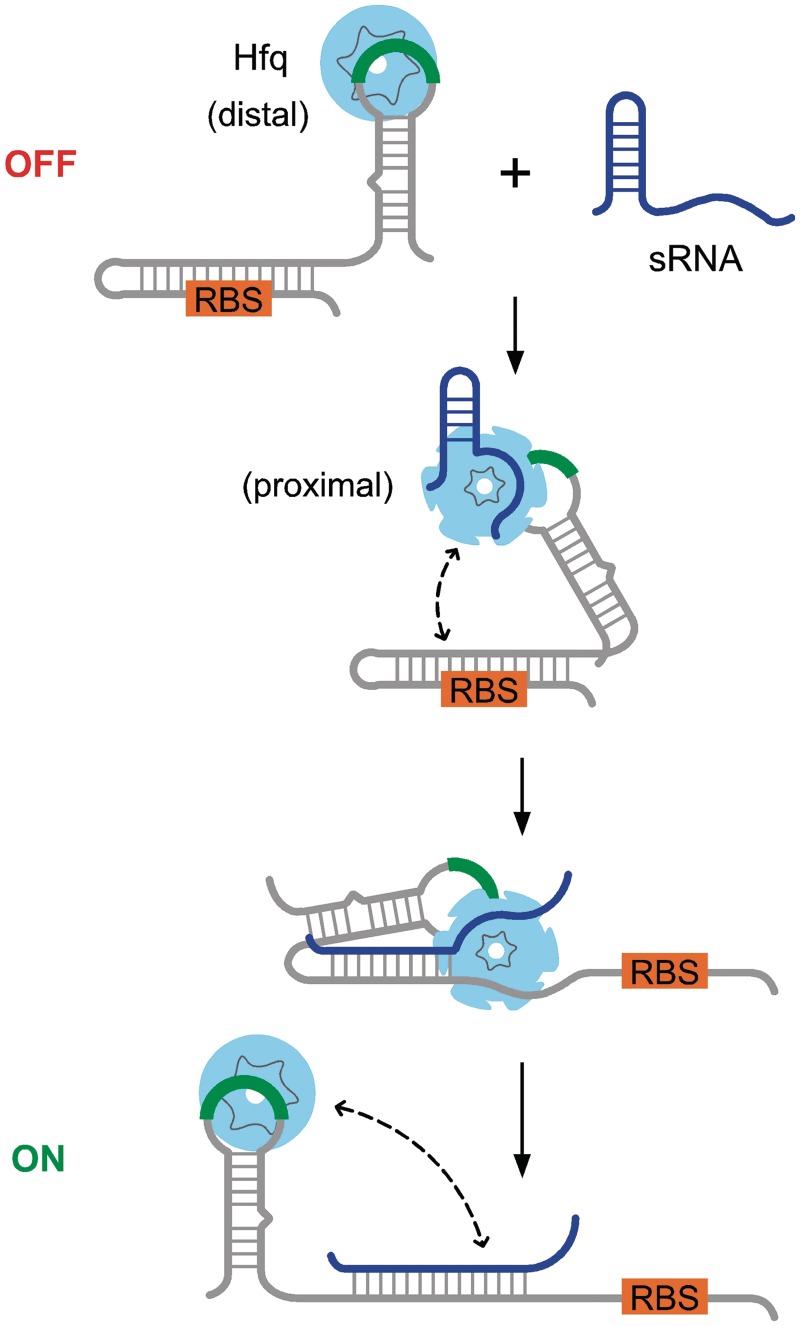
Figure 1.
Positive regulation of gene expression by Hfq and sRNAs. Secondary structure in the mRNA leader (gray) masks the ribosome-binding site and prevents initiation of translation. Expression of a complementary sRNA (blue) up-regulates translation by opening the mRNA and exposing the RBS. Hfq is recruited to (ARN)n motifs in the mRNA (green) through its distal face, facilitating sRNA binding through its proximal face and by restructuring the leader (31). Hfq cycles off the sRNA–mRNA duplex and may remain bound to the (ARN)n motif or transfer to other sequences in the mRNA. In this scheme, the Hfq-binding site (green) is far from sRNA-binding site in the mRNA.
sRNA–mRNA base pairing is facilitated in vitro by Hfq, a bacterial Sm-like protein that is required for sRNA regulation in Escherichia coli and other Gram-negative bacteria (6–8). The ring-shaped Sm core of the Hfq hexamer (9) forms two distinct single-stranded RNA-binding faces: a proximal face that binds U-rich RNA (10) and a distal face that binds A-rich sequences with a repeated ‘ARN’ motif (11,12). How Hfq facilitates annealing or exchange of RNA strands remains unclear, but experiments with oligonucleotides show that rapid RNA binding and release is necessary for Hfq’s chaperone activity (13–15), consistent with computational models of the reaction flux (16). Natural sRNAs compete for access to Hfq, increasing the rate at which RNA molecules cycle on and off the protein (17), and possibly promoting dissociation of Hfq from the complementary region, which is necessary for completion of sRNA–mRNA base pairing (18).
In addition to its dynamic interactions with the complementary region, there is considerable biochemical and genetic evidence that Hfq must be recruited to mRNA regulatory targets through (ARN)n motifs, which bind the distal face. ARN sequence motifs are present in many mRNAs regulated by sRNAs and Hfq (12,19). Experiments on rpoS mRNA showed that specific binding of Hfq to an upstream (AAN)4 binding site (Figure 1) is required for Hfq-dependent base pairing of DsrA (20) and RprA sRNAs (21) with the rpoS leader and up-regulation of rpoS translation in E. coli (22).
Although Hfq typically binds sRNAs near the mRNA complementary region (23–26), in mRNAs the distance between A-rich Hfq and sRNA-binding sites varies considerably. In sodB and shiA mRNAs, Hfq binds <20 nt upstream or downstream of RhyB sRNA (24,27). Beisel et al. (28) showed recently that mRNAs repressed by Spot 42 sRNA require an A-rich Hfq-binding site, preferably near the region complementary to Spot 42. By contrast, Hfq binds ARN motifs 40–60 nt upstream of the sRNA-binding site in fhlA mRNA (29) and 60–80 nt upstream of the sRNA-binding site in rpoS mRNA (20). The variable spacing between the sRNA- and Hfq-binding sites raises questions about the evolution of these genetic interactions and whether the physical proximity and orientation of Hfq is important for sRNA binding and posttranscriptional regulation.
To test whether Hfq location is important for its annealing activity, we constructed a series of ‘toy’ RNA substrates that mimic the functional elements of natural sRNAs and mRNAs. These short artificial substrates can be engineered to bind Hfq at defined locations, allowing us to control Hfq binding, sequence context and RNA secondary structure more systematically and rigorously than is possible in natural RNAs. To mimic natural mRNAs, strong Hfq-binding sequences (A18 or U6) were added at different distances from a common GC-rich target sequence, and the rate of strand annealing was measured in the presence of Hfq.
Our results show that recruitment of Hfq to the target RNA greatly increases the rate of strand hybridization even at very low Hfq concentrations and that this effect is greatest when an A-rich Hfq-binding site is placed immediately 3′ of the complementary region. We also show that RNA secondary structure can mitigate this proximity requirement. Finally, we validate these findings by showing that the insertion of (A)18 adjacent to the sRNA-binding site in a minimal rpoS mRNA increases the binding kinetics of DsrA sRNA 30–100 times in the presence of Hfq. From these results, we suggest that the RNA binding to the distal face serves to present the proximal face to the target and that the annealing rate decreases progressively as Hfq binds further from the target.
MATERIALS AND METHODS
Molecular beacon and RNA substrates
The rMBDss RNA molecular beacon (Supplementary Table S1) was synthesized and purified by reverse-phase high-performance liquid chromatography (Trilink Biotechnologies) as previously described (14). The beacon was modified with 6-FAM (5′) and C6-NH-DABCYL (3′). Substrate oligonucleotides (Supplementary Table S1) were either purchased (Invitrogen) or transcribed from DNA templates (IDT) and purified by 8% polyacrylamide gel electrophoresis. Concentrations were determined by absorption at 260 nm using the manufacturer’s extinction coefficient. DsrA, rpoS138, rpoS138-5′A18 and rpoS138-3′A18 RNA were transcribed in vitro as previously described (18) from plasmids pUCT7DsrA, pUCT7RpoS138, pUCT7RpoS138A18 and pUCT7RpoS138(486A18). The latter were constructed by inverse PCR of pUCT7RpoS138.
Hfq purification
Wild-type Hfq was over-expressed and purified using a Hi-Trap Co2+ column as previously described (22). RNA was not detected by absorption at 260 and 280 nm.
Beacon-target annealing kinetics
The association kinetics between beacon and target RNA in TNK buffer (10 mM Tris–HCl, pH 7.5, 50 mM NaCl and 50 mM KCl) at 30°C was measured using an Applied Photophysics SX 18MV stopped-flow spectrometer as previously described (30). Reactions contained 50 nM molecular beacon (final), 50 nM target RNA and 5–5000 nM Hfq (monomer) unless stated otherwise. The normalized change in fluorescence intensity, ΔF(t), was fit to a double exponential rate equation,
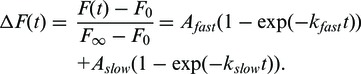 |
The observed rate constants for five or more trials were averaged.
Association kinetics of DsrA and rpoS RNA
The association kinetics of DsrA and a minimal rpoS mRNA leader (rpoS138) was measured by native gel mobility shift as previously described (18,31). Reactions contained either 32P-labeled DsrA and 200 nM rpoS RNA or the reverse, with or without 1 µM Hfq monomer. All reactions were performed at 30°C. The relative counts in bands corresponding to DsrA•rpoS (D•R) and DsrA•Hfq•rpoS (D•H•R) were plotted against time and fit to the biphasic rate equation above.
RESULTS
Basal Hfq activity on engineered RNAs
Hfq is thought to act by transiently binding two RNA strands in a ternary complex and cycling off the RNA as the new duplex is formed (13,14,16,18). Therefore, to measure Hfq’s basal annealing activity, we first designed a pair of complementary RNAs that lack specific binding sites for Hfq, so that Hfq would readily dissociate from the double-stranded product. Our basal 16 nt target (oligo C) was single-stranded and GC-rich (62.5%; Supplementary Table S1). The annealing activity of Hfq was measured using an RNA molecular beacon (rMBDss) complementary to oligo C. Molecular beacons and other fluorescent assays have been shown to reliably report the strand annealing and exchange activity of Hfq (13,30,32).
Competitive binding experiments showed that Hfq binds both oligo C and the RNA beacon poorly, as intended, with Kd = 1 µM Hfq monomer or 170 nM Hfq hexamer (Figure S1 and Supplementary Table S2). This ‘non-specific’ binding was much weaker than Hfq dissociation from natural RNAs [0.1–10 nM Hfq6; (33)] and coincided with assembly of the Hfq hexamer around 1 µM protomer in our experiments (34). We separately measured the stability of the oligo C-beacon duplex (Kd = 3.4 nM) and found that Hfq reduced the final extent of base pairing (Figure S2) as previously observed for a different RNA–beacon pair (30). This suggested that Hfq binds the double-stranded RNA even less well than the single strands, favoring product release.
Consistent with its binding affinity for oligo C and the molecular beacon, the annealing kinetics peaked around 1 µM Hfq (Figure 2b; black line). The base pairing kinetics was measured by stopped-flow fluorescence at 30°C in 50 nM oligo C and 50 nM beacon. In <100 nM protomer, Hfq had no effect on the annealing rate. At 300 nM Hfq, however, the rate of annealing began to increase, reaching a maximum 6-fold acceleration in 1 µM Hfq relative to the no Hfq background. Thus, in the absence of specific RNA interactions, Hfq catalyzed a modest but measurable acceleration of helix formation. Since oligo C binds Hfq weakly, more Hfq is required to see an effect. Furthermore, oligo C is more likely to dissociate from Hfq before base pairing with the complementary beacon commences (14), resulting in more futile cycles of binding and release.
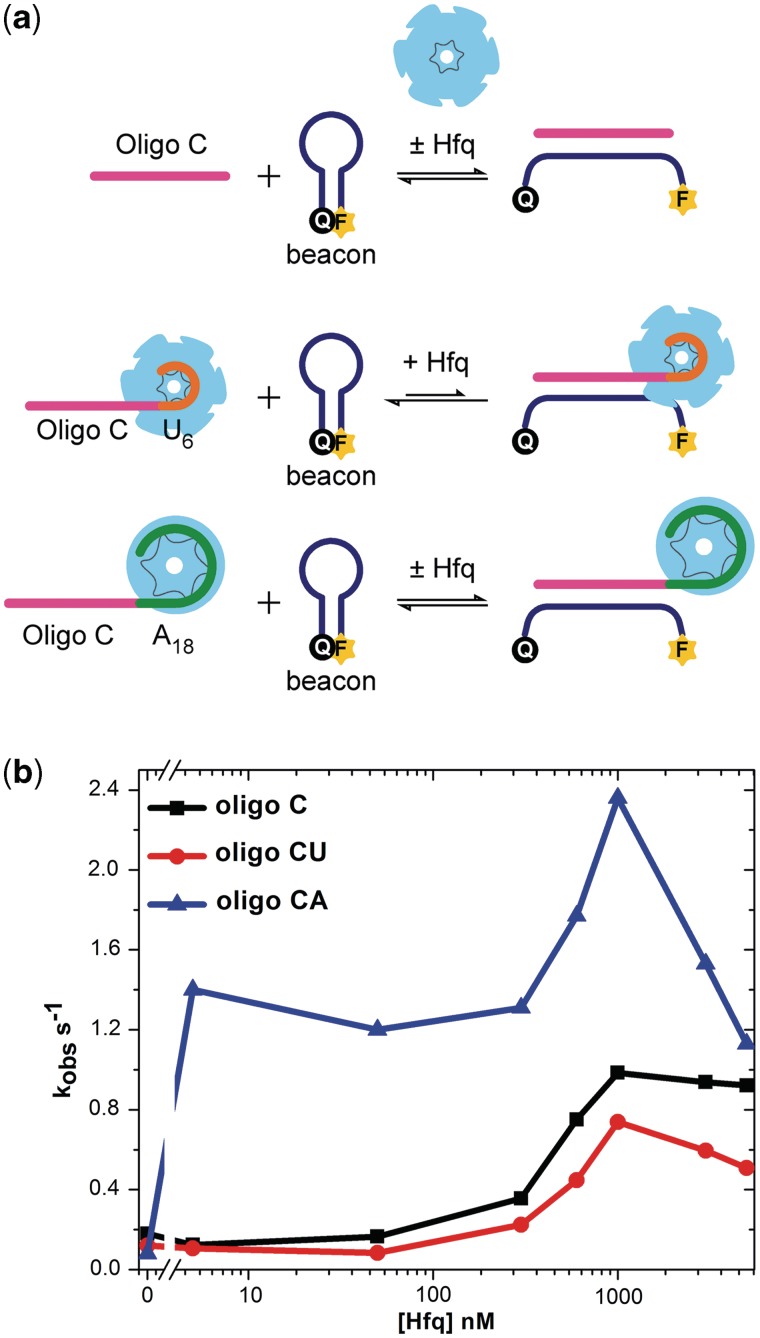
Figure 2.
A-rich Hfq-binding site stimulates RNA annealing. (a) Toy RNAs for Hfq-annealing assays. Base pairing of a molecular beacon (blue) with 16 nt oligo C target RNA (magenta) increases FAM fluorescence intensity. Oligo CA and oligo CU contain A18 and U6 Hfq-binding sites, respectively. (b) Rates of RNA annealing were measured by stopped-flow fluorescence in TNK buffer at 30°C, using 50 nM beacon (rMBDss), 50 nM target RNA and 5–5000 nM Hfq monomer. Observed rate constants from five independent experiments were averaged and plotted against Hfq concentration.
A-rich-binding site for Hfq accelerates RNA annealing
We next asked whether Hfq could facilitate annealing more efficiently if the ‘toy’ RNA contains a strong Hfq-binding site that could recruit the protein to the complementary region. A-rich sequences interact specifically with the distal face of Hfq (11), whereas rU6 forms a tight complex with the proximal face of Hfq (35). When A18 or U6 were appended to the 3′-end of oligo C (oligo CA and oligo CU, respectively), the affinity for Hfq increased dramatically as expected, with Kd’s of 2.7 and 0.65 nM, respectively (Supplementary Figure S1 and Supplementary Table S2). The terminal A’s had little effect on the stability of base pairs between the complementary region and the beacon in the absence of Hfq (Kd = 4.5 nM; Figure S2).
When A18 was appended to the 3′-end of the target, the RNA annealing rate increased strongly in the presence of Hfq, compared to reactions with oligo C (Figure 2b; blue line). This was particularly noticeable in 5 nM Hfq, at which the annealing kinetics for oligo CA were 18-fold faster than the no Hfq control (Figure 2b) and 15-fold faster than oligo C with Hfq. A peak in activity, corresponding to a 30-fold enhancement compared with the no Hfq background, was again observed around 1 µM Hfq. Therefore, specific interactions between Hfq and A18 increased the RNA annealing kinetics, at low and high Hfq concentrations, showing that recruitment of Hfq to the target RNA is important for its chaperone function as observed in mRNAs. Above 1 µM Hfq, the rate of RNA annealing decreased rapidly, presumably due to formation of inert multi-hexamer complexes (34) (Figure S2).
In contrast to these results, addition of U6 to the 3′-end of the target failed to increase the RNA annealing rate in the presence of low amounts of Hfq (Figure 2; red line), despite the fact that U6 binds Hfq very tightly. Thus, although both U6 and A18 sequences increased the affinity of the RNA for Hfq, only A18 resulted in faster annealing between the target region and the RNA beacon. This result suggested that the orientation of Hfq with respect to the target region is important.
Hfq activity depends on target proximity
Among bacterial mRNAs known to interact with Hfq, the Hfq-binding sites are located both 5′ and 3′ and at variable distances from the sRNA-binding site. To address whether the location of bound Hfq affects its ability to facilitate RNA base pairing, we inserted a poly C spacer (S) between the complementary (C) region and the A18 Hfq-binding site (Figure 3a and Supplementary Table S1). Hfq binds cytidine more weakly than other nucleotides (12,19), and thus the spacer is expected to be neutral with respect to Hfq binding. The annealing rate of each RNA substrate with the fluorescent molecular beacon was measured at different Hfq concentrations as before (Figure S3).
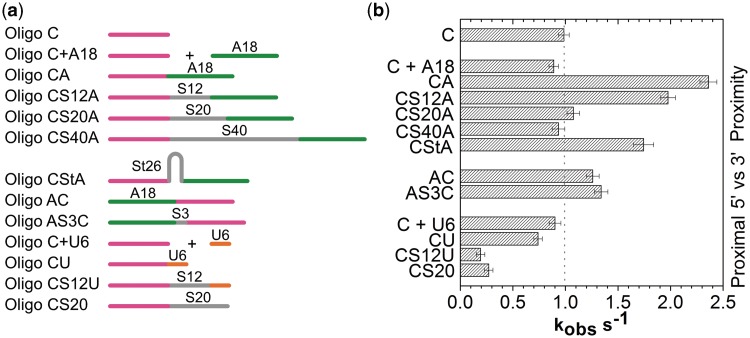
Figure 3.
Position of Hfq-binding site is important for RNA annealing. (a) Scheme of target RNAs listed in Supplementary Table S1. (b) Observed rate constants (kobs) obtained from the binding kinetics between the molecular beacon and different RNA targets (left) in 1 µM Hfq monomer. Rate constants from five independent experiments were averaged and standard deviations indicated by error bars. See Figures S3–S5 for additional data.
We observed a strong correlation between the proximity of the A18-binding site and the rate of RNA annealing (Figure 3b), demonstrating that the catalytic ‘reach’ of a bound Hfq is limited. As the spacer was increased from 0 to 12 nt (oligo CS12A), the annealing kinetics in 1 µM Hfq dropped from 2.4 to 2.0 s−1 and was similar to the basal rate for oligo C (1 s−1) when the spacer was 20–40 nt long. A similar but even more pronounced trend was observed in 5 nM Hfq (Figure S3). The annealing rate depended on having A18 in the substrate, as appending only a 20-nt spacer (Figure. 3; CS20) or adding A18 separately to the reaction (C + A18; Figure 3) had no effect. Thus, recruitment of Hfq through its distal face is needed for its full activity, but this activity decreases to zero when Hfq is located >20 nt from the target region.
We next tested whether secondary structure can overcome this distance effect by shortening the spatial distance between Hfq and the target. A 26-nt stem loop inserted between the complementary target sequence and A18 resulted in faster annealing (oligo CStA, 1.7 s−1; Figure 3a) compared with the unstructured 20 nt linear spacer (oligo CS12A, 1.1 s−1), but was less effective than placing A18 immediately next to the target (oligo CA, 2.4 s−1). Thus, an increase in physical proximity caused by RNA secondary structure partially overcomes the effect of moving the Hfq-binding site further away in the RNA sequence. Curiously, the stem loop did not improve the annealing kinetics in 5 nM Hfq (Figure S3). As discussed below, Hfq may reach the target differently when the RNA has intervening secondary structure.
Orientation of Hfq binding
As A-rich Hfq-binding sites are often upstream of the sRNA-binding site in natural mRNAs, we next addressed whether Hfq’s activity is directional. In one substrate, we appended A18 to the 5′-end with no spacer (oligo AC) and in the other, we inserted a 3-nt spacer between A18 and the target (oligo AS3C; Figure 3a). In 1 µM Hfq, both RNAs base paired with the molecular beacon slightly more rapidly than with oligo C (1.3 vs. 1 s−1), but less rapidly than oligo CA. Thus, in this context, Hfq can stimulate annealing when bound either upstream or downstream of the target, but is more effective when located 3′ of the sRNA-binding site.
Inhibition by U-rich RNA
Finally, we tested whether the annealing activity changed when Hfq was recruited through interactions between its proximal face and a U6-binding site. As noted above, when U6 was placed immediately 3′ of the target, there was no improvement in Hfq activity. When we introduced a 12-nt spacer (oligo CS12U; Figure 3b and Figure S5), RNA annealing became slower compared with oligo C alone, or when U6 was added in trans (C + U6). One explanation for this unexpected result is that U6 strongly sequesters the proximal face of Hfq. As the distance to the target increases, the probability that Hfq can interact productively with the target sequence and molecular beacon is lowered, thereby inhibiting the annealing reaction. Thus, the geometry and orientation of Hfq, as well as its proximity to the RNA-complementary region, are important for annealing.
Distal face binding required for Hfq action
The results above show that an A-rich Hfq-binding site near the target sequence increases the rate of RNA annealing in the presence of Hfq by recruiting the protein to the target RNA via interactions with the distal face of Hfq. By contrast, a U-rich sequence that interacts with the proximal face of Hfq inhibited annealing. To further test the binding orientation of Hfq, we compared the effects of proximal and distal face mutations in Hfq on its annealing activity. As predicted, the Y25D mutation on the distal face of Hfq severely reduced its ability to facilitate annealing of oligo CA with the RNA beacon (Figure 4). The loss of activity was apparent in both 5 nM and 1 µM Hfq, but was most severe in 5 nM Hfq in which assembly of Hfq on the A18-binding site is essential. By contrast, a proximal face mutant (K56A) retained some activity on these substrates, particularly at 1 µM protein (Figure 4). These results were consistent with previous results showing that recruitment of Hfq to an A-rich site in rpoS mRNA is essential for sRNA binding and up-regulation of rpoS translation, while sequence-specific proximal face interactions with DsrA are helpful but dispensable (22,31).
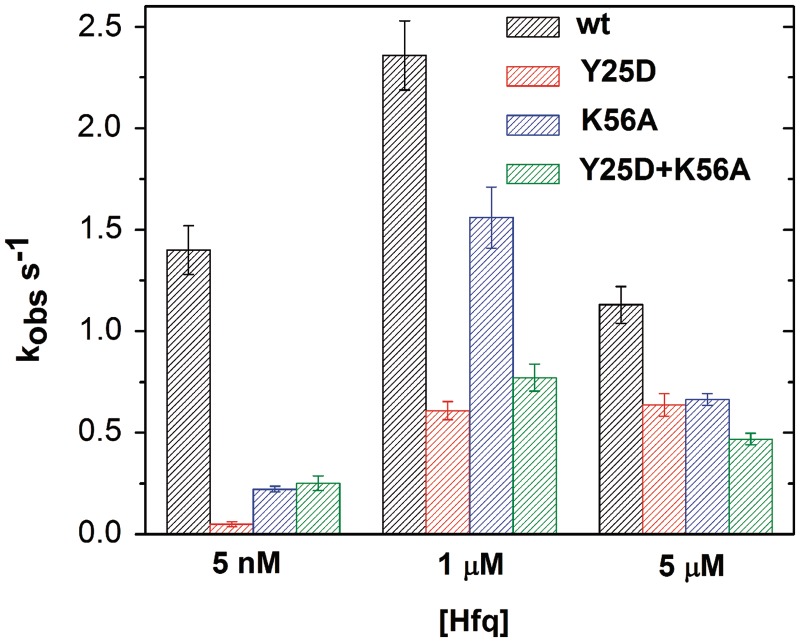
Figure 4.
Hfq distal face interaction is required. Observed annealing rates for 50 nM molecular beacon and 50 nM oligo CA in Hfq. The Y25D mutation on the distal face (red) was more deleterious than the K56A mutation on the proximal face (blue). To test for complementation, Y25D and K56A Hfq subunits were mixed 1:1 (green). Total Hfq concentrations were 5 nM, 1 µM or 5 µM monomer.
We next asked whether the distal and proximal face mutations could compensate each other, by mixing Y25D and K56A Hfq subunits in equal proportion. In 5 nM total Hfq, the mixture was slightly more active than either individual mutant, possibly because assembly of mixed Hfq hexamers at this low concentration (34) recovered partial function of each binding surface. At 1 µM Hfq (total), the mixture had similar activity as Hfq:Y25D, and at 5 µM, the mixture was least active. The failure of mutant subunits to compensate each other was consistent with RNA binding to both faces of a single hexamer (11,15).
Ectopic A-rich-binding sites stimulate sRNA binding to minimal rpoS leader
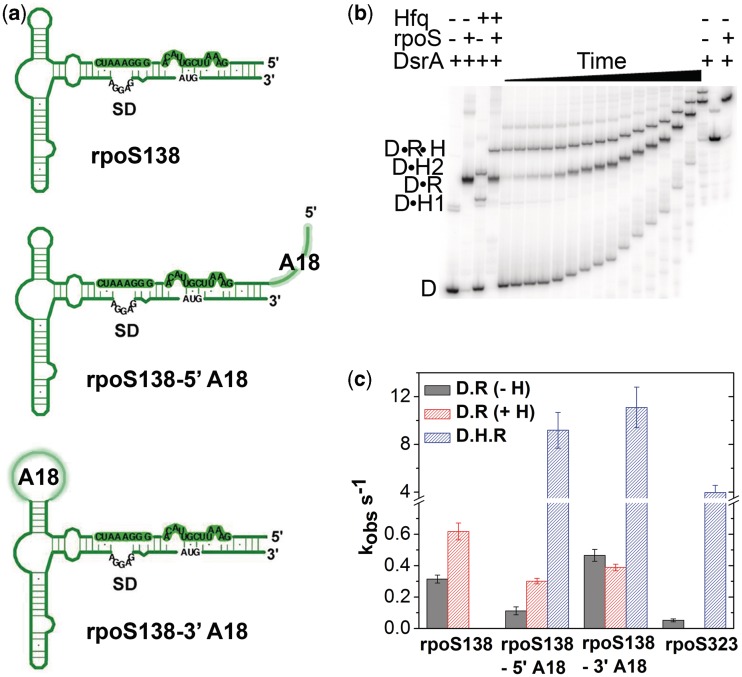
We next asked whether the locations of A-rich binding sites could predict the activity of Hfq on annealing of large RNA substrates. To address this question, we measured binding of DsrA sRNA to a minimal (138 nt) form of the rpoS mRNA leader (Figure 5a) that lacks upstream A6 and (AAN)4 Hfq-binding sites needed to facilitate sRNA binding (20). Native gel mobility shift assays in the presence or absence of 1 µM Hfq confirmed that annealing between DsrA sRNA and minimal rpoS138 increased only 2-fold (Figure S6), as previously reported (18).
Figure 5.
Close Hfq distal binding sites improve sRNA binding to rpoS mRNA. (a) Secondary structure of minimal rpoS138 leader sequence with and without A18 insertions. (b) DsrA and rpoS binding at 30°C was measured using radiolabeled DsrA and 200 nM rpoS138-5′A18 RNA in 1 µM Hfq. Samples were loaded on a 6% native gel from 0.1 to 60 min; gels were run continuously during the experiment. Control reactions with DsrA only, DsrA plus 200 nM rpoS, DsrA plus 1 µM Hfq and DsrA plus rpoS plus Hfq were incubated at 30°C for 2 h and loaded as indicated in the figure. D, free DsrA; D•H1, DsrA bound to one Hfq hexamer; D•H2, DsrA with two Hfq hexamers; D•R, DsrA•rpoS; D•R•H, DsrA•rpoS•Hfq ternary complex, which persists when the mRNA contains an A-rich-binding site for Hfq. (c) Observed rate constants for annealing; no Hfq (D•R, gray); binary complexes with Hfq (D•R, red); ternary complexes with Hfq (D•R•H, blue). Error bars indicate the standard deviation from the average of three trials.
We next attempted to rescue the minimal rpoS138 RNA by adding A18 to the 5′-end of rpoS138 (rpoS138-5′A18) or inserting A18 in the loop at position 486, just 3′ of the sRNA-binding region (rpoS138-3′A18; Figure 5a). Neither of these positions contain many A residues in wild-type rpoS. Native gel mobility shift assays showed that 32P-labeled DsrA formed both binary and ternary complexes with the enhanced rpoS138 and Hfq, indicating that Hfq binds this RNA more tightly than its binds minimal rpoS138 (Figure 5b and Figure S7–S8).
When A18 was placed 5′ of the DsrA-binding site, the rate of annealing based on appearance of the ternary complex was 80 times faster in 1 µM Hfq (kobs = 9.2 min−1) than without Hfq (0.11 min−1; Figure 5c). The same experiment with 32P-labeled rpoS138-5′A18 (data not shown) resulted in 150-fold faster annealing in the presence of Hfq (17.7 vs. 0.13 min-1). Therefore, an artificial Hfq-binding site accelerates annealing in a long, structured RNA as well as a short, unstructured RNA.
To test whether Hfq can also function when bound downstream of the sRNA-binding site in rpoS, we replaced the UUAUUU motif at nt 486–491 with A18 (Figure S8). Once again, Hfq stimulated annealing between 32P-labeled DsrA and rpoS138-3′A18, increasing the kinetics 24-fold (11.1 vs. 0.47 min−1; Figure 5c). A similar rate enhancement was obtained with labeled rpoS138-3′A18 and unlabeled DsrA (42 vs. 0.45 min−1; data not shown). Thus, Hfq is again more potent when bound 3′ of the sRNA target.
Remarkably, both rpoS138-A18 variants base paired with DsrA more readily than a longer rpoS mRNA (rpoS323) containing the natural upstream A-rich motifs (Figure 5c). This is likely because the structure of the rpoS leader inhibits sRNA binding in the absence of Hfq (20). Nonetheless, these results demonstrate that proximity of Hfq to the sRNA predicts its ability to facilitate sRNA–mRNA pairing.
DISCUSSION
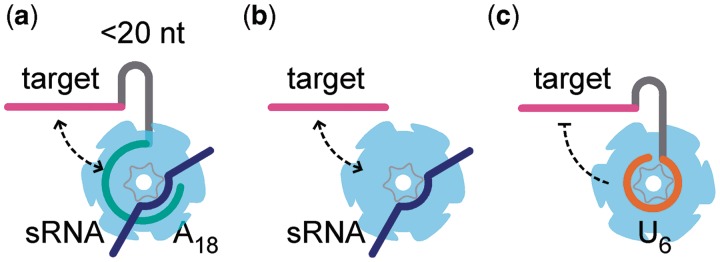
Our results with simple ‘toy’ RNA targets show that the proximity of an A-rich site greatly enhances Hfq’s ability to act on the target region. This agrees with genetic and biochemical evidence that Hfq must be recruited to the mRNA via its distal face for sRNA pairing in vitro and sRNA-dependent regulation in vivo (20,21,28). This is not a simple ‘tethering’ effect, as we observe a preference for 3′-orientation and find that tight binding to a U6 site interferes with annealing. Instead, we propose that binding of Hfq’s distal face to an A-rich sequence not only increases the local concentration of Hfq but also orients the protein so that the complementary target region has maximum opportunity to interact with the proximal face (Figure 6). This idea is consistent with evidence that the complementary strands compete for binding to the proximal face (15).
Figure 6.
Model for Hfq recruitment to sRNA complementary region. Binding of Hfq’s distal face to an (ARN)n motif facilitates interactions between the target region and the proximal face of Hfq by looping around the hexamer (a) or possibly by direct transfer from one site to another (b). Stable interactions between the proximal face and U6 inhibit annealing (c).
The benefit from recruiting Hfq to a specific RNA-binding site diminishes rapidly as the distance to the complementary region lengthens, so that by 20 nt, it is barely different from Hfq’s basal activity on non-specifically bound oligo C RNA (Figure 3). This is surprising insofar as the distance between the Hfq- and sRNA-binding sites is much greater than 20 nt in certain natural mRNAs such as rpoS. Nonetheless, we find this proximity principle for annealing also applies to long, structured RNAs. First, we observed a similar recruitment effect and 3′-bias when an ectopic A-rich Hfq-binding site was inserted immediately 5′ or 3′ of the sRNA-binding site in rpoS mRNA (Figure 5). Second, the distance dependence we observe is consistent with the results of Beisel et al. (28), who found that engineered regulation by Spot 42 ideally requires an A-rich Hfq-binding site within 14 nt of the sRNA target sequence.
The dependence on Hfq concentration suggests that Hfq can reach the complementary target region through at least two paths, which depend on the structure of the mRNA (Figure 6). In one path, binding of Hfq’s distal face to an A-rich sequence adjacent to the target region allows the target region to interact simultaneously with a complementary strand on the proximal face (Figure 6a). Because Hfq binds A18 with Kd = 3 nM, this path functions even at low (5 nM) Hfq (Figure 2b) and is likely to be particularly important when Hfq is limiting. Due to the entropic penalty for looping unstructured RNA, this path becomes less effective as the spacer RNA grows longer, although the ability of the RNA to engage both faces of Hfq likely also depends on interactions with the flexible C-terminal domain (36).
In a second path, the proximal face of Hfq binds the complementary region directly, with little or no specificity (Figure 6b). In our experiments, this path requires 1 µM Hfq, because oligo C and its complementary beacon bind Hfq weakly. Less Hfq may be needed for RNAs that bind Hfq’s proximal face with some selectivity, such as DsrA. Recruitment of Hfq to a specific A-rich-binding site may contribute to non-specific interactions with the target region if Hfq can slide or hop along the RNA, as observed for some DNA-binding proteins. Similarly, a stem loop in the spacer (oligo CStA) may stabilize interactions with Hfq’s proximal face (37) or facilitate hopping from a specific to a non-specific site in 1 µM Hfq (cyan bar; Figure S3).
Although Hfq remains bound to the (AAN)4 motif in rpoS mRNA, resulting in accumulation of the sRNA•mRNA•Hfq ternary complex in vitro, previous biochemical data suggest that Hfq cycles off the sRNA–mRNA anti-sense duplex (18), releasing its binding site in DsrA which is dispensable for forming a ternary complex in vitro (31). Based on these results, we previously concluded that Hfq acts mainly by restructuring rpoS mRNA (31). Although restructuring may indeed be an important component of rpoS regulation, our results here show that Hfq also becomes a more potent annealer when recruited to the target RNA. In other words, the orientation and location of Hfq on the mRNA is important because it positions the proximal face to transiently engage and anneal complementary regions in DsrA and rpoS mRNA. This recruitment effect allows Hfq to act on specific biological targets, even in a large background of nonspecific RNAs.
Experiments to test the importance of Hfq proximity in vivo are in progress. Preliminary results showing that an artificial A18 sequence placed next to the sRNA binding region can rescue up-regulation of rpoS–lacZ fusions that lack the natural (AAN)4 Hfq-binding motif, corroborating the main results here. However, Hfq plays many roles in sRNA regulation—besides facilitating sRNA–mRNA base pairing, it also binds and protects sRNAs and recruits other proteins to the mRNA (6). Thus, although in vitro experiments can dissect individual steps, there is more to learn about how Hfq’s various activities are coordinated in vivo.
Why are Hfq-binding sites located far upstream of their regulatory targets in certain mRNAs? First, mRNAs with distant Hfq-binding motifs, such as rpoS, are highly structured, and the mRNA structure may bring Hfq close in space to the sRNA-binding site. Competitive dissociation experiments indicate that fhlA mRNA binds the distal and proximal faces of Hfq simultaneously, suggesting mRNAs can wrap around the Hfq hexamer (29). Second, the structure of the RNA may favor transfer of Hfq from one site to another in the mRNA. Third, the structure of the mRNA structure may optimize aspects of post-transcriptional regulation other than sRNA annealing. For example, our in vitro results suggest that Hfq could facilitate sRNA binding even better if it bound further downstream. However, the full-length rpoS leader with its upstream (AAN)4 binding site inhibits sRNA annealing more than short leaders, providing tighter control over basal rpoS expression in the absence of Hfq and sRNAs (20,22,38). Although more work is needed to understand how these factors contribute to sRNA regulation in bacteria, our results provide a framework for understanding how the location and orientation of Hfq binding controls its annealing activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–8.
FUNDING
National Institutes of Health [R01 GM46686]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Storz and Gottesman laboratories for helpful discussion.
REFERENCES
- 1.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaberdin VR, Blasi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao Y, Zhang ZJ, Erickson DW, Huang M, Huang Y, Li J, Hwa T, Shi H. Quantifying the sequence-function relation in gene silencing by bacterial small RNAs. Proc. Natl Acad. Sci. USA. 2011;108:12473–12478. doi: 10.1073/pnas.1100432108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron MP, Lafontaine DA, Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7:140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 8.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 10.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins JF, Panja S, Woodson SA. Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res. 2011;39:5193–5202. doi: 10.1093/nar/gkr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang W, Arluison V, Hohng S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011;39:5131–5139. doi: 10.1093/nar/gkr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson DN, Lim HN. Essential requirements for robust signaling in Hfq dependent small RNA networks. PLoS Comput Biol. 2011;7:e1002138. doi: 10.1371/journal.pcbi.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–3808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 22.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Wu J, Liu Q, Zhao Y, Ying X, Cha L, Wang L, Li W. sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments. RNA. 2010;16:2051–2057. doi: 10.1261/rna.2193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 2007;64:1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 28.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS One. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 34.Panja S, Woodson SA. Hexamer to Monomer Equilibrium of E. coli Hfq in Solution and Its Impact on RNA Annealing. J. Mol. Biol. 2012;417:406–412. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Blasi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunning C, Brown L, Elliott T. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 1998;180:4564–4570. doi: 10.1128/jb.180.17.4564-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.