Abstract
The bHLH transcription factor MyoD, the prototypical master regulator of differentiation, directs a complex program of gene expression during skeletal myogenesis. The up-regulation of the cdk inhibitor p57kip2 plays a critical role in coordinating differentiation and growth arrest during muscle development, as well as in other tissues. p57kip2 displays a highly specific expression pattern and is subject to a complex epigenetic control driving the imprinting of the paternal allele. However, the regulatory mechanisms governing its expression during development are still poorly understood. We have identified an unexpected mechanism by which MyoD regulates p57kip2 transcription in differentiating muscle cells. We show that the induction of p57kip2 requires MyoD binding to a long-distance element located within the imprinting control region KvDMR1 and the consequent release of a chromatin loop involving p57kip2 promoter. We also show that differentiation-dependent regulation of p57kip2, while involving a region implicated in the imprinting process, is distinct and hierarchically subordinated to the imprinting control. These findings highlight a novel mechanism, involving the modification of higher order chromatin structures, by which MyoD regulates gene expression. Our results also suggest that chromatin folding mediated by KvDMR1 could account for the highly restricted expression of p57kip2 during development and, possibly, for its aberrant silencing in some pathologies.
INTRODUCTION
The muscle regulatory factor MyoD (1) plays a key role in myogenesis by coordinating the induction of muscle-specific genes with the activation of growth arrest pathways during terminal differentiation of skeletal muscle cells (2,3). MyoD is a bHLH transcription factor recognizing E-box elements and acting in cooperation with other muscle-specific and general transcription factors (4). MyoD regulates a large number of genes with diverse biological functions, by activating or repressing their expression at different times during myogenesis (5). Some targets are directly activated by MyoD as early myogenic events. Others, in contrast, require the induction of intermediate factors, in some cases cooperating with MyoD itself in a feed-forward circuitry (6). The MyoD property to function as a master regulator of myogenesis is also based on its ability to access chromatin and to induce its reorganization by recruiting chromatin-remodeling complexes and histone-modifying enzymes (7–10).
The up-regulation of genes coding for cyclin-dependent kinase inhibitors (CKI), in particular p21cip1/waf1 and p57kip2 (p57), plays a crucial role in MyoD-induced growth arrest (11,12). Although genetic analysis indicated that p21 and p57 are individually dispensable during myogenesis, it is still unclear whether the two CKIs act by controlling the same process or their redundancy involves compensatory mechanisms. p57 participates in myogenesis not only by controlling cell cycle but also by supporting MyoD function in a positive feed-back loop (13,14), a unique property not shared with p21.
p57 is a critical regulator in many tissues, and its deficiency causes severe developmental anomalies both in humans and in mice (15). Genetic studies suggested a role for p57 in Beckwith–Wiedemann syndrome (BWS), a human growth disorder characterized by multiple developmental defects and predisposition to cancer (16). Moreover, genetic ablation of p57 in mice recapitulates many aspects of the syndrome (17,18).
Unlike other CKIs, p57 shows a highly cell-specific, temporal and spatial expression pattern, probably related to its ability to exert specific functions (19). The expression of p57 is subject to a complex epigenetic regulation. p57 is located within a conserved cluster of imprinted genes on mouse chromosome 7 and human chromosome 11p15 and is expressed only by the maternal allele in both species (20,21). This cluster, involved in growth regulation and development, is organized into two independently imprinted sub-domains, both associated with BWS in humans: the first contains Igf2 and H19 genes; the second includes, among other genes, p57, kcnq1 and kcnq1ot1 (22). A differentially methylated region, KvDMR1, located about 150 kb downstream of p57, controls in cis the imprinting of several genes of the sub-domain (23,24). Two mechanisms of action have been proposed to explain the cis-acting repressive effect of KvDMR1 (25). The first one could involve the enhancer-blocking activity of this region. The second one could involve chromatin silencing by a long non-translated ribonucleic acid (RNA). This transcript is coded by the maternally imprinted gene Kcnq1ot1, which promoter is comprised within KvDMR1. Through some complex mechanism not yet clarified, the activity of KvDMR1 is associated with hypermethylation of p57 promoter in the repressed paternal allele (26).
We have previously reported that, during muscle differentiation, p57 is induced at the transcriptional level by MyoD, through an indirect mechanism requiring new-synthesized factors (27). In addition, we have defined a p57 promoter proximal region as responsive to MyoD trans-activation in reporter assays. We have also reported that the MyoD-dependent induction of p57 shows a cell-type restriction. In particular, we observed that different cell types, equally capable to undergo muscle conversion on exogenous-MyoD expression, are differentially responsive regard the induction of p57 (11,27,28). Taking advantage of this experimental tool, we found that the up-regulation of p57 requires, besides the induction of intermediate factors, also the removal of a cis-acting constraint associated with promoter deoxyribonucleic acid (DNA) methylation (28). In this study, we provide evidence that the release from the epigenetically repressed state of p57 involves a functional interaction of MyoD with a distant cis-element located within KvDMR1. We show that MyoD binds in vivo to this region in responsive cells and that this interaction causes the removal of a chromatin loop between KvDMR1 and p57 promoter.
MATERIALS AND METHODS
Cell cultures
Mouse C3H10T1/2, C57BL/6 and (C57BL/6 X SD7)F1 fibroblasts and mouse C2.7 myoblasts were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). To trigger differentiation, myoblasts and MyoD-infected fibroblasts were grown to confluence, shifted to differentiation medium (DMEM-0.5% FCS) and analyzed at the indicated times. K562 cells were grown in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FCS.
Production of empty pBabe and pBabe-MyoD-expressing retroviruses, retroviral infections and MyoD-induced differentiation were performed as described previously (28).
Gene expression analysis
Total cellular RNA was extracted with ‘High Pure RNA Isolation Kit’ (Roche Diagnostics); 1 μg of total RNA was reverse-transcribed at 42°C for 1 h in the presence of MMLV reverse transcriptase (Promega).
Semiquantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analyses were performed as described previously (28). p57, kcnq1 and Hprt (used as reference gene) transcripts were amplified with the primers reported in Supplementary Methods.
Quantitative Real Time-PCR (qRT-PCR) reactions were performed in 20 μl of reaction buffer containing 1 µl of diluted complementary DNA (cDNA), 10 µl of “Go Taq qPCR Master Mix” (Promega) and each primer at the optimized final concentration. The primer sequences are reported in Supplementary Methods. The reaction was performed in the thermocycler ‘MiniOpticon Real-Time PCR detection system’ (Bio-Rad). The primer pair efficiency, the normalized expression [ΔΔC(t)] and the standard error of the mean were determined with CFX Manager™ software (Bio-Rad). 18S rRNA and Hprt were used as reference genes.
To assay the allelic expression status of p57 and Kcnq1, cDNAs from MyoD-infected (C57BL/6 x SD7)F1 fibroblasts were amplified by RT-PCR. Maternal and paternal products were distinguished by restriction fragment length polymorphism (RFLP) analysis of previously described polymorphic restriction sites (Supplementary Methods).
Chromatin immunoprecipitation and allele-specific chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out as described (28). Chromatin was immunoprecipitated with anti-MyoD antibody (sc-760). Genomic regions of interest were amplified using the same DNA quantity for each sample, the primer pairs reported in Supplementary Methods and the following protocol. After 5-min incubation at 95°C, 30 cycles of PCR (95°C for 30 s, 56–58°C for 30 s and 72°C for 30 s) were performed. Quantitation of immunoprecipitated F3 and myogenin (myog) promoter fragments was performed in triplicate using 5 ng of DNA, Go Taq qPCR Master Mix (Promega) and the specific qRT-PCR primer pairs (Supplementary Methods) at the final concentration of 150 nM. The reaction was performed in the real-time thermocycler. The primer pair efficiency, the relative quantity of each immunoprecipitated and no antibody [ΔC(t)] respect to input sample and the standard deviation of the relative quantity were determined with CFX Manager™ software (Bio-Rad). The percentage of the relative quantity of each immunoprecipitated sample was normalized to the percentage of the relative quantity of each no antibody sample.
The parental alleles were distinguished by single-strand conformation polymorphism (SSCP)-PCR using F1 or F3 primers (Supplementary Methods) labeled at their 5′-ends with [γ-32P] Adenosine 5′′-triphosphate (γ-[32P]ATP). After denaturation, PCR products were resolved by non-denaturing acrylamide gel electrophoresis and detected by autoradiography.
Enhancer-blocking assays
Plasmids are described in Supplementary Methods. Four micrograms of each Not-I-linearized plasmid were co-transfected with 4 μg of the plasmid containing unique EcoRI cloning site and Moloney sarcoma virus long terminal repeat (EMSV) empty vector or EMSV MyoD vector and with 0.4 μg of the plasmid Renilla Luciferase-Thymidine Kinase (pRL-TK) plasmid (Promega), coding for renilla-luciferase and used to control the transfection efficiency, into K562 cells. Plasmids were introduced into 0.8 ml of cell suspension (5 × 106 cells/ml) in a 4-mm-gap cuvette (Bio-Rad Laboratories) by electroporation using a Gene Pulser II (Bio-Rad Laboratories) at 250 V and 960 μF. Following electroporation, cells were cultured in 10 ml of RPMI-10% FCS for 48 h. Renilla activities were measured using a Dual-Luciferase Reporter Assay System kit (Promega); and 3 × 105 transfected cell were plated on 6-cm cell culture dishes in 7.5 ml of soft-agar plating medium containing 800 μg/ml active G418 (Calbiochem). Antibiotic-resistant colonies were counted after 3 weeks. In a given experiment, each construct was tested in triplicate, and experiments were repeated three times.
Chromosome Conformation Capture (3C) assay
The 3C assay was performed as described previously (29). Briefly, chromatin was crosslinked with 1% formaldehyde, and nuclei were isolated with lysis buffer (10 mM Tris-HCl, pH 7.5; 10 mM NaCl; 5 mM MgCl2; 0.1 mM Ethylene Glycol Tetraacetic Acid (EGTA); 1X complete protease inhibitor, 11836145001 Roche). DNA was digested with 400 units of NcoI restriction enzyme and ligated in 1X ligation buffer New England Biolabs (NEB). Ligation products were extracted with phenol–chloroform, precipitated with sodium acetate and ethanol, washed with 70% (v/v) ethanol and re-suspended in 150 μl of distilled water. To maximize template accessibility, an additional digestion step with EcoRI was performed. After phenol–chloroform extraction, DNA was precipitated with sodium acetate and ethanol, washed with 70% (v/v) ethanol and re-suspended in 150 μl of distilled water; 10 ng of each sample were used for PCR analysis performed in triplicate. The amplification products were verified by sequencing (data not shown). Primer sequences are available on request.
The positive control plasmid, containing a ligation product of p57 promoter and KvDMR1 sequences, was realized as described in Supplementary Methods. Digested but not-ligated chromatin and un-digested and not-ligated chromatin were used as negative controls.
RESULTS
MyoD co-induces the expression of p57 and kcnq1 during muscle differentiation
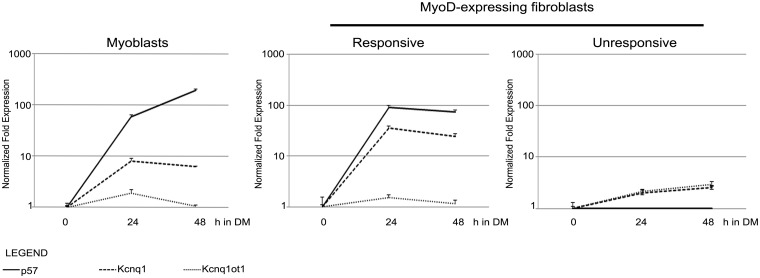
On the basis of our finding that, in unresponsive and in undifferentiated cells, p57 promoter shows epigenetic features, such as DNA hypermethylation, similar to those associated with imprinting, we hypothesized that KvDMR1 could be involved in determining the accessibility of p57 to MyoD-dependent regulation. We first asked whether other genes of the p57 sub-domain, coordinately regulated during imprinting, were also coordinately regulated during myogenesis. In particular, we determined the expression pattern of kcnq1, co-imprinted with p57, and of kcnq1ot1, oppositely imprinted, during muscle differentiation. For this purpose, we used two well-established models of in vitro myogenesis. The first one consists of spontaneously differentiating C2.7 myoblast cells, a satellite-derived myoblast cell line able to undergo a well-characterized process of differentiation that recapitulates many features of in vivo myogenesis. The second one consists of fibroblasts converted to the myogenic lineage by exogenous MyoD expression, a cell system recognized to be highly representative of both in vivo and in vitro myoblasts for many MyoD-dependent functions (5,30). Importantly, we previously found that different fibroblast cell types, equally competent to undergo MyoD-induced differentiation, are differentially responsive regard the induction of p57 (11,27,28). We took advantage of these cell types, which we name as ‘responsive’ and ‘unresponsive’, to dissect the molecular mechanisms underlying the induction and restriction of p57 expression. Mouse C57BL/6 (responsive) and C3H10T1/2 (unresponsive) fibroblasts were infected with a MyoD-retroviral vector and, 48 h later, induced to differentiate. RNA was extracted from C2.7 myoblasts and MyoD-expressing fibroblasts at different times of differentiation and analyzed by qRT-PCR. As shown in Figure 1, kcnq1 is significantly up-regulated during muscle differentiation, both in myoblasts and in MyoD-expressing fibroblasts, in parallel with p57. kcnq1ot1 expression, on the contrary, does not show a significant modulation. Interestingly, cells that are unresponsive to MyoD for p57 are also unable to up-regulate kcnq1 at the same level as responsive cells. Similar results were obtained using fibroblasts from hybrid mice (C57BL/6 female X SD7 male), as responsive cells, and Swiss 3T3 fibroblasts as unresponsive cells (Supplementary Figure S1 and unpublished observations). These findings strongly suggested that MyoD could function by targeting a regulatory element shared by p57 and kcnq1 and that this common element, likely KvDMR1, could be also involved in restricting the expression of these genes in unresponsive cells.
Figure 1.
Expression of p57, Kcnq1 and Kcnq1ot1 during muscle differentiation. C2.7 myoblasts and MyoD-expressing fibroblasts were analyzed by qRT-PCR at different times (hours) after the shift to differentiation medium (h in DM). The expression levels are relative to the T0 value for each transcript. The results shown are representative of three independent experiments. Error bars indicate standard error of the mean for each sample analyzed in triplicate.
MyoD binds in vivo to KvDMR1 and relieves its repressive activity
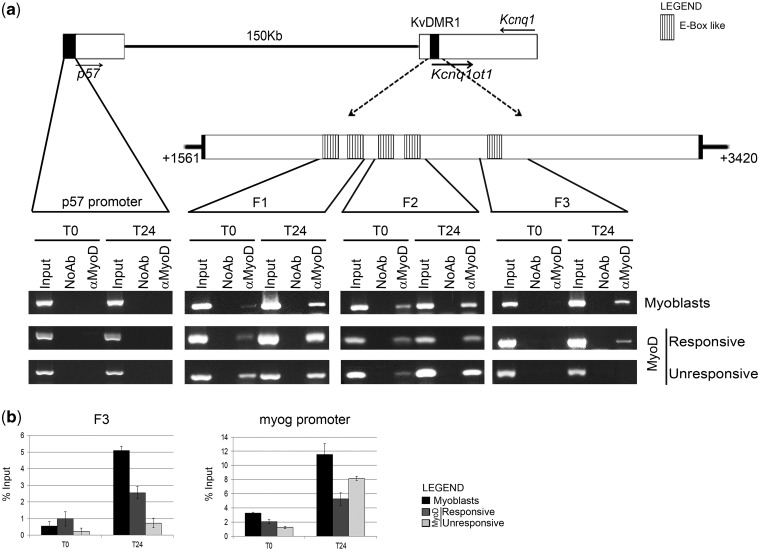
Mouse KvDMR1 is composed of multiple cis-acting modules, including an element showing repressive activity in enhancer-blocking assays and a promoter and a putative enhancer driving the expression of the long non-coding RNA kcnq1ot1 (31). Our observation that the expression of kcnq1ot1 was not significantly affected by MyoD prompted us to focus our attention on the repressive element. Using MatInspector software to scan putative MyoD-binding sites within KvDMR1 sequence, we detected the presence of five E-box-like elements, as outlined in Figure 2a.
Figure 2.
MyoD interacts in vivo with KvDMR1. (a) Top: schematic diagram showing the reciprocal location of p57, Kcnq1 and Kcnq1ot1 genes and KvDMR1 region. Filled arrows indicate the direction of transcription. Dashed arrows point to an enlargement of the repressive KvDMR1 element previously described (31) extending from nt +1561 to +3420 of AF119385 sequence. The positions of the putative MyoD binding sites are also indicated. A further enlargement indicates the location of the three fragments (F1, F2 and F3) amplified in the ChIP assay shown in the bottom. Bottom: In vivo binding activity of MyoD during differentiation. Chromatin from C2.7 myoblasts and MyoD-expressing fibroblasts (responsive and unresponsive) kept either in growth (T0) or in differentiation medium for 24 h (T24) was immunoprecipitated using a specific antibody to MyoD or in the absence of antibody (NoAb). Input represents non-immunoprecipitated cross-linked chromatin. The indicated fragments were amplified by PCR as described in ‘Materials and Methods’ section. A p57 promoter fragment, which we have previously demonstrated not to be bound by MyoD (28), was used as a negative control. The results obtained with 30 amplification cycles are shown. (b) qRT-PCR analysis and quantitation of the ChIP assay for MyoD binding to fragment 3 (F3) and myogenin (myog) promoter. Values derived from three independent experiments were normalized for background signals (NoAb) and expressed as percentage of input chromatin (% input). Data are shown as mean ± standard deviation.
To address the hypothesis that MyoD could physically interact with KvDMR1, we performed ChIP assays using primers surrounding the putative binding sites. MyoD-bound chromatin was immunoprecipitated from undifferentiated and differentiated cultures of C2.7 myoblasts and of MyoD-expressing fibroblasts, both responsive and unresponsive. As shown in Figure 2a, MyoD binds to all three KvDMR1 fragments targeted. However, although MyoD binding to fragments 1 (F1) and 2 (F2) occurs regardless of differentiation and responsiveness, the binding to fragment 3 (F3) takes place exclusively in differentiated and responsive cells. This finding suggests that the transcriptional activation of p57 is functionally correlated with the recruitment of MyoD to the F3 sub-region of KvDMR1 repressive element. qRT-PCR analysis of ChIP products, reported in Figure 2b, confirmed that MyoD binding to F3 fragment relates with both differentiation and responsiveness. The binding to myogenin promoter, used as a positive control, validated MyoD immunoprecipitation and activity even in unresponsive cells. The results of electrophoretic mobility shift assays, performed with fragments containing either the wild type or a mutated recognition sequence, demonstrated that MyoD binding to F3 is direct and is mediated by the E-box-like element (Supplementary Figure S2). It has been recently reported in a genome-wide analysis of MyoD binding in muscle cells (30). By inspecting the ChIP-seq raw data deposited, we noted that the MyoD binding sites that we have detected within KvDMR1 through ChIP assays only show very weak signals, if any, in the ChIP-seq study. This discrepancy could be explained by some critical difference in the two experimental approaches, such as, for example the differentiation times at which MyoD binding was assessed (24 h in our ChIP assays and 72 h in ChIP-seq experiments). Indeed, in our preliminary assays, we observed that MyoD binding to KvDMR1 peaks at 24 h and then declines until becoming almost undetectable at 72 h (data not shown).
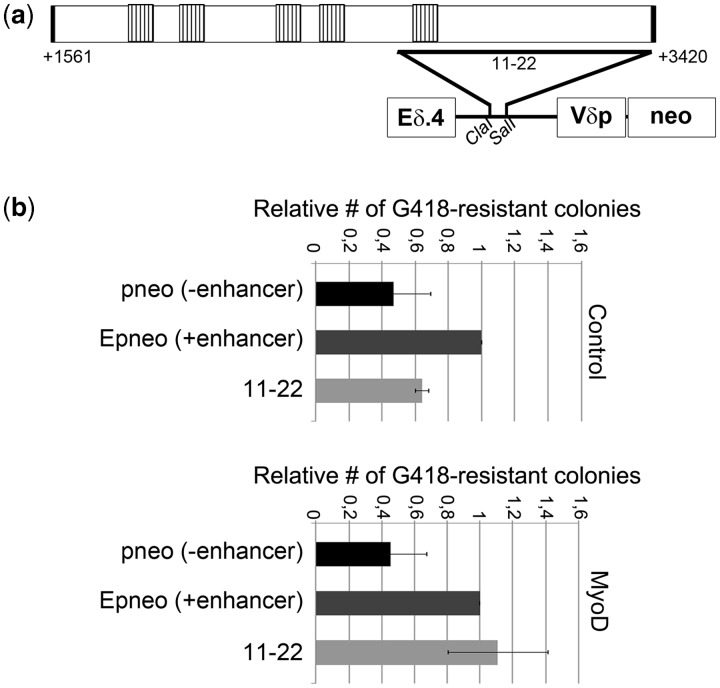
It has been previously reported that different KvDMR1 sub-regions show insulator or silencer activity in enhancer-blocking assays (31,32). In this kind of assays, any enhancer-blocking element inserted between an enhancer and a promoter for a neomycin resistance gene prevents the activation of the reporter gene and thus reduces the number of G418-resistant colonies formed by stably transfected cells. The E-p-neo vector, which contains the Eδ TCR enhancer and Vδ TCR promoter driving the expression of the selectable marker, demonstrated to be suitable to reveal the enhancer-blocking potential of DNA elements from different loci and species (31,33–35). To test the hypothesis that MyoD could promote the up-regulation of p57 by interfering with the described repressive function of KvDMR1, we took advantage of one of the E-p-neo-derived constructs previously used to demonstrate the enhancer-blocking activity of this imprinting control region (31,32). As outlined in Figure 3a, this construct contains a fragment comprising the F3 sub-region placed between the enhancer and the promoter. We determined the possible MyoD effects on its activity by measuring the colony-forming efficiencies of cells co-transfected with a MyoD-expressing vector and the indicated constructs, in the presence of G418 antibiotic. As a cell background for the assay, we chose the K562cell line, because it demonstrated to be competent not only for enhancer activity but also for MyoD stability and functionality. In fact, we found that MyoD was capable to activate a reporter gene driven by muscle creatine kinase E-box elements in K562 cells (data not shown), indicating its ability to functionally interact with its targets in this cell context. Remarkably, the results of the enhancer-blocking assays, shown in Figure 3b, indicated that MyoD expression significantly reduces the enhancer-blocking activity of the F3-containing test fragment. This suggests that MyoD induces p57 by counteracting a repressive function of KvDMR1.
Figure 3.
MyoD relieves the enhancer-blocking activity of a KvDMR1 sub-region. (a) Schematic representation of the plasmid construct used for the enhancer-blocking assay, indicating the extent of the [11–22] test fragment (31) with respect to the KvDMR1 repressive element reported in Figure 2a. (b) Relative enhancer-blocking activity of the 11–22 fragment of KvDMR1 in K562 cells co-transfected with the MyoD expression vector (MyoD) or with the empty vector (Control). For each co-transfection, values, normalized relative to luciferase activity, were determined by dividing the number of G418-resistant colonies obtained with the indicated constructs by the colony number obtained with the Epneo construct. The results are the mean of three independent transfection experiments.
MyoD binding causes the release of a chromatin loop between KvDMR1 and p57 promoter
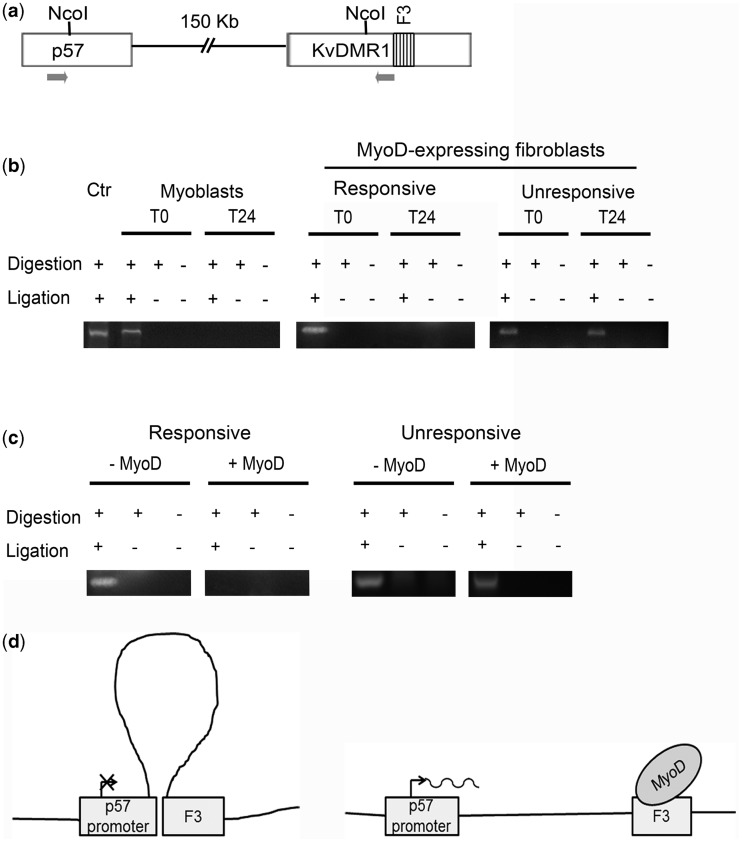
The enhancer sequence(s) driving p57 expression, and possibly affected by KvDMR1, have not been identified yet. Enhancer-blocking insulators interact with each other, with enhancers or with promoters forcing these elements into ineffective chromatin conformations (36). To determine whether KvDMR1 physically associated with p57 promoter and whether the supposed interaction was related to responsiveness and/or differentiation, we performed 3C assays. Crosslinked chromatin, extracted from differentiating C2.7 myoblasts and MyoD-expressing fibroblasts, both responsive and unresponsive, was subjected to digestion and ligation. PCR reactions were performed, as outlined in Figure 4a, to amplify hybrid fragments deriving from the ligation of p57 promoter and KvDMR1 sequences. The results reported in Figure 4b and the sequencing of the amplified fragment clearly indicated that the two regions are close to each other in undifferentiated cells and that their association is lost on differentiation but exclusively in responsive cells. To demonstrate the causal relationship between MyoD binding to KvDMR1 and disruption of the three-dimensional structure occurring on differentiation, we compared the status of the chromatin loop in the presence and in the absence of MyoD. To this end, we performed further 3C assays in responsive and unresponsive fibroblasts expressing either MyoD or the empty vector. The results reported in Figure 4c indicate that, in the absence of MyoD, the chromatin loop is maintained even in differentiation-promoting conditions and even in responsive cells, thus confirming that it is just MyoD that prompts the rearrangement. This analysis also highlights that the same chromatin loop is present in both responsive and unresponsive fibroblasts, before MyoD expression, thus supporting the conclusion that the differential accessibility of KvDMR1 to MyoD binding does not depend on differential chromatin looping but most likely on other epigenetic differences (see also ‘Discussion’ section).
Figure 4.
KvDMR1 physically interacts with p57 promoter in undifferentiated and in unresponsive cells. (a) Schematic representation of the p57-KvDMR1 locus showing the locations of the NcoI sites and of the PCR primers (arrows) used for 3C analysis. (b) 3C analysis of the p57-KvDMR1 locus in C2.7 myoblasts and in responsive (C57BL) and in unresponsive (C3H10T1/2) mouse embryo fibroblasts expressing MyoD kept either in growth (T0) or in differentiation medium for 24 h (T24). Ctr consists of a plasmid construct containing a ligation product of p57 promoter and KvDMR1 sequences and represents a positive control for the pair of primers used. The results shown are representative of three independent experiments. (c) 3C analysis of the p57-KvDMR1 locus in responsive and unresponsive fibroblasts infected with either the MyoD retroviral vector (+MyoD) or with the empty vector (–MyoD) and analyzed 24 h after the shift to differentiation medium. The results shown are representative of three independent experiments. (d) Schematic model of MyoD effect on the putative chromatin looping between KvDMR1 and p57 promoter.
All together, these results strongly support a picture in which a chromatin loop between KvDMR1 and p57 promoter represses its activity in undifferentiated myoblasts. On differentiation, MyoD recruitment to KvDMR1/F3, enabled only in responsive cells, causes the removal of this constraint, allowing p57 expression (Figure 4d). Consistently with the well-recognized property of MyoD to recruit chromatin modifying enzymes to its targets, we found that MyoD binding to F3 correlates with increased histone H3 acetylation (Supplementary Figure S3). This suggests that a regional chromatin modification of KvDMR1, consequent to MyoD binding, may play a role in the spatial reorganization of the chromatin loop.
MyoD affects a non-allelic function of KvDMR1
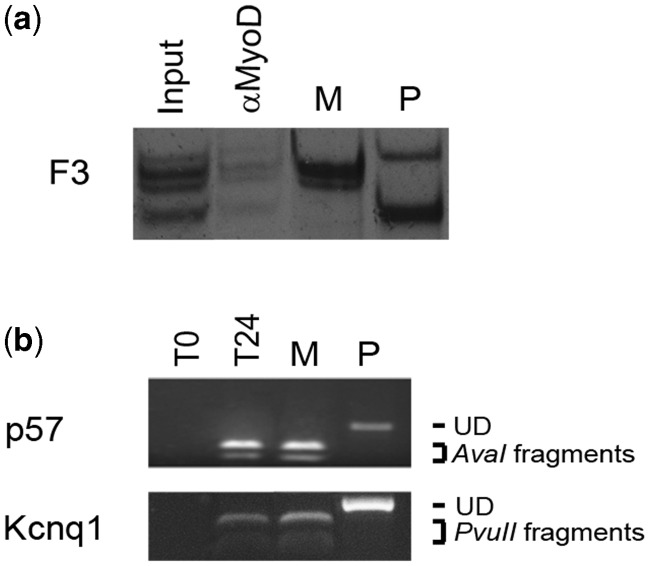
KvDMR1, similar to most imprinting control regions, shows allele-specific epigenetic marks, including DNA methylation, histone modifications and transcription factor binding (37,38), which participate in the molecular mechanism driving allele-specific expression of the imprinted genes of the sub-domain. To further explore how MyoD affects KvDMR1 function, we first evaluated the possible allele-specific binding of MyoD to the KvDMR1 repressive element during differentiation. To distinguish allele-specific sequences, we took advantage of mouse fibroblasts derived from hybrid mice (C57BL/6 female X SD7 male), carrying allele-specific single-nucleotide polymorphisms (SNPs) in the distal part of chromosome 7. As reported in Supplementary Figure S1, these fibroblasts are responsive to exogenous MyoD expression for the induction of p57. ChIP was performed after MyoD-induced differentiation. Allele-specific binding was examined by exploiting different SNPs, present in the amplification target comprising F3 fragment, and revealed through SSCP analysis. As reported in Figure 5a, MyoD binds to both maternal and paternal alleles of KvDMR1/F3.
Figure 5.
MyoD binds biallelically to KvDMR1 but does not affect the imprinting status of the locus. (a) Allele-specific chromatin immunoprecipitation of MyoD. Hybrid mouse fibroblasts expressing MyoD, kept 24 h in differentiation medium, were treated for ChIP assay as described earlier. MyoD immunoprecipitates were then analyzed by radioactive PCR specific for F3, followed by SSCP analysis. M and P show the electrophoretic mobility of maternal- and paternal-specific bands, respectively. (b) Semiquantitative RT-PCR analysis of allele-specific expression of p57 and kcnq1. Hybrid mouse fibroblasts expressing MyoD were kept either in growth (T0) or in differentiation medium for 24 h (T24). Maternal and paternal alleles were distinguished by RFLP analysis as described in ‘Materials and Methods’ section. UD indicates the electrophoretic mobility of the undigested paternal-specific fragments (P). AvaI and PvuII fragments indicate that of digested maternal-specific fragments (M).
In light of the observed biallelic binding, we queried whether the MyoD-dependent induction of p57 and kcnq1 involved the up-regulation of the maternal or the de-repression of the paternal imprinted alleles. Hybrid mouse fibroblasts were infected with a MyoD-encoding retrovirus and induced to differentiate. p57 and Kcnq1 RNAs were then examined by combining RT-PCR with RFLP analysis. The results reported in Figure 5b clearly showed that during MyoD-induced differentiation, both genes are up-regulated uniquely from the normally active maternal alleles. This finding allows us to exclude that the up-regulation of p57 and kcnq1 could be ascribed to loss of imprinting and supports the conclusion that KvDMR1 plays an additional role, other than imprinting control, in restricting the expression of these genes in muscle cells.
DISCUSSION
The cdk inhibitor p57 is a fundamental regulator of development and differentiation of many tissues in mammals (19). Its expression levels must be strictly regulated, as deduced by the aberrant phenotypes resulting from imprinting perturbations, inactivating mutations or over-expression (17,18,39,40). Increasing evidence has been accumulated indicating that epigenetic control plays a key role in p57 silencing during imprinting (41,42) and tumorigenesis (43). In this work, we report that a completely novel epigenetic mechanism is involved in the MyoD-dependent induction of p57 during myogenesis.
We previously suggested that the accessibility of p57 promoter was prevented by a repressive, cis-acting mechanism associated with promoter hypermethylation in undifferentiated myoblasts and in cell types unable to induce p57. In this study, we report that this epigenetic constraint involves a long-distance regulatory element, KvDMR1, known to participate in the imprinting process. We show that the induction of p57 expression is associated with the in vivo binding of MyoD to KvDMR1. The analysis by ChIP assays of different KvDMR1 sub-regions showed that MyoD is constitutively bound to F1 and F2 in both responsive and unresponsive cells, whereas it is recruited to F3 only after differentiation and only in responsive cells. This suggested that the binding to F3 represents a limiting factor for p57 de-repression. We also show that MyoD is capable to relief the enhancer-blocking activity of this sub-region. Several models, involving complex three-dimensional changes in chromatin architecture, have been proposed to explain long-distance gene repression by enhancer-blocking insulators (36,44). The results of our 3C assay demonstrated that KvDMR1 sequences contact p57 promoter in undifferentiated and in unresponsive cells. Interestingly, we have noticed that both F3 region and p57 promoter contain putative recognition sites for CCCTC-binding factor (CTCF), the prototypical organizer of long-range chromatin interactions (45). The possible involvement of CTCF in establishing the observed chromatin contact is currently under investigation. Whatever the molecular mechanism mediating chromatin looping, the dynamics of this structure is consistent with a role in preventing p57 expression. Further work will be required to understand how this structure is functionally correlated with promoter hypermethylation and p57 silencing.
The picture emerging from our previous and present results is that MyoD plays a dual function in the induction of p57 expression. One consists in positively regulating p57 promoter, through the up-regulation of the intermediate factors p73, Sp1 and Egr1 (27,28). The other one consists in counteracting the repression exerted by a long-distance element, located within KvDMR1, through direct binding to an E-box-like sequence. This interaction is required to allow the disruption of the chromatin loop, the release of p57 promoter and then its trans-activation. It is conceivable that the ability of MyoD to cause spatial reorganization of chromatin at KvDMR1 is linked with its ability to recruit chromatin modifying complexes to F3, as inferred by the increased H3 acetylation observed in this region (Supplementary Figure S3). However, we do not exclude that the binding to F1 and F2, although not sufficient for p57 de-repression, could participate in creating a chromatin environment that facilitates the disruption of the chromatin loop triggered by binding to F3. A comprehensive analysis of the histone modifications and of the changes in the DNA methylation status of KvDMR1 during muscle differentiation will provide important insights into this issue. Recent analysis of genome-wide MyoD binding revealed that this myogenic factor binds to thousands of sites outside of promoter regions, where it induces histone acetylation (30). The functional significance of this widespread binding is not yet clear. Our data support the hypothesis that a possible function of MyoD interaction with at least some of these sites consists in causing long-range chromatin changes. This suggestion opens new perspectives in understanding how a pioneer transcription factor may open repressive chromatin and coordinate gene expression during determination and differentiation.
Notably, we found that in unresponsive cells, MyoD is unable to bind to KvDMR1 and, hence, to cause the same chromatin conformation change triggered in responsive cells. This failure could be explained by the presence of a distinct, non-accessible chromatin status at KvDMR1 or, alternatively, by the presence (or absence) of MyoD-interacting factor(s) preventing (or promoting) its binding. As unresponsive cells represent a useful model system to gain insights into the epigenetic mechanisms responsible for the restriction of p57 expression to specific tissues and cell types, this issue deserves to be investigated.
We observed that MyoD also induces the expression of kcnq1, co-imprinted with p57. Kcnq1 codes for a sub-unit of a voltage-dependent potassium channel. Although its best established role is the control of cell excitability, kcnq1 has been suggested to be also involved, by regulating membrane potential and cell volume, in muscle cell proliferation and differentiation (46). Interestingly, several imprinted genes, belonging to independent imprinting domains, were shown to be up-regulated during muscle regeneration (47). In this regard, it has been reported the existence of a network of co-regulated imprinted genes involved in growth promotion or restriction and including p57 (48). The significance of this co-regulation could be to assure an equilibrium in growth control during development. It has been proposed that these imprinted genes are regulated, directly or indirectly, downstream of two members of the network, Zac1 and H19 (48,49). Our present results suggest that additional epigenetic mechanisms, involving the functional interaction of differentiation factors with imprinting control elements, may participate in the developmental co-regulation of imprinted genes. The observation that MyoD binding, despite biallelic, does not induce expression from the paternal imprinted alleles also highlights the existence of a repressive function of KvDMR1, distinct from and hierarchically subordinated to the imprinting control.
In summary, this work uncovered a novel mechanism by which MyoD coordinates gene expression during muscle differentiation. Not less important, the data obtained could be of general relevance to clarify the regulation of a CKI that exerts unique functions in development and differentiation. It is worth to mention that other bHLH transcription factors, such as Mash2 and E47, have been shown to induce p57 transcription in neural cells (50,51). It is likely that a molecular mechanism similar to that acting in muscle cells can also operate in other differentiation systems. Moreover, it is reasonable to hypothesize that epigenetic constraints, analogous to those underlying the cell type restriction of p57 expression, can also participate in the aberrant p57 silencing observed in cancer and in some developmental pathologies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3, Supplementary Methods and Supplementary References [52–54].
FUNDING
‘Sapienza Università di Roma’ [C26A082MNW]; Tavola Valdese – fondi OPM by way of ‘Associazione per lo Sviluppo delle Scienze Pasteuriane’; ‘Fondazione Adriano Buzzati-Traverso’ (fellowship to A.B.) and ‘Consorzio Interuniversitario Biotecnologie’ (fellowship to M.C.). Funding for open access charge: Tavola Valdese — fondi OPM by way of “Associazione per lo Sviluppo delle Scienze Pasteuriane”.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Paolo Amati for critical reading of the manuscript. They also thank Michael Higgins for the constructs used in enhancer-blocking assays, Andrea Riccio for hybrid mouse fibroblasts bearing genetic polymorphisms in the p57 locus, Stefano Gabriele for advice in qRT-PCR assays and Silvia Gioiosa for help in database analysis.
REFERENCES
- 1.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 2.Kitzmann M, Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell. Mol. Life Sci. 2001;58:571–579. doi: 10.1007/PL00000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maione R, Amati P. Interdependence between muscle differentiation and cell-cycle control. Biochim. Biophys. Acta. 1997;1332:M19–M30. doi: 10.1016/s0304-419x(96)00036-4. [DOI] [PubMed] [Google Scholar]
- 4.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 5.Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 6.Tapscott SJ. The circuitry of a master switch: MyoD and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 7.Albini S, Puri PL. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: it's time to exchange! Exp. Cell Res. 2010;316:3073–3080. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Sem. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr. Opin. Genet. Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yahi H, Philipot O, Guasconi V, Fritsch L, Ait-Si-Ali S. Chromatin modification and muscle differentiation. Exp. Opin. Ther. Targets. 2006;10:923–934. doi: 10.1517/14728222.10.6.923. [DOI] [PubMed] [Google Scholar]
- 11.Figliola R, Maione R. MyoD induces the expression of p57Kip2 in cells lacking p21Cip1/Waf1: overlapping and distinct functions of the two cdk inhibitors. J. Cell. Physiol. 2004;200:468–475. doi: 10.1002/jcp.20044. [DOI] [PubMed] [Google Scholar]
- 12.Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborn DP, Li K, Hinits Y, Hughes SM. Cdkn1c drives muscle differentiation through a positive feedback loop with Myod. Dev. Biol. 2011;350:464–475. doi: 10.1016/j.ydbio.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynaud EG, Leibovitch MP, Tintignac LA, Pelpel K, Guillier M, Leibovitch SA. Stabilization of MyoD by direct binding to p57(Kip2) J. Biol. Chem. 2000;275:18767–18776. doi: 10.1074/jbc.M907412199. [DOI] [PubMed] [Google Scholar]
- 15.Swanger WJ, Roberts JM. p57KIP2 targeted disruption and Beckwith-Wiedemann syndrome: is the inhibitor just a contributor? BioEssays. 1997;19:839–842. doi: 10.1002/bies.950191002. [DOI] [PubMed] [Google Scholar]
- 16.Hatada I, Nabetani A, Morisaki H, Xin Z, Ohishi S, Tonoki H, Niikawa N, Inoue M, Komoto Y, Okada A, et al. New p57KIP2 mutations in Beckwith-Wiedemann syndrome. Hum. Genet. 1997;100:681–683. doi: 10.1007/s004390050573. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y, Frisen J, Lee MH, Massague J, Barbacid M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Liegeois NJ, Wong C, Finegold M, Hou H, Thompson JC, Silverman A, Harper JW, DePinho RA, Elledge SJ. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature. 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 19.Pateras IS, Apostolopoulou K, Niforou K, Kotsinas A, Gorgoulis VG. p57KIP2: “Kip”ing the cell under control. Mol. Cancer Res. 2009;7:1902–1919. doi: 10.1158/1541-7786.MCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 20.Hatada I, Mukai T. Genomic imprinting of p57KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat. Genet. 1995;11:204–206. doi: 10.1038/ng1095-204. [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka S, Thompson JS, Edwards MC, Bartletta JM, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP. Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc. Natl Acad. Sci. USA. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher ER, Reik W. Beckwith-Wiedemann syndrome: imprinting in clusters revisited. J. Clin. Invest. 2000;105:247–252. doi: 10.1172/JCI9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 24.Horike S, Mitsuya K, Meguro M, Kotobuki N, Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y, Oshimura M. Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum. Mol. Genet. 2000;9:2075–2083. doi: 10.1093/hmg/9.14.2075. [DOI] [PubMed] [Google Scholar]
- 25.Shin JY, Fitzpatrick GV, Higgins MJ. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 2008;27:168–178. doi: 10.1038/sj.emboj.7601960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yatsuki H, Joh K, Higashimoto K, Soejima H, Arai Y, Wang Y, Hatada I, Obata Y, Morisaki H, Zhang Z, et al. Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region. Genome Res. 2002;12:1860–1870. doi: 10.1101/gr.110702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaccarello G, Figliola R, Cramerotti S, Novelli F, Maione R. p57Kip2 is induced by MyoD through a p73-dependent pathway. J. Mol. Biol. 2006;356:578–588. doi: 10.1016/j.jmb.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Figliola R, Busanello A, Vaccarello G, Maione R. Regulation of p57(KIP2) during muscle differentiation: role of Egr1, Sp1 and DNA hypomethylation. J. Mol. Biol. 2008;380:265–277. doi: 10.1016/j.jmb.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 30.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzpatrick GV, Pugacheva EM, Shin JY, Abdullaev Z, Yang Y, Khatod K, Lobanenkov VV, Higgins MJ. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Mol. Cell. Biol. 2007;27:2636–2647. doi: 10.1128/MCB.02036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakur N, Kanduri M, Holmgren C, Mukhopadhyay R, Kanduri C. Bidirectional silencing and DNA methylation-sensitive methylation-spreading properties of the Kcnq1 imprinting control region map to the same regions. J. Biol. Chem. 2003;278:9514–9519. doi: 10.1074/jbc.M212203200. [DOI] [PubMed] [Google Scholar]
- 33.Zhong XP, Krangel MS. An enhancer-blocking element between alpha and delta gene segments within the human T cell receptor alpha/delta locus. Proc. Natl. Acad. Sci. USA. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanduri C, Fitzpatrick G, Mukhopadhyay R, Kanduri M, Lobanenkov V, Higgins M, Ohlsson R. A differentially methylated imprinting control region within the Kcnq1 locus harbors a methylation-sensitive chromatin insulator. J. Biol. Chem. 2002;277:18106–18110. doi: 10.1074/jbc.M200031200. [DOI] [PubMed] [Google Scholar]
- 35.Gombert WM, Farris SD, Rubio ED, Morey-Rosler KM, Schubach WH, Krumm A. The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol. Cell. Biol. 2003;23:9338–9348. doi: 10.1128/MCB.23.24.9338-9348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raab JR, Kamakaka RT. Insulators and promoters: closer than we think. Nat. Rev. Genet. 2010;11:439–446. doi: 10.1038/nrg2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis A, Reik W. How imprinting centres work. Cytogenet. Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 38.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv. Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 39.Andrews SC, Wood MD, Tunster SJ, Barton SC, Surani MA, John RM. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev. Biol. 2007;7:53. doi: 10.1186/1471-213X-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature. 1991;351:667–670. doi: 10.1038/351667a0. [DOI] [PubMed] [Google Scholar]
- 41.Lewis A, Green K, Dawson C, Redrup L, Huynh KD, Lee JT, Hemberger M, Reik W. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- 42.Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 43.Kavanagh E, Joseph B. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim. Biophys. Acta. 2011;1816:50–56. doi: 10.1016/j.bbcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 45.Zlatanova J, Caiafa P. CCCTC-binding factor: to loop or to bridge. Cell. Mol. Life Sci. 2009;66:1647–1660. doi: 10.1007/s00018-009-8647-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soldovieri MV, Miceli F, Taglialatela M. Driving with no brakes: molecular pathophysiology of Kv7 potassium channels. Physiology. 2011;26:365–376. doi: 10.1152/physiol.00009.2011. [DOI] [PubMed] [Google Scholar]
- 47.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 2003;278:8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 48.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 50.Kury P, Greiner-Petter R, Cornely C, Jurgens T, Muller HW. Mammalian achaete scute homolog 2 is expressed in the adult sciatic nerve and regulates the expression of Krox24, Mob-1, CXCR4, and p57kip2 in Schwann cells. J. Neurosci. 2002;22:7586–7595. doi: 10.1523/JNEUROSCI.22-17-07586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothschild G, Zhao X, Iavarone A, Lasorella A. E Proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol. Cell. Biol. 2006;26:4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caspary T, Cleary MA, Baker CC, Guan XJ, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc. Natl. Acad. Sci. USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.