Abstract
Site-specific recombination catalyzed by tyrosine recombinases follows a common pathway consisting of two consecutive strand exchanges. The first strand exchange generates a Holliday junction (HJ), which is resolved by a second strand exchange. In integrons, attC sites recombine as folded single-stranded substrates. Only one of the two attC site strands, the bottom one, is efficiently bound and cleaved by the integrase during the insertion of gene cassettes at the double-stranded attI site. Due to the asymmetry of this complex, a second strand exchange on the attC bottom strand (bs) would form linearized abortive recombination products. We had proposed that HJ resolution would rely on an uncharacterized mechanism, probably replication. Using an attC site carried on a plasmid with each strand specifically tagged, we followed the destiny of each strand after recombination. We demonstrated that only one strand, the one carrying the attC bs, is exchanged. Furthermore, we show that the recombination products contain the attC site bs and its entire de novo synthesized complementary strand. Therefore, we demonstrate the replicative resolution of single-strand recombination in integrons and rule out the involvement of a second strand exchange of any kind in the attC × attI reaction.
INTRODUCTION
Integrons are bacterial recombination systems acting as genetic platforms able to capture, stockpile and rearrange gene cassettes through a site-specific recombination mechanism (1). They are responsible for the gathering of antibiotic resistance genes in mobile elements (2). Integrons are composed of three key elements: an integrase gene (IntI) which encodes a tyrosine recombinase performing the site-specific recombination reaction, a primary recombination site (attI) where the incorporation of gene cassettes occurs and a strong resident promoter (Pc) which ensures the expression of the first cassettes of the array (3–5). Indeed, gene cassettes are generally promoterless and mostly correspond to a single open reading frame (ORF) immediately followed by an attC recombination site (called 59-base element) (6). Cassette insertion mostly occurs through intermolecular recombination between an attC site and the attI site, while excision principally occurs through intramolecular recombination between two attC sites (7,8). Such excision and reinsertion events result in cassette array reordering.
The length of natural attC sites varies from 57 to 141 bp. They include two regions of inverted homology, R″-L″ and L′-R′, which are separated by a central region that is highly variable in length and sequence (Supplementary Figure S1) (9). It has been shown using a DNA-binding assay that IntI1 strongly and specifically binds the bottom strand (bs) of single-stranded (ss) attC sites and has no affinity for the double-stranded (ds) form (10). We have previously shown in vivo, using a conjugation assay, that the attC site recombines as a ssDNA folded structure, in contrast with the attI site, which is recombined as a canonical ds form (Supplementary Figure S1) (11). In contrast to canonical core recombination sites, the genetic information required for proper recombination is not entirely contained in the primary sequences of attC sites but mostly in specific features of their secondary structures (Supplementary Figure S1) (12,13). Indeed, the attC sites display a strikingly conserved palindromic organization (9,14,15) that can form secondary structures through the self-pairing of DNA strands (Supplementary Figure S1). Upon folding, ss attC sites present a structure resembling canonical core site consisting of R and L boxes with specific structural features (defined in (12)). The annealing of the R″-L″ and L′-R′ arm sequences, which contain two non-complementary spacer regions, leads to the formation of the first structural element, the unpaired central spacer. The second structural features, the extra-helical bases (EHB), correspond to the single bases always located on the R″-L″ arm of the symmetrical attC sequence that have no complementary nucleotides on the R′-L′ arm (Supplementary Figure S1). Depending on the attC sites, there are two or three EHB (12). The last structural feature is defined as the stem terminal structure or variable terminal structure (VTS) (12). The VTS varies in length among the various attC sites, going from three predicted unpaired nucleotides as in attCaadA7, to a complex branched secondary structure in the larger sites such as the VCRs (Vibrio cholerae attC sites, Supplementary Figure S1, (16)).
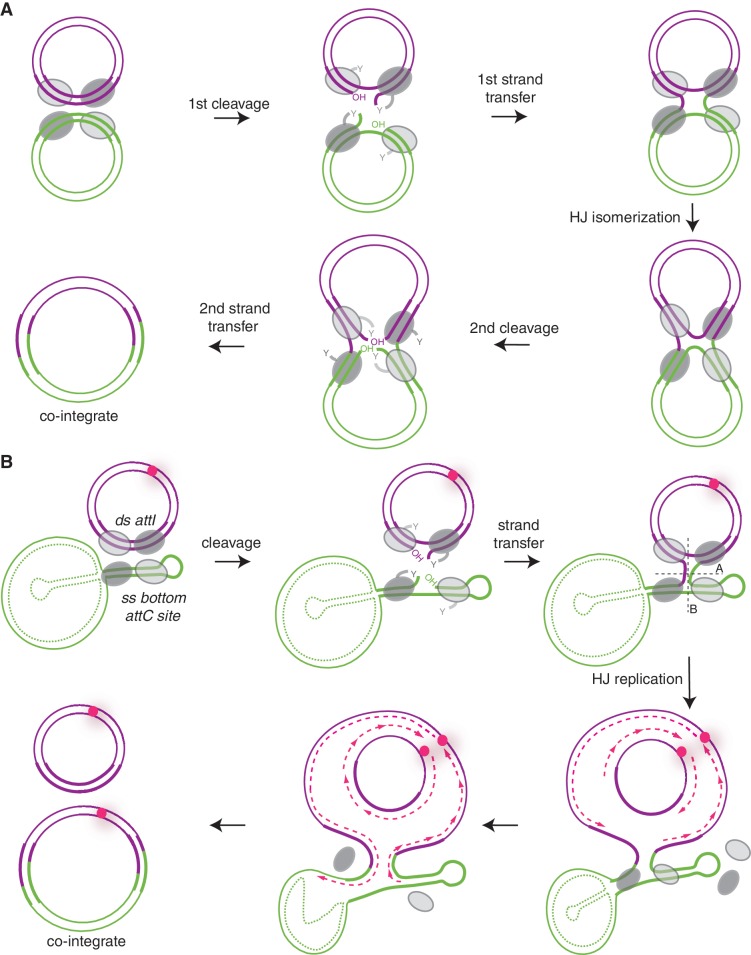
The integron integrase is a member of the tyrosine recombinase (Y-recombinase) family. Other members of this family, such as the lambda phage integrase or the P1 phage Cre recombinase, generally recombine dsDNA (for review, see (17)). Y-recombinase recombination is considered to conform to the following model. A synaptic complex consisting of two recombination substrates and four recombinases is formed. Initially, only two opposing monomers are active and recombination is initiated through the cleavage of one strand of each substrate by their nucleophilic tyrosine. Covalent phosphotyrosine bounds are formed between the attacking monomers and the 3′-ends of DNA, while the 5′-OH ends remain free. The next step consists in the 5′-ends attacking the opposing 3′-phosphodiester bonds, resulting in the first strand exchange forming a Holliday junction (HJ). The complex can then isomerize, the inactive monomers becoming active and vice versa. A second strand exchange can then proceed following the exact same mechanism. This last strand transfer resolves the HJ, frees the proteins and the recombination reaction is achieved (Figure 1A) (17).
Figure 1.
Two models of site-specific recombination. (A) Classical site-specific recombination catalyzed by Y-recombinases. The bold strands on plasmids represent the recombination sites. The synaptic complex comprises two DNA duplexes bound by four recombinases protomers. The bending of DNA determines which protomer is activated and therefore which strand will be cleaved first. In this scheme, the first two activated protomers are represented by dark gray color. One strand from each duplex is cleaved, exchanged and ligated to form a HJ. Isomerization of this junction alternates the catalytic activity between the two pairs of protomers (dark and light-gray ovals) ensuring the second strand exchange and the recombination product formation. (B) Replicative model of site-specific recombination in insertion of gene cassettes. The model shows the recombination between a gene cassette containing the ss bottom attC site and a molecule carrying the ds attI site. The attI and attC recombination sites are represented by bold purple and green lines, respectively. The top strand of the attC site is represented as dotted green lines as we do not exactly know the nature of the gene cassettes (ss or ds) and the role of the top strand in cassette insertion. Steps are identical to classical site-specific recombination steps catalyzed by other Y-recombinases up to the HJ intermediate (see A). Classical resolution through the A axis reverses the recombination to the original substrates, while resolution through the B axis, giving rise to covalently closed linear molecules, is abortive. Our model suggests that the non-abortive resolution implies a replication step. The origin of replication is represented by a pink circle and the newly synthesized leading and lagging strands by dotted pink lines.
In the case of integrons, the fact that this genetic system has evolved unique recombination processes involving non-canonical substrates, such as ss attC sites, could lead to a mechanistic problem. Indeed, if one of the recombination partners is ds (attI) and the other ss (bottom attC), it generates, after the first strand exchange, an atypical HJ (aHJ) in which a second strand exchange of the bottom attC strand would form linearized abortive and potentially lethal recombination products (Figure 1B, see B axis). Hence, a second strand exchange in the bs attC sites must somehow be avoided. Accordingly, the crystal structure of the attC × attC/IntI synaptic complex has shown that binding to the attC extra-helical ‘T’ acts to pull the catalytic tyrosine away from the phosphate link in the bound IntI monomer (13). Still, the aHJ has to be resolved somehow.
It was thus proposed that replication is involved in this last step (Figures 1B and 2B (11)). According to this hypothesis, after the passage of a replication fork through an aHJ, the ds recombination product would arise from replication of the strand carrying the integrated bottom attC site, while on the opposite strand the initial substrate would be reconstituted (Figures 1B and 2B). The recombination would thus be semi-conservative. Nevertheless, a second strand exchange involving the top strand could hypothetically also lead to productive integration. However, this scenario seems improbable and would need to be integrase independent. Still, we have recently shown that the integration reaction can take place in the presence of the top strand through cruciform extrusion (18), and thus the model needs further proof.
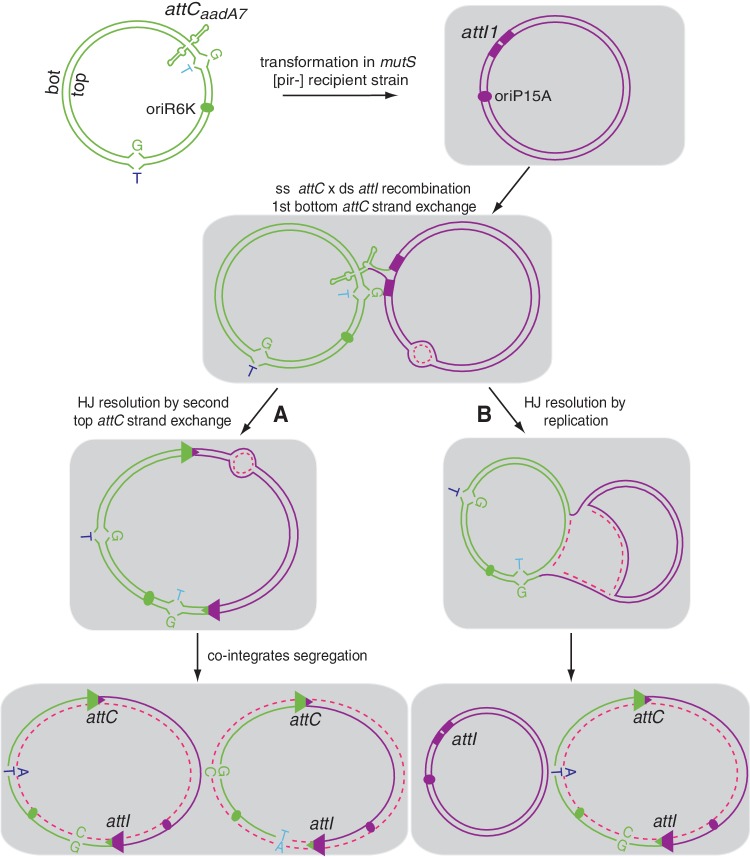
Figure 2.
Schematic representation of the non-replicative recombination assay used for the integron cassette integration reaction. The attC-containing mismatched circles are represented in green and the target pSU38Δ::attI1 in purple. The attC site is shown as a ss folded structure and the core site in ds attI1 site is represented by two purple boxes. Green and purple ovals indicate the oriR6K and oriP15A origin of replication, respectively. T nucleotides of the G-T mismatches contained in the SacII and NarI restriction sites are, respectively, indicated in cyan and blue bold letters. Both pathways, HJ resolution by second strand cleavage in the top attC site (A) and by replication (B), are shown. De novo synthesized strands are shown in pink dotted lines.
In this article, we precisely studied the resolution of this aHJ formed by attC-bs and attI-ds recombination. We developed a genetic setup in which we independently tagged the top and bottom strands (bs) of the attC-containing molecule by introducing two point mutations. Once both strands hybridized, it generates mismatched covalent circles containing an attC site. The analysis of the segregation of these point mutations in recombined DNA molecules allowed us to follow the destiny of each entire strand. For the first time, we demonstrate that only one of the two attC-donor strands, the bottom reactive strand, is found in the recombined molecule arising from a replication process. Moreover, since the reaction involved the entire bs, we can rule out a second strand exchange of any kind. To confirm this model, we tested the involvement of the RuvABC and RecG host proteins. Usually, when HJs are formed between homologous sequences, the RecG proteins or the RuvABC complexes are implicated in the sliding, branch migration and strand cleavage leading to the HJ resolution. As expected, we showed that these proteins are not involved in integron recombination confirming the exclusive implication of the replication pathway to resolve the aHJ.
MATERIALS AND METHODS
Media
Escherichia coli strains were grown in Luria–Bertani broth at 37°C and, for single-strand DNA production in yeast extract and tryptone broth. Antibiotics were used at the following concentrations: ampicillin, 100 µg/ml; chloramphenicol (Cm), 25 µg/ml; kanamycin (Km), 25 µg/ml; spectinomycin, 15 µg/ml; and erythromycin 200 µg/ml. Thymidine and diaminopimelic acid (DAP) were supplemented when necessary to a final concentration of 0.3 mM. Glucose and l-arabinose were added at 10 and 2 mg/ml final concentration, respectively.
Bacterial strains, plasmids and primers
Bacterial strains and plasmids are described, respectively, in Tables 1 and 2 and primers in Supplementary Table S1.
Table 1.
Bacterial strains used in this study
| Strain number | Relevant Escherichia coli genotypes or description | References |
|---|---|---|
| Basic strains | ||
| ω6706 | AB1157 thr1 ara14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 | Laboratory collection |
| ω6789 | AB1157 ruvB52 | RG Lloyd |
| ω6790 | AB1157 ruvC53 | RG Lloyd |
| ω2779 | AB1157 ruvA60::Tn10 | RG Lloyd |
| ω8080 | AB1157 ΔrecG263::Kan | RG Lloyd |
| ω72 | β2163 (F-) RP4-2-Tc::Mu ΔdapA::(erm-pir) [KmR EmR] | (36) |
| ω1628 | П1 DH5α ΔthyA::(erm-pir) [EmR] | (36) |
| ω2580 | MG1655 mutS215::Tn10 [NalR] | Laboratory collection |
| ω4446 | β2150 ΔdapA:(erm-pir) thrB1004 pro thi strA hsdS lacZ DM15, (F’/lacZ DM15 lacIq, traD36, proA+, proB+) | (36) |
| ω2484 | GM48 (F-) thr leu thi lacY galK galT ara fhuA tsx dam dcm glnV44 | (37) |
| Transformed strains used in conjugation assay | ||
| ω1881 | β2163 (p1880) | (11) |
| ω4137 | β2163 (p4136) | (11) |
| ω8101 | AB1157 (p3938) (p4884) | This study |
| ω8106 | AB1157 ruvB52 (p3938)(p4884) | This study |
| ω8107 | AB1157 ruvC53 (p3938) (p4884) | This study |
| ω8109 | AB1157 ruvA60 (p3938) (p4884) | This study |
| ω8122 | AB1157 ΔrecG263 (p3938)(p4884) | This study |
| Transformed strains used in mismatched circle preparation | ||
| ω7790 | β2150 (p7770) | This study |
| ω7791 | β2150 (p7771) | This study |
| ω2533 | GM48 (p2396) | This study |
| Transformed strains used in non replicative assay | ||
| ω7120 | MG1655mutS215 (p1177) | This study |
| ω7994 | MG1655mutS215 (p3938) (p929) | This study |
| ωA266 | MG1655mutS215 (p929) | This study |
Table 2.
Plasmids used and constructed in this study
| Plasmid number | Plasmid description | Relevant properties and construction |
|---|---|---|
| p4136 | pSW23T::attC (B) | pSW23T::attCaadA7, oriVR6Kγ [CmR], (11) |
| p1880 | pSW23T::VCR2/1 (B) | pSW23T::VCR2/1, oriVR6Kγ [CmR], (20) |
| p3938 | pBADIntI1 | pBAD::intI1, oriColE1 [ApR], (38) |
| p929 | pSU38Δ-Km-attI1 | pSU38Δ::attI1, orip15A, [KmR], (20) |
| p4884 | pSU38Δ-Sp-attI1 | pSU38Δ::attI1, orip15A, [SpR], this study |
| p1177 | pSB118::pir116 | pSB118::pir116, oriColE1, [ApR], (36) |
| p421 | pSW24 | pSW23::oriFd1, oriVR6Kγ, [CmR], (36) |
| p902 | pNOTΔ::pTac::dfrB1 | pNOTΔ::pTac::dfrB, oriColE1, [ApR], (20) |
| p3634 | pSW24::attC (B) | pSW24::attCaadA7, oriVR6Kγ, [CmR], (36) |
| p4919 | pSW24::attC-dfrB-oriFd1 | pSW24::attCaadA7-BglII-dfrB-oriFd1, oriVR6Kγ, [CmR], this study |
| p4997 | pSW24::attC-dfrB-oriFd2 | pSW24::attCaadA7-oriFd2, oriVR6Kγ, [CmR], this study |
| p7770 | p4919 NarI mutated | pSW24::attCaadA7-oriFd1, SacIIWt, Narm, oriVR6Kγ, [CmR], this study |
| p7771 | p4997 SacII mutated | pSW24::attCaadA7-oriFd2, SacIIm, NarWt, oriVR6Kγ, [CmR], this study |
| p2396 | pUC18::metAD | pUC18::metAD, oriColE1, [ApR], (39) |
DNA procedures
Standard techniques were used for DNA manipulation and cloning (19). Restriction and DNA-modifying enzymes were purchased from New England Biolabs and Roche. DNA was isolated from agarose gels using the QIAquick gel extraction kit (Qiagen). Plasmid DNA was extracted using the miniprep or midiprep kits (Macherey-Nagel, Qiagen).
Polymerase chain reaction (PCR) was performed with GoTaq Flexi DNA polymerase (Promega) according to the manufacturer’s instructions. PCR products were purified using the QIAquick PCR purification kit (Qiagen). 1% agarose electrophoresis gels were used to visualize DNA. When necessary, the sequence of each constructed plasmid was verified using an ABI BigDye Terminator v.3.1 sequencing kit and an ABI Prism 3100 Capillary GeneticAnalyzer (Applied Biosystem).
Mismatched covalent circles preparation
Construction of the p7770 and p7771 phagemid vectors
Introduction of the NarI site
NarI-containing PCR products were obtained by mixing a p3634 core PCR product (using primers 3634BclI and dfrBcat2) with a p902 core PCR product (using primers dfrBcat1 and dfrBBclI) and by performing a final PCR amplification with 3634BclI and dfrBBcII primers. The expected product was then gel-purified, digested by BclI and self-ligated to create pSW24-attCaadA7-BglII-dfrB-oriFd1 (p4919).
M13 oriFd inversion (+ and – ssDNA production)
In order to invert the oriFd region of the p4919, we amplified the oriFd of the p421 plasmid (pSW24) using primers Fd2-1 and Fd2-2. The expected product was digested by KpnI and SalI and ligated into p4919 digested by the same enzymes.
Finally, we obtained two pSW24 derivative plasmids, pSW24-attCaadA7-BglII-dfrB-oriFd1 (p4919) and pSW24-attCaadA7-BglII-dfrB-oriFd2 (p4997), carrying the oriFD in both orientations ensuring the production of the bottom (oriFd1) and top strand (oriFd2), respectively.
Introduction of NarI and SacII mutations
We introduced a mutation in the NarI restriction site of the p4919 plasmid and in the SacII restriction site of p4997, in order to generate, after single-strand production and hybridization, G-T mismatched nucleotides in each restriction site.
The SacII mutant site was constructed by the annealing of two sets of complementary partially overlapping primers (5′-7560-1 and 3′-7560-1, and 5′-6978-2 and 3′-6978-2). After annealing, the primers’ ends reconstitute the EcoRI and SacI enzyme restriction sites. The product is ligated with similarly digested p4997 plasmid to obtain the p7771 plasmid. The NarI mutation was introduced by PCR using the 5′-NarIm and 3′-NarIm primers. The PCR products were digested by SacI and FspI and ligated with similarly digested p4919 plasmid to obtain the p7770 plasmid.
The presence of wild-type SacII and mutated NarI restriction sites in the p7770 plasmid and mutated SacII and wild-type NarI restriction sites in the p7771 plasmid was confirmed by digestion (Supplementary Figure S4A, left and central panels).
Single-strand DNA production
In order to produce single-strand DNA, we used the M13K07 Helper Phage, which is an M13 bacteriophage containing the origin of replication from p15A and the Km resistance gene from Tn903. Both phagemid vectors (p7770 and p7771) are introduced by transformation into F′ carrying pir strain cells (β2150 and ω4446) to obtain the ω7790 and ω7791 strains, respectively. The growth and M13 infection of F′ cells containing phagemid vectors for preparation of ssDNA were performed as described by the Invitrogen protocol (M13K07 Helper Phage). The single-strand M13 DNA was purified following the protocol provided by Qiagen (QIAprep Spin M13 kit).
Hybridization, oriFd elimination and ligation
The complementary single-strand DNA molecules were annealed, digested by both EcoRI and MfeI restriction enzymes in order to eliminate oriFd and self-ligated (since these enzymes generate ‘compatible cohesive ends’). We confirmed the loss of both wild-type NarI and SacII sites in the EcoRI–MfeI linearized mismatched substrate through resistance to NarI and SacII digestion (Supplementary Figure S4A, right panel).
Methylation test
To determine the methylation status of the mismatched covalent circles, we digested the EcoRI–MfeI linearized mismatched substrates with MboI (blocked by dam methylation) and Sau3A (insensitive to dam methylation) restriction enzymes (Supplementary Figure S3, right panel). A control was performed by digesting the p2396 non-methylated plasmid (prepared from the GM48 dam- strain; Supplementary Figure S3, left panel).
Non-replicative recombination assay
Mismatched covalent circles are used in the non-replicative recombination assay developed previously in the laboratory (18). This assay supplies the attC site on a ds plasmid that cannot replicate once introduced into the recipient cell by transformation. In this context, the attC site is extruded from ds circles as cruciform structures containing both top and bottom strands (18). Briefly, we transformed the MG1655mutS215 strain (ω2580) containing the pBAD::intI1 (p3938) and the pSU38Δ::attI1 (p929) plasmids with 200 ng of mismatched covalent circles. Note that competent cells were prepared in the presence of 0.2% arabinose to allow integrase expression. Transformants were selected on Cm-containing plates (the mismatched circles marker). As these circles cannot replicate in the recipient strain (ω7994), CmR clones correspond to attC × attI recombination events. The attC × attI cointegrate formation was checked by PCR with appropriate SWbeg and MFD primers (on 107 clones) and by SpeI digestion (Supplementary Figure S4B; see pattern examples, R1 and R2). The recombination point of eight randomly chosen clones was checked by sequencing using the SWbeg primer. After that, the 107 recombination products were analyzed for their resistance or sensitivity to both SacII and NarI restriction sites (Supplementary Figure S4C; see pattern examples, R1 and R2) and/or by sequencing (using SeqNarI and SeqSacII primers).
As a control, we performed the same experiment by transforming the MG1655mutS215 strain (ω2580) containing the pSU38Δ::attI1 but lacking pBAD::intI1. To establish a recombination frequency, we in parallel transformed the covalent mismatched circles in the same mutS strain lacking both the pSU38Δ::attI1 and pBAD::intI1 plasmids but containing a Pir-expressing plasmid (pSB118::pir116, p1177) ensuring the replication of the incoming circle. The recombination activity corresponds to the ratio of CmR clones obtained in pir– conditions (with and without integrase) to those obtained in pir+ conditions. Note that the efficiency of transformation of each strain was determined beforehand and used to adjust the final ratio and normalize the results. We obtained a recombination frequency of 2.2 × 10−3 in presence of integrase, while we did not obtain any recombination events (<1.1 × 10−5) in absence of integrase.
Moreover, as a supplementary control, we analyzed the CmR clones obtained in pir+ context. After pooling of ∼350 clones, plasmids were extracted and transformed in П1 pir+ competent cells (ω1628). Then, 101 transformants were analyzed for the resistance or sensitivity to the SacII and NarI digestion and/or by sequencing (using SeqNarI and SeqSacII primers).
Suicide conjugation assay
We used the ω1881 and ω4137 strains as donor and the ω8101, ω8106, ω8107, ω8109 and ω8122 recipient strains (Tables 1 and 2).
This conjugation assay was based on that of Biskri et al. and was previously implemented in Bouvier et al. (11,20). Briefly, the attC sites provided by conjugation are carried on a suicide vector from the R6K-based pSW family that is known to use the Pir protein to initiate its own replication. This plasmid also contains an RP4 origin of transfer (oriTRP4). The donor strain β2163 carries an RP4 integrated in its chromosome, requires DAP to grow in rich medium and can sustain pSW replication through the expression of a chromosomally integrated pir gene. The AB1157 recipient strain, which contain the pBAD::intI1 [ApR] (expressing the IntI1 integrase) and the pSU38Δ::attI1 [SpR] (carrying the attI1 site), lacks the pir gene and therefore cannot sustain replication of the attC-containing suicide vector. The only way for the pSW vector to be maintained in the recipient cell is to form a cointegrate by attC × attI recombination. The recombination frequency is calculated as the ratio of transconjugants expressing the pSW marker [CmR] to the total number of recipient clones [ApR and SpR]. Recombination frequencies correspond to the average of at least three independent trials. The attC × attI co-integrate formation was checked by PCR with the appropriate SWbeg and MFD primers (on eight randomly chosen clones per experiment). The attC × attI recombination point was checked by sequencing using the SWbeg primer.
RESULTS
aHJs are resolved by a replicative mechanism
We developed a genetic setup to precisely understand the mechanism of the resolution of the aHJ. We tagged both the top and bottom strands of the attC-containing molecule by introducing point mutations at two different positions, one in the vicinity of the attC site (at ∼30 bp) and the other in a distal position (at ∼730 bp; see the exact positions in Supplementary Figure S2). The choice of the mutation’s position allows us to follow the destiny of each entire strand separately and to verify if only one or both strands of the attC-containing molecule are involved in recombination. Both mutation-containing top and bottom strands were produced as single-strand DNA phagemid using a M13 Helper Phage, annealed and covalently ligated to obtain circles containing two mismatched positions (‘Materials and Methods’ section). These circles carry the attC sites, a Pir-dependant origin of replication (oriR6K) and a Cm resistance marker (CmR). We tested these circles in the previously developed non-replicative recombination assay (‘Materials and Methods’ section). Briefly, these Pir-dependent circles containing the attC sites were introduced by transformation into pir-deficient strains. In these strains, circles can only be maintained upon recombination with the attI site carried by a Pir-independent replicon. Transformants were selected on Cm (the circle resistance marker). The selection of CmR clones directly corresponds to attC × attI recombination events. If the HJ is resolved by a second strand exchange of the top attC strand, we expect that after plasmid segregation, half of the recombined molecules will carry the paired nucleotides corresponding to the mismatch introduced in the top strand and the other half will carry those corresponding to the mismatch introduced in the bs (see ‘Introduction’; Figures 2A and 3). Alternatively, if the HJ is directly resolved by replication fork passage, only the attC-reactive strand will be copied to generate the recombined molecule. Therefore, only the paired nucleotides corresponding to the mutation introduced in the reactive strand will be found on the recombined molecule (Figures 2B and 3).
Figure 3.
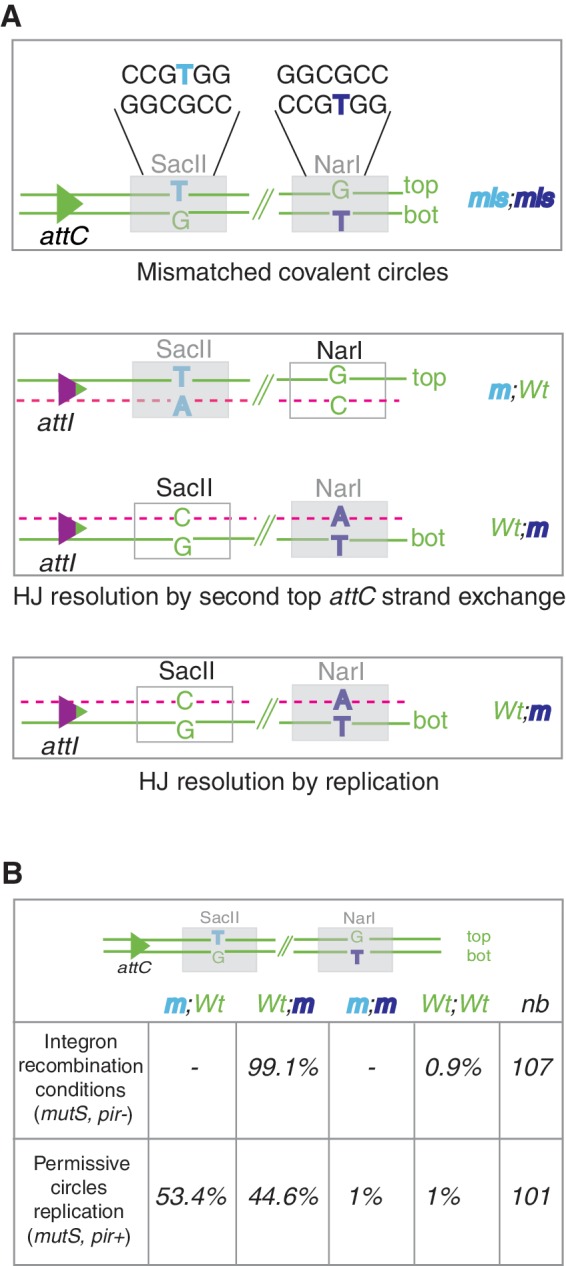
Strategy for the analysis of recombination products. (A) Sequences of the mismatched (mis)-containing regions in duplex substrate are represented. The mutated NarI and SacII restriction sites (m) are indicated by gray boxes. NarI and SacII restriction sites become cleavable (Wt) (white boxes) only following replication of the top strand (NarI) and the bottom (bot) strand (SacII). HJ resolution by top attC strand exchange followed by cointegrate segregation is expected to generate 50% of each product (m;Wt and Wt;m). HJ resolution by replication of the bottom recombined strand is expected to generate 100% of cleavable SacII and non-cleavable NarI products (Wt;m). (B) Segregation analysis in integron recombination conditions and permissive mismatched circles replication conditions are shown. The percent of each obtained product (m;Wt, Wt;m, m;m and Wt;Wt) is indicated. ‘nb’ represents the number of analyzed recombination events.
To easily detect the presence of mutations in the newly synthesized recombined molecules, we precisely introduced mutations in both SacII and NarI restriction sites so that mutated SacII and wild-type NarI sites will be representative of top strand replication, while wild-type SacII and mutated NarI sites will be representative of bs replication (Figure 3A). The results showed that a large majority (106/107 = 99.1%) of recombination events corresponds to a bs replication product ((Wt; m); Figure 3B and Supplementary Figure S4; clone R1). The remaining clone presents a Wt; Wt phenotype. This is probably because of a repair of the GT mismatch contained in the NarI site into a GC pair of nucleotides before the aHJ resolution (Supplementary Figure S4, clone R2).
To confirm that these results are not biased by a repair process such as methyl-direct mismatch repair (MMR) and are directly related to the attI × attC HJ resolution, we performed several controls. First, we demonstrated that the mismatched covalent circles were highly methylated (Supplementary Figure S3; ‘Materials and Methods’ section) and therefore refractory to MMR (21). Still, in order to prevent excision repair of the lesion and mismatch correction, all of the experiments were performed in a mutS genetic context where MMR is abolished (21). As an additional control, we transformed the mismatched circle in the same mutS strain expressing the Pir protein from a plasmid to ensure the replication of the incoming DNA circle out of the recombination reaction context (no integrase expression; ‘Materials and Methods’ section). In these conditions, we obtained a completely different segregation of the mutations in the replicated circles (close to 50/50 segregation as expected; Figure 3B). These results show an insignificant rate of repair of the GT mismatches contained in the covalent circles (1% of m; m and 1% of wt; wt) and confirm that the obtained rate of 99.1% (Wt; m products) in the recombination assay is not biased. Note that the 1% (1/101) of double mutant restriction sites (m; m and wt; wt) is the result of a MutS-independent repair mechanism. In any case, this control allows us to rule out the influence of repair processes in our recombination results.
Finally, these results are coherent with previous data showing that the bs of the attC site is the recombined strand. Moreover, we showed for the first time that only one strand, the one carrying the bottom attC site, is found in the recombined product. We proved that the obtained recombination products do not arise from a HJ resolution involving second strand exchange of the top attC strand, but result from replication of the full aHJ produced by the strand exchange of the bottom attC strand. As stated above, this mode of recombination also implies that the original substrate is replicated (Figure 2B).
Integron aHJs are not processed by RuvABC and RecG proteins
In E. coli, the ruvA, ruvB and ruvC gene products are required for recombinational repair of DNA damage. The RuvABC complex processes HJs made by RecA protein-mediated strand exchange (22). RuvA protein binds the four-way HJ with high affinity and acts as a specificity factor that directs RuvB (an ATPase helicase) to the junction (23,24). RuvB promotes ATP-dependent branch migration of the junction leading to the formation of heteroduplex DNA. The third protein, RuvC, is a nuclease that cleaves the HJ, thereby resolving it into two duplex DNA (25).
RecG, like RuvAB, is a branched DNA-specific helicase. RecG is unable to process model HJs in vitro, but can bind and unwind a variety of branched substrates including three-strand junctions, D-loops or R-loops to form four-stranded HJs as substrates for the RuvABC complex or to convert branched structures in suitable substrates for PriA-directed reloading of the replication machinery (see review (26)). In vitro, RecG has a higher affinity for substrates mimicking replication forks in which synthesis on the leading strand terminates before synthesis on the lagging strand (27,28). These substrates therefore include a ss region such as the atypical integron HJs.
We tested the involvement of these host factors mediating branch migration and/or potential second strand cleavage in integron aHJ resolution. We used our previously described ‘suicide’ conjugation assay (11) in ruvABC and recG mutant recipient strains. We studied the recombination of both attCaadA7 and VCR2/1 natural attC sites. This assay involves conjugation and thus proceeds exclusively via ssDNA transfer to deliver the attC site in ss form to a recipient cell expressing the IntI1 integrase and carrying the attI1 partner recombination site. As described in the Introduction, in this assay we cannot exclude before the first cleavage of the bottom attC strand a synthesis of the top strand and therefore a HJ resolution by branch migration followed by a second cleavage of the top attC strand. The obtained results are shown in Table 3. We did not find any significantly different recombination frequencies for both tested attC sites, no matter the genetic background of the recipient strains used (Table 3). Thus, as expected from a replicative resolution of the aHJ, neither RecG nor the RuvABC complex affects integron recombination. Hence, we confirmed that the integron HJ resolution does not imply a second strand exchange of the top attC strand but rather the complete bs replication.
Table 3.
Recombination frequencies of the attCaadA7 and VCR2/1 sites obtained in ruvABC and recG-deficient strains
| Genetic background | VCR2/1 | attCaadA7 |
|---|---|---|
| WT | 4.5 × 10−3 ± 1.4 × 10−3 | 7.4 × 10−3 ± 5.0 × 10−3 |
| ruvA60 | 6.4 × 10−3 ± 3.9 × 10−3 | 3.4 × 10−3 ± 1.5 × 10−3 |
| ruvB52 | 1.3 × 10−2 ± 8.7 × 10−3 | 4.0 × 10−3 ± 2.3 × 10−3 |
| ruvC53 | 1.2 × 10−2 ± 7.2 × 10−3 | 5.0 × 10−3 ± 1.8 × 10−3 |
| ΔrecG263 | 3.1 × 10−3 ± 1.9 × 10−3 | 4.7 × 10−3 ± 1.3 × 10−3 |
DISCUSSION
Resolving the junction
Studies of the recombination reactions performed in integrons revealed that this genetic system has evolved unique recombination processes involving non-canonical substrates such as ss attC sites. The fact that recombination involves a ss substrate puts forward a unique model in which recombination must stop after the first exchange contrarily to the classical site-specific recombination catalyzed by other Y-recombinases (Figure 1A). Indeed, a second strand exchange of the bottom attC strand would generate linearized abortive products with covalently closed ends. We suggested that proper resolution is thus dependent on unidentified host factors and a model relying on replication (Figures 1B and 2B). According to this hypothesis, cassette insertion would only affect one of the daughter DNA molecules upon replication. In this article, we first performed experiments which demonstrated the replicative nature of integron recombination. Indeed, we performed experiments involving the transformation and recombination of an attC-containing suicide vector where two mismatched nucleotides were introduced. This allowed us to follow the destiny of each strand separately and to verify if only one or both strands of the plasmid are involved in recombination. When an attC-containing suicide vector is transformed, the result is clear-cut. Indeed, we demonstrated that only one strand is involved: the one carrying the bottom attC strand. This, first, confirms previous results which showed that IntI1 has a single-strand preference for the bottom attC site. Indeed, it has been demonstrated that the recombination frequency obtained following delivery of the bottom attC strand by conjugation in a suitable recipient strain carrying the integron platform and expressing the integrase was 1000-fold higher than that obtained following delivery of the top strand. Second, our results indicate that the obtained recombination products do not result from a second strand exchange of any kind but arise from the replication of the full molecule produced by the first strand exchange of the bottom attC site. As stated above, this mode of recombination also implies that the original substrate is replicated. Thus, we conclude on the replicative nature of the integron recombination process (Figure 2B). We confirmed this result by showing that RecG protein and the RuvABC complex implicated in HJ resolution during recombination repair processes do not influence attI × attC integron recombination. These results are complementary to Messier and Roy’s studies, which demonstrated that the attC × attC integron recombination is RuvC-independent (29).
Although these results demonstrate that integron recombination involves a replicative process, they do not reveal the precise mechanism nor the nature of the host enzymes involved. Two pathways involving different sets of enzymes seem plausible. In the first scenario, the aHJ would remain unresolved until the replication fork assembled at the origin of replication of the host molecule arrives and resolves it. For E. coli, the set of enzymes involved in this pathway would include DnaA (the replication initiation factor, which promotes the unwinding or denaturation of DNA), DnaE (the catalytic subunit of the polIII polymerase essential for processive replication) and DnaN (the clamp, presumably essential for all kinds of replication). On the other hand, a second scenario in which the aHJ could mimic an arrested replication fork would involve the local recruitment of the replication complexes capable of restarting a halted fork. In E. coli, as in most bacteria, the key protein in this process is PriA. Depending on the cause of replication arrest, the PriA helicase can directly act upon branched DNA structures, such as Y-forks, which are structurally similar to the integron aHJ (30,31). Thus, one could potentially discriminate between these two different replication machineries for the resolution of the aHJ by using different set of mutants.
Explaining integron cassette duplication and transposon spreading
This atypical mechanism of recombination may be crucial for cassette ‘duplication’. Here, we demonstrated that the insertion of cassettes (attC × attI recombination) involves a replicative process. Since this mechanism seems directly linked to the ss nature of the attC site, we can easily consider that this mechanism is also involved in cassette excisions (attC × attC recombination). This mode of recombination which implies that the original substrate is replicated could account for cassette duplications. Indeed, a cassette that is excised and reintegrated at the attI site of the conserved integron (deriving from the replication of the top strand) would be duplicated in this molecule. Notably, large integrons, such as Vibrio cholerae’s superintegron, contain duplicated cassettes. Moreover, the divergence of duplicated cassettes may then increase the cassette diversity [e.g. aadA1 and aadA2 which are 89.3% identical over the whole cassette gene and attC site and likely come from duplication (32)].
Coupling of recombination and replication is also an efficient process of dissemination used by transposons. For example, bacteriophage Mu, Tn3 transposon family or IS6 insertion sequence family use a replicative mechanism resulting in the presence of one copy of the transposon in both the donor and the target DNA after transposition (reviewed in (33)). Indeed, simply maintaining transposon copy number by ‘passive’ replication would seem to be a poor strategy to promote their spreading within and between genomes.
ssDNA integration strategy
Other mobile DNA elements or viruses are known to use ssDNA forms that until recently were believed to be converted into dsDNA prior to their integration. But, recently, it has been demonstrated that secondary structures formed by ssDNA can also be the substrate for recombination reactions. This is the case for integrons, and also the CTX phage (34) and the IS200/605 insertion sequence family (35). We demonstrated here that the consequence of this ssDNA integration strategy in integrons is that only one pair of exchanges is performed creating an aHJ assisted by a host replication process to generate recombined products.
Finally, these results provide yet another example of host process involvement in integron recombination directly related to the special ss features of attC sites. The apparent requirement for a range of host-encoded factors in integron recombination illustrates how integrated biological systems integrons are.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
The Institut Pasteur, Centre National de la Recherche Scientifique [CNRS-UMR 3525]; French National Research Agency [ANR-08-MIE-016]; LABEX IBEID. The European Union Seventh Framework Programme [FP7-HEALTH-2011-single-stage under agreement no. 282004 to J.A.E.]; EvoTAR. Funding for open access charge: The Institut Pasteur.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Alfonso Soler Bistué, Zeynep Baharoglu and Michael Jason Bland for critical reading of this article.
REFERENCES
- 1.Cambray G, Guerout AM, Mazel D. Integrons. Annu. Rev. Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 2.Martinez E, de la Cruz F. Transposon Tn21 encodes a RecA-independant site-specific integration system. Mol. Gen. Genet. 1988;211:320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- 3.Levesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 4.Jove T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 2010;6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazel D. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 6.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 7.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 8.Collis CM, Hall RM. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 9.Stokes HW, O'Gorman DB, Recchia GD, Parsekhian M, Hall RM. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 10.Francia MV, Zabala JC, de la Cruz F, Garcia-Lobo JM. The IntI1 integron integrase preferentially binds single-stranded DNA of the attC site. J. Bacteriol. 1999;181:6844–6849. doi: 10.1128/jb.181.21.6844-6849.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. Embo. J. 2005;24:4356–4367. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouvier M, Ducos-Galand M, Loot C, Bikard D, Mazel D. Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS Genet. 2009;5:e1000632. doi: 10.1371/journal.pgen.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA specificity in integron recombination. Nature. 2006;440:1157–1162. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 14.Hall RM, Brookes DE, Stokes HW. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 15.Rowe-Magnus DA, Guerout AM, Biskri L, Bouige P, Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the vibrionaceae. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazel D, Dychinco B, Webb VA, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 17.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 18.Loot C, Bikard D, Rachlin A, Mazel D. Cellular pathways controlling integron cassette site folding. Embo. J. 2010;29:2623–2634. doi: 10.1038/emboj.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbour, USA: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- 20.Biskri L, Bouvier M, Guerout AM, Boisnard S, Mazel D. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 2005;187:1740–1750. doi: 10.1128/JB.187.5.1740-1750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983;104:571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West SC. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 23.Parsons CA, Stasiak A, Bennett RJ, West SC. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, West SC, Egelman EH. Structure and subunit composition of the RuvAB-Holliday junction complex. J. Mol. Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 25.Eggleston AK, West SC. Cleavage of holliday junctions by the Escherichia coli RuvABC complex. J. Biol. Chem. 2000;275:26467–26476. doi: 10.1074/jbc.M001496200. [DOI] [PubMed] [Google Scholar]
- 26.McGlynn P, Lloyd RG. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002;18:413–419. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- 27.McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- 29.Messier N, Roy PH. Integron integrases possess a unique additional domain necessary for activity. J. Bacteriol. 2001;183:6699–6706. doi: 10.1128/JB.183.22.6699-6706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka T, Mizukoshi T, Sasaki K, Kohda D, Masai H. Escherichia coli PriA protein, two modes of DNA binding and activation of ATP hydrolysis. J. Biol. Chem. 2007;282:19917–19927. doi: 10.1074/jbc.M701848200. [DOI] [PubMed] [Google Scholar]
- 31.Grompone G, Ehrlich SD, Michel B. Replication restart in gyrB Escherichia coli mutants. Mol. Microbiol. 2003;48:845–854. doi: 10.1046/j.1365-2958.2003.03480.x. [DOI] [PubMed] [Google Scholar]
- 32.Gestal AM, Stokes HW, Partridge SR, Hall RM. Recombination between the dfrA12-orfF-aadA2 cassette array and an aadA1 gene cassette creates a hybrid cassette, aadA8b. Antimicrob. Agents. Chemother. 2005;49:4771–4774. doi: 10.1128/AAC.49.11.4771-4774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turlan C, Chandler M. Playing second fiddle: second-strand processing and liberation of transposable elements from donor DNA. Trends. Microbiol. 2000;8:268–274. doi: 10.1016/s0966-842x(00)01757-1. [DOI] [PubMed] [Google Scholar]
- 34.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, Barre FX. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell. 2005;19:559–566. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B. In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol. Cell. 2008;29:302–312. doi: 10.1016/j.molcel.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marlière P, Mazel D. A new family of mobilizable suicide plasmids based on the broad host range R388 plasmid (IncW) or RP4 plasmid (IncPa) conjugative machineries and their cognate E. coli host strains. Res. Microbiol. 2005;156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Palmer BR, Marinus MG. The dam and dcm strains of Escherichia coli—a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 38.Demarre G, Frumerie C, Gopaul DN, Mazel D. Identification of key structural determinants of the IntI1 integron integrase that influence attC × attI1 recombination efficiency. Nucleic Acids Res. 2007;35:6475–6489. doi: 10.1093/nar/gkm709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto-Mashimo C, Guerout AM, Mazel D. A new family of conditional replicating plasmids and their cognate Escherichia coli host strains. Res. Microbiol. 2004;155:455–461. doi: 10.1016/j.resmic.2004.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.