Abstract
The abortive activity of topoisomerases can result in clastogenic and/or lethal DNA damage in which the topoisomerase is covalently linked to the 3′- or 5′-terminus of a DNA strand break. This type of DNA damage is implicated in chromosome translocations and neurological disease and underlies the clinical efficacy of an important class of anticancer topoisomerase ‘poisons’. Tyrosyl DNA phosphodiesterase-1 protects cells from abortive topoisomerase I (Top1) activity by hydrolyzing the 3′-phosphotyrosyl bond that links Top1 to a DNA strand break and is currently the only known human enzyme that displays this activity in cells. Recently, we identified a second tyrosyl DNA phosphodiesterase (TDP2; aka TTRAP/EAPII) that possesses weak 3′-tyrosyl DNA phosphodiesterase (3′-TDP) activity, in vitro. Herein, we have examined whether TDP2 contributes to the repair of Top1-mediated DNA breaks by deleting Tdp1 and Tdp2 separately and together in murine and avian cells. We show that while deletion of Tdp1 in wild-type DT40 cells and mouse embryonic fibroblasts decreases DNA strand break repair rates and cellular survival in response to Top1-induced DNA damage, deletion of Tdp2 does not. However, deletion of both Tdp1 and Tdp2 reduces rates of DNA strand break repair and cell survival below that observed in Tdp1−/− cells, suggesting that Tdp2 contributes to cellular 3′-TDP activity in the absence of Tdp1. Consistent with this idea, over-expression of human TDP2 in Tdp1−/−/Tdp2−/−/− DT40 cells increases DNA strand break repair rates and cell survival above that observed in Tdp1−/− DT40 cells, suggesting that Tdp2 over-expression can partially complement the defect imposed by loss of Tdp1. Finally, mice lacking both Tdp1 and Tdp2 exhibit greater sensitivity to Top1 poisons than do mice lacking Tdp1 alone, further suggesting that Tdp2 contributes to the repair of Top1-mediated DNA damage in the absence of Tdp1. In contrast, we failed to detect a contribution for Tdp1 to repair Top2-mediated damage. Together, our data suggest that Tdp1 and Tdp2 fulfil overlapping roles following Top1-induced DNA damage, but not following Top2-induced DNA damage, in vivo.
INTRODUCTION
DNA strand breaks are induced by a variety of endogenous or exogenous genotoxic agents. Accumulation of these breaks threatens genome stability and underlies the clinical utility of several anti-cancer strategies including radiotherapy and chemotherapy. DNA breaks can arise directly from attack of endogenous reactive oxygen species or by exposure to ionizing radiation. They can also arise indirectly through the abortive activity of DNA topoisomerases, which undergo a rapid catalytic cycle in which the topoisomerase becomes covalently attached to the 3′- or 5′-terminus of a DNA strand break (1). The hydrolytic removal of the covalently linked topoisomerase from DNA termini is required for repair of the break and thus for restoration of genetic integrity. The prototype example of this hydrolytic activity was first identified by Nash and coworkers (2) in 1996 and was named tyrosyl DNA phosphodiesterase-1 (TDP1). Homozygous mutation of TDP1 underlies the cerebellar degeneration observed in a human hereditary disease with spinocerebellar ataxia and axonal neuropathy (3). Consistent with its role during repair of Top1-linked DNA breaks, cellular depletion of TDP1 results in accumulation of Top1-linked DNA breaks and sensitizes cells to Top1 poisons such as camptothecin (CPT) (4–6).
Although TDP1 is the main 3′-tyrosyl DNA phosphodiesterase (3′-TDP) in human cells, cells lacking TDP1 exhibit residual Top1-DNA processing activity, suggesting that other cellular mechanisms may be able to compensate for TDP1 (7,8). In a hunt for such activities, we recently identified TTRAP/EAPII, a metal-dependent phosphodiesterase of unknown function that associates with CD40, tumor necrosis factor (TNF) receptor, TNF receptor-associated factors (9), and that can influence TGF-β signalling (10,11). We demonstrated that TTRAP possesses both weak 3′-TDP activity and robust 5′-tyrosyl DNA phosphodiesterase (5′-TDP) activity in vitro, and consequently denoted this protein TDP2 (12). Consistent with its 5′-TDP activity, TDP2 is required for cellular resistance to Top2 poisons, which induce DNA strand breaks in which Top2 is covalently linked to 5′-DNA termini through a 5′-phosphotyrosyl bond (13). However, cells depleted or lacking Tdp2 are not hypersensitive to Top1 poisons, suggesting that the relatively weak 3′-TDP activity of this protein is not required for repair of Top1-induced DNA damage (12,13). Herein, we have addressed this question in detail and examined whether TDP2 is required for repair of Top1-induced DNA damage in the absence of TDP1. Our data demonstrate that while TDP1 is the primary source of cellular 3′-TDP activity and resistance to Top1-induced DNA strand breaks, TDP2 promotes the repair of Top1-induced DNA damage in cells in which TDP1 is absent.
MATERIALS AND METHODS
Generation of Tdp1/Tdp2 double knockout mice and mouse embryonic fibroblasts
Tdp1−/− mice were generated, maintained and genotyped as described previously (14) in an outbred mixed 129Ola and C57BL/6 background. TTRAP (Tdp2−/−) mice were generated by targeted deletion of exons 1–3 using a Cre/Lox system and maintained as an outbred 129Ola and CD1 background (see Supplementary Figure S6 for the targeting strategy and confirmation of knockout alleles). A detailed description of these mice will be described elsewhere. To generate Tdp1−/−/Tdp2−/− mice, Tdp1+/− and Tdp2+/− mice were interbred to generate Tdp1+/−/Tdp2+/− double heterozygote mice, which were then interbred to generate the appropriate F2 progeny. Genotyping for the wild-type Tdp2 allele was conducted using primers 5′-CCTTCATTACTTCTCGTAGGTTCTGGGTC-3′ (LV043) and 5′-ACCCGCTCTTCACGCTGCTTCC-3′ (A183). Primers LV103 and 5′-TACACCGTGCCATAATGACCAAC-3′ (A185) were used to amplify the mutant Tdp2 allele deficient in exons 1–3. Polymerase chain reaction (PCR) conditions were 94°C for 30 s, 60°C for 1 min and 72°C for 1 min, for 35 cycles, resulting in the amplification of a 429-bp fragment from the wild-type allele or a 561-bp fragment from the mutant allele. All animals were housed within the School of Life Sciences and maintained in accordance with the institutional animal care and ethical committee at the University of Sussex. For the generation of WT, Tdp1−/−, Tdp2−/− and Tdp1−/−/Tdp2−/− mouse embryonic fibroblasts (MEFs), embryos from appropriate matings were harvested 14-d.p.c and were dissociated, trypsinized and plated in Dulbecco's modified Eagle's medium (DMEM) with 10% foetal calf serum (FCS). Primary MEFs were maintained in DMEM media supplemented with 10% FCS, at 37°C and maintained at 5% oxygen. Genotypes were confirmed by PCR and loss of Tdp1 and/or Tdp2 protein was confirmed by activity assays on cell lysates (see below). Xrcc1−/− MEFs were a kind gift from Prof Larry Thompson.
Generation of Tdp1−/− and Tdp1−/−/Tdp2−/−/− DT40 cells
DT40 chicken cells were maintained in RPMI 1640 medium containing 10−5 M β-mercaptoethanol, penicillin, streptomycin, 10% FCS and 1% chicken serum (Sigma) at 39°C. The generation of Tdp2−/−/− DT40 cells was described previously (13). To generate Tdp1−/− cells, genomic Tdp1 sequences were PCR amplified from DT40 genomic DNA (clone 18) to generate left and right arms for the targeting constructs using the primers 5′-CCCAAGCTTGCACAAGCACGCCCTTTTGAG-3′ and 5′-CGCGGATCCCATTCCTTGAGCACAGGAGAAC-3′ for the left arm and 5′-CGCGGATCCGCCTGTTGTGGGACAGTTCTCAAGC-3′ and 5′-AAGGAAAAAAGCGGCCGCCACAGCTGTTTCTGTGCGGTCTG-3′ for the right arm. The PCR amplified products were subcloned into pCR2.1-TOPO vector (Invitrogen) and confirmed by sequencing. Fragments encoding the left arm (2.9-kb) and right arm (2.2-kb) were recovered from the above pCR2.1-TOPO constructs using HindIII/BamH1 and BamH1/NotI, respectively, and subcloned into pCR2.1-TOPO vector. A BamH1 fragment encoding the puromycin (Puro) or hygromycin (Hyg) selection cassette was then inserted into the pCR2.1-TOPO constructs at the BamH1 site separating the left and right arms, completing the Puro-resistant (Puror) and Hyg-resistant (Hygr) TDP1-targeting constructs. To generate Tdp1−/− cells, 2 × 107 cells (clone 18) were first electroporated (Bio-Rad) with 30 μg of NotI-linearized Puror targeting construct (to disrupt the first allele) followed by, after confirmation of successful targeting as described below, the Hygr targeting construct (to disrupt the second allele). Following each round of transfection, transfected clones were selected for 8–10 days in the presence of medium containing 0.5 μg/ml of puromycin hydrochloride (Sigma) and/or 2.5 mg/ml hygromycin B (Sigma), as appropriate. To detect successful targeting of Tdp1 alleles, genomic DNA was isolated from drug-resistant clones, digested with NcoI and subjected to Southern blot analysis using a 0.55-kb probe amplified from genomic DNA clone 18 using the primers 5′-CGCAAGCCTAAATCAAAAGC-3′ and 5′-CCTGTTGCTCAACGCTGATA-3′. Following the two consecutive rounds of Tdp1 gene targeting, two Tdp1−/− clones were recovered (denoted clone 10 and clone 12) and characterized further, as indicated.
To generate Tdp1−/−/Tdp2−/−/− DT40 cells, the puromycin (Puro) resistance cassette present in Tdp2−/−/− DT40 cells (clone 8) (13) was first removed by transient transfection of a tamoxifen-inducible Cre recombinase expression construct (pANcreMer-Neo) (a gift from Helfrid Hochegger). Cre expression was induced in transiently transfected cell populations by incubation in growth medium containing 50 nM 5-hydroxytamoxifen (Sigma) for 1 day and single clones were selected by serial dilution to 1 cell/ml and amplified in 96-well plates for 5–7 days. Successful excision of the Puro-resistance cassette was confirmed by assessing the sensitivity of single clones to 0.5 μg/ml puromycin hydrochloride (Sigma). Tdp1 was then disrupted in one of the Puro-sensitive Tdp2−/−/− clones (clone 3) by sequential transfection with Puro-resistance and Hyg-resistance Tdp1 targeting constructs, as described above. Disruption of Tdp1−/− was confirmed in relevant clones by Southern blot analysis as described above and disruption of Tdp2−/−/− as reported previously (13). For complementation with human TDP2 (hTDP2), Tdp1−/−/Tdp2−/−/− mutant DT40 cells (clone 14) were electroporated with pcDNA3.1-HisC-TDP2 or pcDNA3.1-HisC empty vector as previously described (13) and pooled populations of transfected cells selected in medium containing 1.5 mg/ml G418 (Invitrogen) for 6 days.
Cell culture and western blotting
For detection of hTDP2 expression in DT40 cells, cells were lysed in 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 50 mm Tris–Cl, pH 8.0 and 1 mm phenylmethanesulfonylfluoride, complete protease inhibitor mixture (Roche) and analyzed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting using anti-TDP2 polyclonal antibody (SY1340).
Alkaline single-cell agarose gel electrophoresis assays
MEFs or DT40 cells were incubated with 20 μM CPT for 60 min at 37°C or exposed to 20 Gy ionizing radiation. Where indicated, cells were subsequently incubated in drug-free complete media for the indicated repair periods. DNA strand breakage was quantified by alkaline comet assays essentially as described (15). Briefly, cells were suspended in pre-chilled phosphate buffered saline (PBS) and mixed with equal volume of 1.2% low-gelling-temperature agarose (Sigma, Type VII) maintained at 42°C. Cell suspension was immediately layered onto pre-chilled frosted glass slides (Fisher) pre-coated with 0.6% agarose and maintained in the dark at 4°C until set, and for all further steps. Slides were immersed in pre-chilled lysis buffer (2.5 M NaCl, 10 mM Tris–HCl, 100 mM ethylenediaminetetraacetic acid (EDTA) pH8.0, 1% Triton X-100 and 1% dimethylsulfoxide (DMSO); pH 10) for 1 h, washed with pre-chilled distilled water (2× 10 min) and placed for 45 min in pre-chilled alkaline electrophoresis buffer (50 mM NaOH, 1 mM EDTA and 1% DMSO). Electrophoresis was then conducted at 1 V/cm for 25 min, followed by neutralization in 400 mM Tris–HCl pH 7.0 for 1 h. Finally, DNA was stained with Sybr Green I (1:10 000 in PBS) for 30 min. Average tail moments from 50 cells/sample were measured using Comet Assay IV software (Perceptive Instruments, UK). Data are the average ± s.e.m. of three independent experiments. Statistical analyses were conducted using student t-test.
Clonogenic survival assays
Primary MEFs were maintained at low oxygen (5%) and cells at passage 7 were plated in duplicate into 10-cm dishes (2000 cells for untreated cells, 6000 cells for treatment with 2.5 or 5 μM CPT and 10 000 cells for treatment with 7.5 and 10 μM CPT) and incubated at 37°C and 5% oxygen for at least 8 h. DT40 cells were plated in 5 ml medium containing 1.5% wt/vol methylcellulose (Sigma) in 6-well plates at 50, 500 and 5000 cells/well. Cells were then mock-treated or treated with the indicated doses of CPT or etoposide for 1 h at 37°C (MEFs) or during the course of the experiment (DT40). In case of MEFs, following treatment with CPT, cells were washed with PBS (3×) and then incubated for 7–10 days in drug-free medium to form colonies, which were then fixed with 90% ethanol and stained with 1% methylene blue. We typically obtained 60 colonies (a colony defined as ≥25 cells) out of 2000 cells plated (i.e. plating efficiency ≈ 0.03). Since colonies were very small and dispersed, we used a magnifier scope to count (Colony counter, Stuart, model SC6). Survival was calculated by dividing the average number of colonies on treated plates by the average number of colonies on untreated plates. Data are the mean ± s.e.m. of three biological replicates.
DNA substrates and in vitro repair assays
Gel-purified oligonucleotides were labelled with 32P at the 5-terminus or the 3′-terminus essentially as described (13). For the 5′-TDP substrate, a 5′-Y-18-mer [5′-Y-TCC GTT GAA GCC TGC TTT-3′] (Midland Certified Reagent Company, TX) was annealed with a 20-mer [5′-AGAA AGC AGG CTT CAA CGG A-3′] and the resulting 2-bp recessed 3′-terminus filled with ddTTP and [α-32P] dCTP using klenow DNA polymerase. For the 43-mer 3′-phosphotyrosyl SSB (nick) substrate, a radiolabeled 3′-Y-18-mer [32P-5′-TCC GTT GAA GCC TGC TTT-Y-3′] was annealed with a 25-mer [5′-GAC ATA CTA ACT TGA GCG AAA CGG T-3′] and a 43-mer [5′-CCG TTT CGC TCA AGT TAG TAT GTC AAA GCA GGC TTC AAC GGA T-3′]. 32P-labelled oligonucleotide duplexes were incubated at 50 nM with 5–20 μg total cell extract, in 8 μl total volume in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 8.0, 130 mM KCl, 1 mM dithiothreitol and 10 mM MgCl2 unless otherwise indicated. Reactions were incubated for 1 h at 37°C and stopped by addition of formamide loading buffer. Reaction products were fractionated by denaturing PAGE and analysed by phosphorimaging.
Immunostaining for γH2AX
MEFs grown on plastic coverslips were incubated with 1 µM CPT for 30 min at 37°C, washed 3× with PBS and re-incubated in CPT-free medium for 30 or 60 min repair periods. Cells were fixed with 3% paraformaldehyde for 10 min at room temperature (RT). Cells were then incubated with 0.2% Triton for 2 min at RT and subsequently rinsed with 3× PBS and incubated with 2% bovine serum albumin (BSA) for 30 min at RT to block nonspecific binding, followed by incubation with anti-mouse phospho-Histone (S139; Millipore) γH2AX monoclonal antibodies (1:1000 in BSA) for 30 min at RT. Immunopositive signal was finally detected using alexa Fluor 555 goat anti-mouse secondary antibodies (1:600 in BSA). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole and the number of γH2AX foci was counted from 50 non-S-phase cells. Data are the average of three independent experiment ± s.e.m.
CPT sensitivity in vivo
Administration of (S)-(+)-CPT was adapted from (16). Briefly, CPT was dissolved in PBS supplemented with 25% DMSO and diluted to a concentration of 0.25 mg/ml. Conditions were first established that resulted in minimal toxicity to wild type and Tdp1−/− mice. A fixed dose of CPT at 4 μg/g body weight was then administered by intraperitoneal injection of 3-month old littermate wild type, Tdp1−/−, Tdp2−/− and Tdp1−/−/Tdp2−/− mice. Animals were monitored daily for general health and body weight for a period of 10 days, followed by weekly monitoring of survivors for 3 months. Experiments were repeated on four independent littermates, each contained all indicated genotypes.
RESULTS
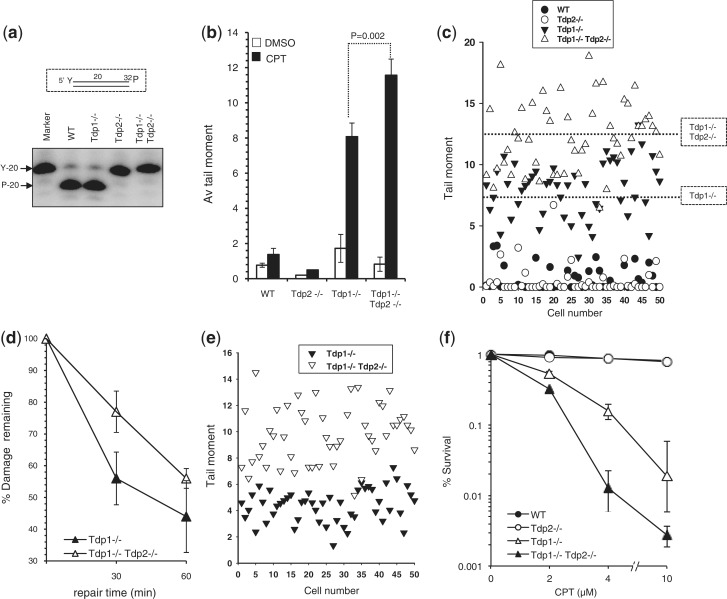
To examine whether Tdp2 contributes to the 3′-TDP activity in vertebrate cells, we generated MEFs in which Tdp1 and Tdp2 were deleted both separately and together. Note that successful inactivation of Tdp1 in the Tdp1−/− mice employed to generate these MEFs has been shown previously (14), and inactivation of Tdp2 is described briefly here (Supplementary Figure S6) and will be described in detail elsewhere. Cell extract from Tdp2−/− and Tdp1−/−/Tdp2−/− MEFs lacked detectable 5′-TDP activity in vitro, confirming Tdp2 disruption in these cells (Figure 1a). To examine whether Tdp2 might compensate for loss of Tdp1 in MEFs, we compared WT, Tdp1−/−, Tdp2−/− and Tdp1−/−/Tdp2−/− cells for their ability to repair DNA damage induced by the Top1 poison CPT, using alkaline comet assays. While Tdp1−/− cells accumulated ∼4-fold more DNA breaks than did wild-type cells, DNA breaks did not increase above background in CPT-treated Tdp2−/− MEFs (Figure 1b). However, co-deletion of Tdp1 and Tdp2 resulted in the accumulation of significantly more DNA breaks than did deletion of either gene alone (Figure 1b and c, P < 0.01) or loss of the critical single-strand break repair scaffold protein, Xrcc1 (Supplementary Figure S1). Moreover, Tdp1−/−/Tdp2−/− MEFs exhibited reduced rate of repair during subsequent incubation in CPT-free medium (Figure 1d and e). Consistent with these data, while Tdp2−/− MEFs exhibited normal levels of clonogenic survival following CPT treatment, Tdp1−/−/Tdp2−/− MEFs were more sensitive to CPT than were Tdp1−/− MEFs (Figure 1f). We conclude from these experiments that Tdp2 contributes to the repair of Top1-induced DNA damage in murine cells, in absence of Tdp1.
Figure 1.
Murine Tdp2 repairs Top1-mediated DNA damage in the absence of Tdp1. (a) Total cell lysate (10 μg) from WT, Tdp1−/−, Tdp2−/− or Tdp1−/−/Tdp2−/− MEFs was incubated with duplex DNA substrates harbouring the indicated 5′-phosphotyrosine (‘Y’) terminus (inset) and reaction products resolved and detected by denaturing PAGE and phosphorimaging. The positions of oligonucleotide substrate (‘Y-20’) and product (‘P-20’) harbouring 5′-phosphotyrosine and 5′-phosphate termini, respectively, are shown. (b) MEFs of the indicated genotype were incubated with DMSO or 20 μM CPT for 60 min at 37°C and DNA strand breakage quantified by alkaline comet assays. Mean tail moments were quantified for 50 cells/sample/experiment and data are the average of n = 3 biological replicates ± s.e.m. (c) A representative scatter plot of data from one of the experiments in (b) showing comet tail moments of individual cells (50 cells per sample). Dotted lines denote the position of the mean tail moments for the indicated genotypes. (d) MEFs of the indicated genotypes were subjected to CPT treatment as described in (b) followed by subsequent incubation in CPT-free media for a 30- or 60-min repair period. The fraction of DNA breaks remaining was calculated from n = 3 biological replicates and depicted as % damage remaining ± s.e.m. (e) A representative scatter plot of data from the 30 min repair time point from one of the experiments in (d), showing comet tail moments of individual cells (50 cells per sample). (f) MEFs of the indicated genotype were mock-treated or treated with the indicated concentrations of CPT and the number of surviving colonies determined after 7–10 days. Data are from the mean (±s.e.m.) of three independent experiments.
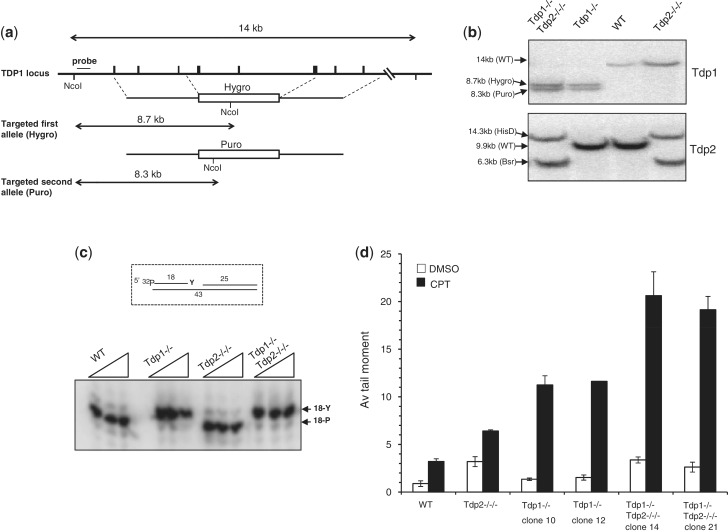
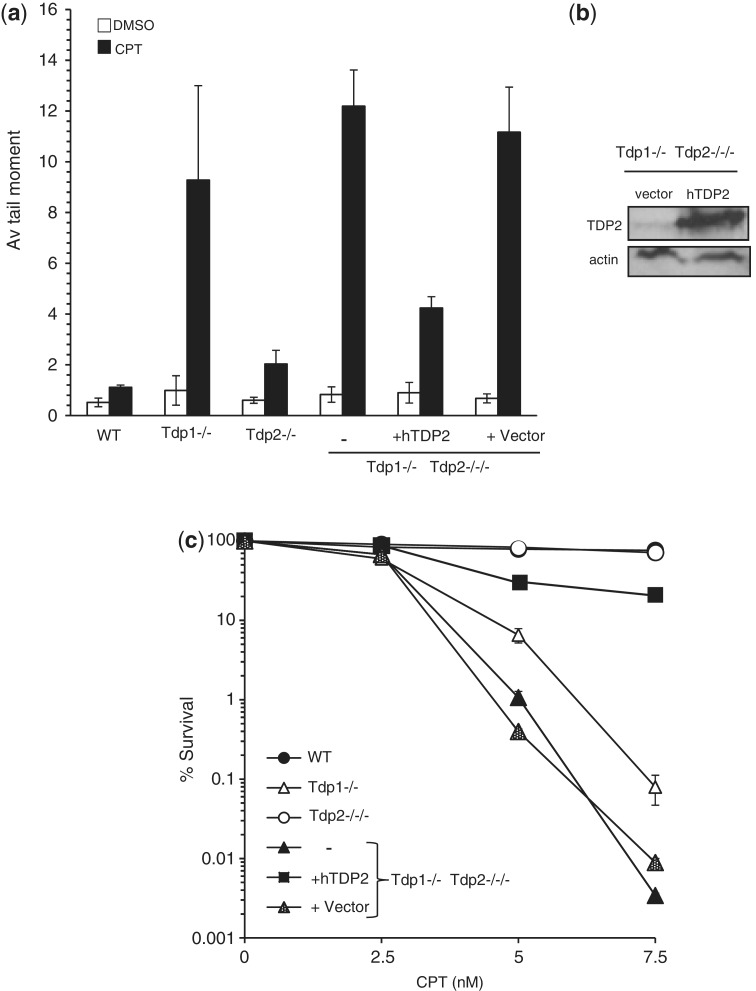
Next, we examined whether Tdp2 also contributes to the repair of Top1-induced DNA damage in avian DT40 cells, by again deleting Tdp1 and Tdp2 separately and together. Note that Tdp2−/−/− DT40 cells have been described previously (13), and the targeting strategy and successful deletion of Tdp1 is shown here (Figure 2a and 2b). Extracts from Tdp1−/− DT40 cells did not exhibit detectable 3′-TDP activity in vitro, even in reactions containing divalent metal and thus under conditions in which TDP2 is active, consistent with Tdp1 being the major source of 3′-TDP activity in DT40 cells (Figure 2c). As expected, Tdp1−/− DT40 cells accumulated ∼3-fold more DNA strand breaks than wild-type cells, following incubation with the Top1 poison CPT (Figure 2d). Tdp2−/−/− cells also accumulated higher levels of DNA breaks than did wild-type DT40 cells, though this difference was not statistically significant (P = 0.92; student’s t-test). Importantly, Tdp1−/−/Tdp2−/−/− cells accumulated significantly higher DNA breaks than Tdp1−/− cells, suggesting that Tdp2 contributes to the repair of Top1 damage in DT40 cells in the absence of Tdp1 (Figure 2d). Indeed, ectopic expression of hTDP2 in Tdp1−/−/Tdp2−/−/− cells reduced the accumulation of DNA strand breaks induced by CPT below that observed in Tdp1−/− cells (Figure 3a and b). Furthermore, Tdp1−/−/Tdp2−/−/− cells were more sensitive than Tdp1−/− or Tdp2−/−/− cells to Top1 breaks induced by CPT (Figure 3c), but not more sensitive to DNA breaks induced by γ-radiation (Supplementary Figure S3). Similar results were observed for the anti-cancer Top1 poisons NSC 724998, NSC 725776 or MJ-III-65 (1) as measured by viability assays (Supplementary Figure S4). In contrast, the reverse was not true since deletion of Tdp1 did not further sensitise Tdp2−/−/− cells to the Top2 poison etoposide, suggesting that Tdp1 is unable to contribute significantly to 5′-TDP activity in DT40 cells, even in the absence of Tdp2 (Supplementary Figures S2 & S3). We conclude from these data that TDP2 can contribute significantly to the repair of Top1-induced DNA damage in the absence of Tdp1, in both avian and murine cells.
Figure 2.
Avian Tdp2 repairs Top1-mediated DNA damage in absence of Tdp1. (a) Schematic representation of the wild type and targeted chicken Tdp1 locus. Closed boxes indicate the exons. ‘Hygro’ is a hygromycin cassette and ‘Puro’ is a puromycin cassette. The schematic representation of the targeting locus of the chicken Tdp2 gene has been described previously (13). (b) Genomic DNA from DT40 cells of the indicated genotype was digested with NcoI and Southern blotting was conducted using the probe depicted in (a) (for Tdp1, top) or as described in ref 13 (for Tdp2, bottom). (c) Total cell lysate (2, 10 and 20 μg protein) from avian cells of the indicated genotype was incubated with a nicked duplex DNA substrate harbouring a 3′-phosphotyrosine (‘18-Y’) terminus (top) for 1 h at 37°C and reaction products were fractionated by denaturing PAGE and detected by phosphorimaging. The positions of oligonucleotide substrate (‘18-Y’) and product (‘P-20’) harbouring 3′-phosphotyrosine and 3′-phosphate termini, respectively, are shown. (d) DT40 cells of the indicated genotype were incubated with DMSO or 20 μM CPT for 60 min at 37°C and DNA strand breakage quantified by alkaline comet assays. Mean tail moments were quantified for 50 cells/sample/experiment and data are the average of n = 3 biological replicates ± s.e.m.
Figure 3.
Over-expression of hTDP2 protects avian DT40 cells lacking Tdp1 from Top1-mediated DNA damage. (a) DT40 cells stably transfected with empty vector or vector encoding full-length hTDP2 ‘hTDP2’ were treated with DMSO vehicle or 20 μM CPT for 1 h at 37°C. DNA strand breaks were quantified by alkaline comet assays in 50 cells/sample/experiment and data are the average of n = 3 biological replicates ± s.e.m. (b) Total cell lysate (30 μg protein) from Tdp1−/−/Tdp2−/−/− cells harbouring empty vector or vector encoding hTDP2 was fractionated by SDS–PAGE and immunoblotted using anti-hTDP2 polyclonal antibody (SY1340). Actin immunoblots were used as a loading control. (c) DT40 cells of the indicated genotype were treated with the indicated concentrations of CPT and the number of surviving colonies was calculated from n = 3 biological replicates ± s.e.m.
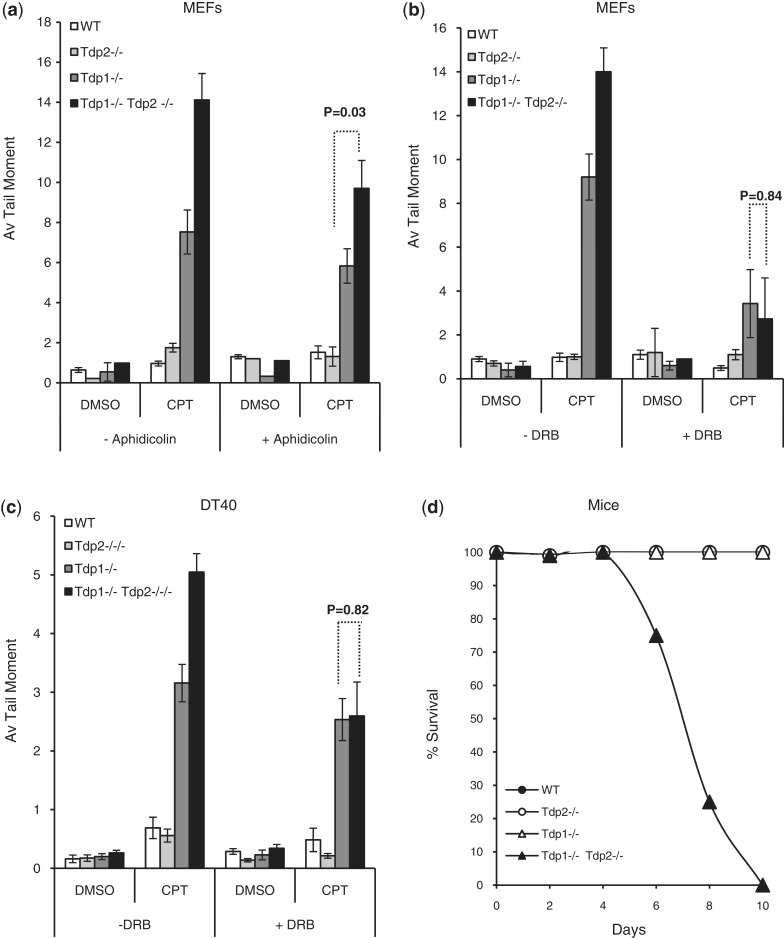
Top1-associated DNA breaks arise by collision of Top1 intermediates with DNA replication forks or elongating RNA polymerases. To examine the source of the Top1-induced DNA breaks that Tdp2 can repair, we used chemical inhibitors of DNA transcription and replication. First, we inhibited DNA replication in MEFs by incubating cells with aphidicolin and then compared the accumulation of DNA strand breaks in Tdp1−/− and Tdp2−/− cells during incubation with CPT. While aphidicolin reduced the level of DNA strand breaks that accumulated in Tdp1−/−/Tdp2−/− cells (Figure 4a), the residual level of DNA breaks was still higher than that in Tdp1−/− cells, suggesting that a significant proportion of the Top1-induced breaks that are repaired by Tdp2 arise independently of DNA replication (Figure 4a, P = 0.03). Consistent with this observation, the transcription inhibitor 5,6-dichloro-1-β-d-ribobenzimidazole (DRB) reduced the level of DNA breaks that accumulated in CPT-treated cells to a similar level in Tdp1−/− and Tdp1−/−/Tdp2−/− cells (Figure 4b, P = 0.8). Moreover, similar results were observed in avian DT40 cells (Figure 4c, P = 0.82). Together, these observations suggest that, in the absence of Tdp1, Tdp2 facilitates the repair of Top1-induced DNA strand breaks that arise, primarily, during transcription.
Figure 4.
Tdp2 preferentially repairs transcription-associated Top1-DNA breaks. MEFs (a and b) or DT40 cells (c) of the indicated genotype were incubated with DMSO vehicle or with 50 μM of either aphidicolin or 5,6-dichlorobenzimidazole 1-b-d-ribofuranoside (DRB) for 2 h at 37°C, followed by an additional incubation with 20 μM ‘CPT’ for 1 h at 37°C. DNA strand breaks were quantified by alkaline comet assays for 50 cells/sample/experiment and data are the average of n = 3 biological replicates ± s.e.m. (d) 3-month-old littermate wild type, Tdp1−/−, Tdp2−/− and Tdp1−/−/Tdp2−/− mice at 4 weeks old were ip-injected with CPT at 4 μg/g body weight. Animals were monitored daily for general health and body weight for a period of 10 days, followed by weekly monitoring of survivors for 3 months. The % of surviving mice was calculated from four independent littermates of each genotype.
Finally, to examine whether the impact of Tdp2 on Top1-induced DNA strand breakage is important physiologically, we examined the response of Tdp1−/−/Tdp2−/− mice to treatment with concentrations of CPT that were non-toxic to WT, Tdp2−/−, and Tdp1−/− mice. Strikingly, while all WT, Tdp2−/−, and Tdp1−/− mice survived for >100 days after CPT treatment, none of the Tdp1−/−/Tdp2−/− mice survived beyond 10 days (Figure 4d). These data suggest that Tdp2 contributes to the repair of Top1-induced DNA damage in vivo, in absence of Tdp1.
DISCUSSION
DNA topoisomerases introduce transient breaks in the genome to release torsional stress. Failure to reseal broken DNA strands results in protein-linked DNA breaks, which in turn are implicated in neurodegeneration and underlie the clinical utility of topoisomerase poisons as chemotherapeutic agents (3,17). TDPs liberate DNA termini from the covalently stalled topoisomerase by cleaving the covalent phosphotyrosyl bond linking the topoisomerase to DNA, a process that is tightly regulated by post-translational protein modifications (18). Eukaryotes possess two distinct TDPs as defined by their enzymatic activities in vitro. These are a metal independent TDP1, which primarily acts on DNA breaks with 3′-phosphotyrosyl termini and a metal-dependent TDP2, which acts on DNA breaks with 5′-phosphotyrosyl termini (2,12,13). TDP2, however, also possesses weak 3′-TDP activity in vitro and in fact was identified in a screen for novel TDP1-like activities that complement the sensitivity of Tdp1-mutant budding yeast cells to Top1-induced DNA damage. Consequently, we have compared in this study the role of TDP2 in processing Top1-induced DNA breaks in the presence and absence of TDP1 activity, by using murine and avian DT40 cells in which Tdp1, Tdp2 or both were deleted.
Avian DT40 and murine cells lacking Tdp2 exhibited normal sensitivity to CPT, whereas cells lacking Tdp1 were hypersensitive. Notably, however, co-deletion of Tdp1 and Tdp2 resulted in greater cellular sensitivity than did deletion of Tdp1 alone, suggesting that Tdp2 contributes to cellular resistance to Top1-induced DNA breaks in cells lacking Tdp1. Analogous results were observed in alkaline comet assays and in γ-H2AX immunostaining assays (Supplementary Figure S5), suggesting that Tdp2 contributes to the repair of Top1-induced DNA breaks in the absence of Tdp1. In agreement with these data, over-expression of hTDP2 is sufficient to complement the hypersensitivity of Tdp1-mutant budding yeast to CPT (12), and in this study to restore cell survival and DNA strand break repair rates in CPT-treated Tdp1−/−/Tdp2−/−/− DT40 cells to a level greater than that observed in Tdp1−/− DT40 cells. The simplest explanation for these data is that Tdp1 is the primary source of 3′-TDP activity in wild-type cells, but that Tdp2 contributes measurably to the repair of some of these breaks if Tdp1 is absent.
Incubation with the transcription inhibitor DRB prevented the appearance of the ‘extra’ Top1-induced breaks that accumulate in Tdp1-deleted cells if Tdp2 is additionally deleted, suggesting that the Top1-induced breaks that Tdp2 repairs in the absence of Tdp1 arise primarily during transcription. Recently, Tdp2 was shown to regulate the level of ribosomal RNA processing, consistent with the possibility that Tdp2 plays a role in removing DNA breaks at sites of transcription (19). In non-cycling tissues active transcription is the main source for Top1-breaks and their progressive accumulation may result in death of post-mitotic neurons. It is thus intriguing to speculate that TDP2 deficiency in human may result in neurodegeneration similar to that observed for TDP1 deficiency.
Structural studies suggest that the peptide-binding pocket in Tdp1 is unable to accommodate full-length Top1 and thus prior degradation of topoisomerase at DNA breaks is required for processing by Tdp1 (20,21). This notion was further supported by cellular and cell-free studies (7,22). While it is possible that the 3′-TDP activity of Tdp2 similarly prefers DNA breaks linked to short peptides, it is also possible that Tdp2 can remove full-length Top1 from DNA 3′-termini. This may explain the specific requirement of Tdp2 for maintaining ribosomal transcription upon proteasome inhibition (18). Whether or not human TDP1 operates at Top2 breaks in the absence of TDP2 is unclear. Studies in yeast, which lack a Tdp2 homologue, suggest that yeast Tdp1 is involved in the repair of Top2-induced DNA damage (23). Similarly, in human and DT40 cells, over-expression of human TDP1 has been reported to protect from Top2-mediated DNA damage (24,25). In agreement with the latter observation, weak activity of recombinant human TDP1 was reported on DSB substrates harbouring a 4-base pair 5′-phosphotyrosine overhang typical of Top2-induced breaks, in vitro (25). However, mice and murine cells lacking Tdp1 have not been reported to be sensitive to Top2-induced DNA damage (5,26). Furthermore, using colony survival assays Tdp1 deletion did not confer measurable sensitivity to Top2 poisons, even in the absence of Tdp2 (Supplementary Figure 2) suggesting that Tdp1 is not involved in the repair of Top2-induced DNA damage in higher eukaryotes.
In summary, we present evidence implicating Tdp2 in the repair of Top1-mediated DNA damage in cultured cells and in vivo, in the absence of Tdp1. Our data suggest that Tdp1 and Tdp2 fulfil overlapping roles following Top1-mediated DNA damage and highlight their possible utility as novel drug targets during cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6 and Supplementary Methods.
FUNDING
Z.Z. and L.J. were funded by MRC grants to K.W.C. [MR/J006750/1]; A.S. was funded by Wellcome trust grants to S.E.K. [085284 and 091043]; Generation of the Tdp2 (Ttrap)−/− mouse in the DH laboratory was supported by the EC FP6 Integrated Project EndoTrack and Interuniversity Attraction Pole IUAP-P6/20 and, in an early phase, VIB7 funding. Center for Cancer Research, the Intramural Program of the US National Cancer Institute, NIH [to J.M. and Y.P.]. Funding for open access charge: Wellcome Trust Fellowship (to S.E.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Shih-Chieh Chiang for technical assistance.
REFERENCES
- 1.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, et al. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 4.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 5.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme—DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou T, Akopiants K, Mohapatra S, Lin PS, Valerie K, Ramsden DA, Lees-Miller SP, Povirk LF. Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair. 2009;8:901–911. doi: 10.1016/j.dnarep.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair. 2007;6:1485–1495. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 8.El-Khamisy SF. To live or to die: a matter of processing damaged DNA termini in neurons. EMBO Mol. Med. 2011;3:78–88. doi: 10.1002/emmm.201000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N, de Boer M, Vandenabeele P, Huylebroeck D, Remacle JE. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J. Biol. Chem. 2000;275:18586–18593. doi: 10.1074/jbc.M000531200. [DOI] [PubMed] [Google Scholar]
- 10.Varady G, Sarkadi B, Fatyol K. TTRAP is a novel component of the non-canonical TRAF6-TAK1 TGF-beta signaling pathway. PLoS One. 2011;6:e25548. doi: 10.1371/journal.pone.0025548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esguerra CV, Nelles L, Vermeire L, Ibrahimi A, Crawford AD, Derua R, Janssens E, Waelkens E, Carmeliet P, Collen D, et al. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development. 2007;134:4381–4393. doi: 10.1242/dev.000026. [DOI] [PubMed] [Google Scholar]
- 12.Ledesma FC, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Z, Cortes-Ledesma F, El Khamisy SF, Caldecott KW. TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J. Biol. Chem. 2011;286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katyal S, El-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Khamisy SF, Katyal S, Patel P, Ju L, McKinnon PJ, Caldecott KW. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair. 2009;8:760–766. doi: 10.1016/j.dnarep.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovanella BC, Hinz HR, Kozielski AJ, Stehlin JS, Jr, Silber R, Potmesil M. Complete growth inhibition of human cancer xenografts in nude mice by treatment with 20-(S)-camptothecin. Cancer Res. 1991;51:3052–3055. [PubMed] [Google Scholar]
- 17.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 18.Hudson JJR, Chiang S-C, Wells OS, Rookyard C, El-Khamisy SF. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat. Commun. 2012;3:733. doi: 10.1038/ncomms1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilotti S, Biagioli M, Foti R, Dal Ferro M, Lavina ZS, Collavin L, Del Sal G, Zucchelli S, Gustincich S. The PML nuclear bodies-associated protein TTRAP regulates ribosome biogenesis in nucleolar cavities upon proteasome inhibition. Cell Death Differ. 2011;19:488–500. doi: 10.1038/cdd.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies DR, Interthal H, Champoux JJ, Hol WG. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Chem. Biol. 2003;10:139–147. doi: 10.1016/s1074-5521(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 21.Davies DR, Interthal H, Champoux JJ, Hol WG. Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes. J. Med. Chem. 2004;47:829–837. doi: 10.1021/jm030487x. [DOI] [PubMed] [Google Scholar]
- 22.Interthal H, Champoux JJ. Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase (TDP1) Biochem. J. 2011;436:559–566. doi: 10.1042/BJ20101841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitiss KC, Malik M, He X, White SW, Nitiss JL. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc. Natl Acad. Sci. USA. 2006;103:8953–8958. doi: 10.1073/pnas.0603455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, Pouliot JJ, Boege F, Marko D. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 25.Murai J, Huang SY, Das BB, Dexheimer TS, Takeda S, Pommier Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damages induced by topoisomerases I and II, and base alkylation in vertebrate cells. J. Biol. Chem. 2012;287:12848–12857. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, Bhattacharjee MB, Arimura K, Umehara F, Izumo S, et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.