Abstract
We present a comprehensive map of over 1 million polyadenylation sites and quantify their usage in major cancers and tumor cell lines using direct RNA sequencing. We built the Expression and Polyadenylation Database to enable the visualization of the polyadenylation maps in various cancers and to facilitate the discovery of novel genes and gene isoforms that are potentially important to tumorigenesis. Analyses of polyadenylation sites indicate that a large fraction (∼30%) of mRNAs contain alternative polyadenylation sites in their 3′ untranslated regions, independent of the cell type. The shortest 3′ untranslated region isoforms are preferentially upregulated in cancer tissues, genome-wide. Candidate targets of alternative polyadenylation-mediated upregulation of short isoforms include POLR2K, and signaling cascades of cell–cell and cell–extracellular matrix contact, particularly involving regulators of Rho GTPases. Polyadenylation maps also helped to improve 3′ untranslated region annotations and identify candidate regulatory marks such as sequence motifs, H3K36Me3 and Pabpc1 that are isoform dependent and occur in a position-specific manner. In summary, these results highlight the need to go beyond monitoring only the cumulative transcript levels for a gene, to separately analysing the expression of its RNA isoforms.
INTRODUCTION
Alterations in 3′ untranslated regions (UTRs) that drive post-transcriptional control of gene expression is a common theme in multiple diseases including cancer (1–3). Recent reports also provide surprising and compelling evidence that in proliferating/cancer cells, genes often switch their expression toward the short 3′ UTR isoforms that correspond to truncated versions of the canonical long isoforms (4,5). However, 3′ UTR isoforms are poorly annotated in databases (6) because common high-throughput technologies such as microarrays, RNA-Seq and quantitative reverse transcriptase-polymerase chain reaction cannot readily distinguish between 3′ UTR isoforms. This limitation arises because the long 3′ UTR isoforms encompass the short isoforms, and therefore the short isoform-specific probes/primers hybridize to both short and long isoforms, leading to inseparable, mixed signals (5,7). Such limitations in precisely identifying and quantifying 3′ UTR variants can lead to erroneous conclusions in not only gene expression studies but also in investigations of posttranscriptional events (4,5). New approaches such as Direct RNA sequencing or DRS (8), 3P-Seq (6), PAS-Seq (9) and others (10,11) are beginning to be adopted to identify 3′ UTR variants, allowing the construction of a near-complete map of 3′ UTR variants of the human genome (9,12).
Polyadenylation can generate many alternative transcripts of a given gene, with important consequences. Alternative polyadenylation (APA) can lead to truncated protein isoforms, abrogate protein-coding capacity, affect transcript stability, alter translation efficiency and affect transcript export (13). Important oncogenes such as p53 and CCND1 are known to have altered polyadenylation that results from simple 3′ UTR mutations (14,15). A related emerging theme in cancer biology is that APA within the same 3′ UTR can enhance production of oncogenes (5) because shorter 3′ UTR isoforms have higher translational efficiency than their respective long isoforms. Although APA within the same 3′ UTR simply results in mRNA isoforms that code for identical proteins, it can alter miRNA targeting regions (5), subcellular localizations and stability (16), and protein production rate (13). Thus, tandem APA seems to provide an alternative mechanism to complement more subtle posttranscriptional regulatory modes such as miRNAs that can cause either translational arrest (17) or destabilize target RNAs (18,19) in human cells (20). The existence of a mechanism that preferentially alters the relative expression ratio of long and short isoforms is also observed in embryonic cells (9,21). The diversity of transcripts produced via polyadenylation and its consequences underscore the need to accurately catalog and study polyadenylation in both normal and diseased cells.
We sought to build a comprehensive polyadenylation map of the human genome across major cancers and their cognate normal tissues, which could also facilitate studies on 3′ UTR isoforms, and enable discoveries of important novel gene isoforms. To reduce artifacts associated with common deep sequencing strategies, we used DRS to construct the polyadenylation landscape and measure gene expression directly without manipulating purified RNA (8). We built a public resource for tissue-specific polyadenylation sites termed Expression and PolyAdenylation Database (xPAD), which represents a comprehensive analysis and discovery platform for the investigation of more than 1 million polyadenylation sites, 3′ UTRs and their cellular usage, across five major organs, their cognate tumor samples, and six different cell types including commonly used ENCODE (22) cell types (Hela-S3, HepG2, K562 and MCF7). Approximately 9000 polyadenylation sites are well expressed (≥10 reads) and are at least 1000 nucleotides (nts) away from annotated gene regions, indicating the presence of many novel polyadenylated gene isoforms and genes. DRS reads also reveal potential polyadenylation in many non-coding RNAs including the lncRNA (23) GAS5 that encodes 10 snoRNAs, which are all unexpectedly polyadenylated. The simultaneous determination of gene isoforms and their levels also opens up additional opportunities in exploring the data, such as using network and motif analysis for studying gene isoforms. Indeed, network analysis of genes that manifest APA suggest that cell–cell and cell–extracellular matrix (ECM) contact mediated signaling cascades are potentially key targets of APA in cancer, particularly involving regulators of Rho family small GTPases that are consistently targeted by these mechanisms across the samples tested. The nucleotide level resolution of the polyadenylation maps also enabled the discovery of novel sequence motifs and regulatory marks (e.g. H3K36Me3) that are highly position specific with respect to polyadenylation sites and are isoform dependent. In summary, the polyadenylation site maps and its usage reported here reveal an extensive landscape of both known and novel polyadenylated genes, their isoforms, and their regulation.
MATERIALS AND METHODS
DRS sequencing and genome mapping
Matched pair (normal/tumor) total RNA from tissues was used for DRS (8,12); all paired RNA samples for sequencing were purchased from Biochain (Hayward, CA), except for Breast (Asterand, MI). For DRS analysis, the 3′ blocking reaction was performed with 3′deoxyATP (Jena Biosciences, Germany) by incubating the reaction mixture at 37°C for 30 min. Raw DRS reads were quality filtered using HeliSphere (http://open.helicosbio.com) and in-house tools (DocS1) to remove low-quality sequences and reads shorter than 25 nts. The filtered reads were mapped to the human genome (GRCh37) with maximum allowed error of 10% (mismatches and indels) by MOSAIK (24) with conservative parameters (-mmp 0.1 -mhp 100 -act 20 -hs 15 -p 8 -bw 13). Although internal priming in DRS data is negligible, we implemented filters for quality control, following a more stringent criterion than the recommended removal of sequences with a stretch of eight consecutive adenosines (25). For quality control, uniquely mapped reads were further filtered to remove all sequences that genomically contained either six consecutive Adenosines or at least seven Adenosines within 10 nts downstream of the end of the mapped DRS reads.
The mapped locations were annotated using the UCSC genome browser tables (26). When a locus could be attributed to multiple possible annotations, the locus was assigned with a single annotation in the following priority order: 3′ UTRs (sense), coding sequences (CDS, sense), 5′ UTRs (sense), intron (sense), non-coding RNAs (ncRNAs, sense), 5′ UTR antisense, CDS antisense, 3′ UTR antisense, intron antisense, promoter antisense, ncRNA antisense and intergenic. Unlike other regions, intergenic regions are not separated into sense and antisense strands. Promoter regions were defined as regions 1000 nts upstream of transcription start sites provided by UCSC genome browser tables. DRS reads mapping to regions that are at least 5000 nts away from the closest gene region (sense/antisense) were annotated as intergenic.
Because polyadenylation sites vary by a few nucleotides in a given isoform, we used the snow-ball method to define polyadenylation sites (27). The snow-ball method iteratively clusters genomic locations that are located within 24 nts, using the genomically mapped 5′ positions of the DRS reads as a reference. For analysis of genes containing tandem polyadenylation sites, polyadenylation sites with less than 10 total reads in the combined set of normal and tumor reads, were removed. The proximal polyadenylation site was defined as the first polyadenylation site after the end of the CDS on the 3′ UTR. The distal polyadenylation site was defined as the last polyadenylation site after the end of CDS on the 3′ UTR. If a gene contains more than two tandem polyadenylation sites on the 3′ UTR, only the first and last sites are used for analysis (5). To account for variations in sample loading, the number of reads from each polyadenylation site was standardized to contain an equal number of reads across all tissues, using the normal breast tissue reads as a reference. To enable more accurate comparisons, the resulting expression levels were further quantile-normalized (R package) between normal and tumor within each tissue. To analyse whether quantile normalizations of the samples are justified, Quantile–quantile plots of raw read numbers (log2 scale) between tumor and normal samples were analysed, which yielded high correlation (R = 0.98−0.99) along the diagonal. The distance between adjacent polyadenylation sites were defined as the distance between their 5′ most cleavage sites. To determine variances in polyadenylation site locations, the mean location of each polyadenylation cluster (≥10 reads) was determined. For a given cluster, polyadenylation sites corresponding to every read was used to determine the mean location of the polyadenylation site. For each polyadenylation site cluster, the distance between the polyadenylation site of each read and the mean location was used to calculate variance.
Motif-enrichment analysis
The total number of genes used for motif-enrichment analysis is identical for both short and long isoforms. Because some of the polyadenylation sites are tissue-dependent, motif analysis was constrained to short and long isoforms detected in a single tissue-type (normal breast). To ensure that the polyadenylation sites of short and long isoforms are well separated, we further constrained the analysis to those sites that are at least 100 nts apart, resulting in a total of 3270 genes. Motifs in regions flanking (±500 nts) the polyadenylation sites of the short and the long isoforms were detected based on the DREME (28) motif discovery algorithm, followed by a statistical assessment (E < 0.05) of the positional preference of the motifs, using CentriMo (29), a position-specific motif-enrichment analysis method. The analysis resulted in a total of 27 motifs, six of which were eliminated because they occur in a limited number of sequences, corresponding to less than 5% of the sequences at those distinguishing positions that are most enriched for the motifs. To focus on isoform-dependent motifs, the observed preference of each motif was further tested by the bootstrapping approach to determine statistical significance, and a final non-redundant set of eight motifs were selected using default parameters of TOMTOM (30).
Statistical tests
Both the Ansari–Bradley and F tests were used to test whether the variances in locations of polyadenylation-sites in short and long isoforms are statistically different (MATLAB). As F-tests yielded more statistically significant numbers, we report only the P values obtained using the most conservative test (one-tailed Ansari–Bradley). For testing the observed increase/decrease in tumor cells for the absolute or relative expression of short and long isoforms, the non-parametric one tailed Wilcoxon signed rank test was used. Bootstrapping analysis for motif detection was performed by randomly sampling (with replacement, n = 3270), the complete set of either short or long isoforms to yield 10 different datasets for each isoform. The resulting distribution of occurrences for each motif at their characteristic location in each of the 10 datasets were calculated for both short and long isoforms, and compared using two-tailed student t test.
RESULTS
Genomic features of polyadenylation sites
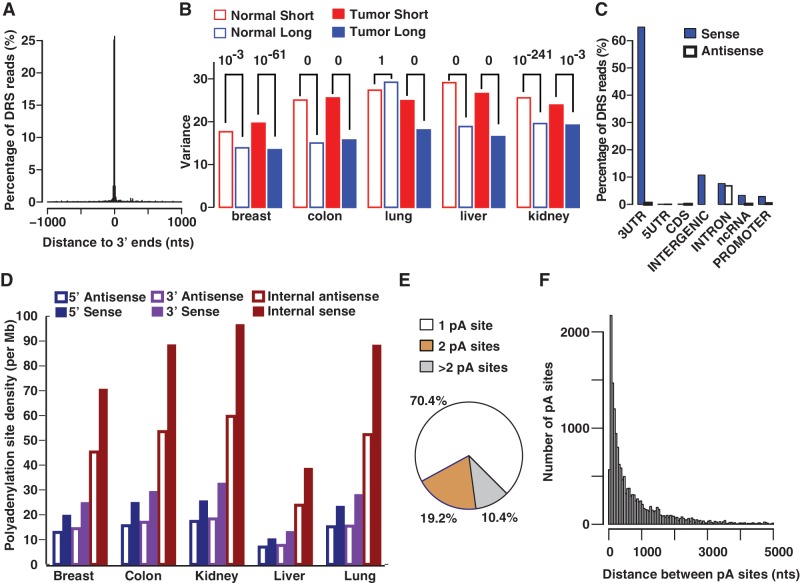
Five matched pairs (10 samples) of tumor and normal tissues from human breast, colon, kidney, liver and lung were used to obtain DRS reads using a total of 10 channels of the Helicos Sequencer. Both normal and tumor tissue samples yielded comparable number of reads. For example, a single channel for breast tissue generated 10 222 436 quality-filtered reads ranging from 25 to 64 nts, of which 2 628 790 reads remained after retaining high confidence reads (see ‘Materials and Methods’). The final reads dominantly mapped within ±10 nts of known 3′ ends of human genes expressed in normal breast tissue (Figure 1A). A similar pattern, in agreement with known 3′ UTR annotations is seen across all other normal and tumor tissues, demonstrating that the inferred polyadenylation maps accurately identify known polyadenylation sites (Supplementary Figure S1A). The total number of genes that were expressed in a given cancer type (reads ≥ 10) and contained tandem polyadenylation sites were approximately the same across organs: 4170 (breast), 4405 (colon), 4268 (lung), 3128 (liver) and 4728 (kidney). Consistent with previous reports, we find that although polyadenylation sites frequently vary (31,32) the average absolute deviation based on all distinct locations within a cleavage site cluster (see Materials and Methods) is small (±2.7 nts), indicating that cleavage is highly precise. Intriguingly, perhaps due to differences in the factors (e.g. motifs) that govern the cleavage of the short and long isoforms, short isoforms manifest a statistically significant higher variance for cleavage sites than long isoforms, across all tissues (Figure 1B).
Figure 1.
Characteristics of polyadenylation sites. DRS reads from normal breast tissue are used for this illustration. (A) DRS reads predominantly map to annotated 3′ ends of known genes (bin size = 10 nts). (B) Cleavage sites of short isoforms in either normal (normal short) or tumor (tumor short), are generally (9/10) more variant than that of the corresponding long isoforms (P values on top). (C) In breast, the majority (∼90%) of DRS reads match to sense strands of transcriptionally active regions and the remaining reads mainly map to intergenic regions and introns. For illustration purposes, intergenic polyadenylation sites are assigned to the sense strand because categorizing (see ‘Materials and Methods’) them into sense and antisense strands separately can be ambiguous. (D) Polyadenylation density (number of sites/Mb) in internal introns is higher than that of terminal (5′ and 3′) introns for both sense and antisense intronic transcripts. (E) A considerable fraction (∼30%) of genes contains tandem polyadenylation (pA) sites within the same 3′ UTR. (F) Distribution of distances between adjacent tandem 3′ UTR pA sites of genes expressed in normal breast (bin size = 50 nts). Because of long 3′ UTRs, the separation between adjacent pA sites within the same UTR can be very large (>5 Kb) in some cases (1%).
In normal breast tissue alone, we identified 401 873 distinct polyadenylation sites of which 36 093 sites generated at least 10 reads. Among a total of 1 287 130 polyadenylation sites that were identified across all tissues, 61 788 sites were sequenced multiple (≥10) times in at least one tissue. The majority (73−90%) of the reads mapped to the sense strands (Figure 1C and Supplementary Figure S1B) of functionally important regions (3′/5′ UTR, coding sequence i.e. CDS, intron, non-coding RNA i.e. ncRNA, and promoter—see Materials and Methods). The observation that sense transcripts constitute the major fraction (∼70%) of polyadenylated total RNA is fully in agreement with other studies (33,34). Notably, compared with other functional regions where polyadenylated antisense transcripts are rare, anti-sense transcripts are enriched in intronic regions across all 10 tissue types (Supplementary Figure S1B). Although most of these antisense transcripts are not abundant, ∼17% of these transcripts occur in clusters separated by less than 2500 nts, suggestive of transcription and subsequent polyadenylation of antisense transcripts in intronic regions. Further analysis of intronic transcripts did not indicate any dependence between the length of the introns and the number of polyadenylation sites, but revealed that in comparison with terminal (5′ or 3′) introns, internal introns are significantly more enriched in both sense and antisense polyadenylated transcripts (Figure 1D). The enrichment is consistently stronger for intronic sense transcripts than antisense transcripts. Although the polyadenylation site density in antisense internal intronic regions is lower than that of the cognate sense strand, it is always higher than that of terminal introns. The observed biases in intronic polyadenylation might be related to nucleosome depletion at internal introns (35) that may be more susceptible to aberrant intragenic transcription (36). Indeed, consistent with notion of aberrant transcription, the polyadenylation sites in these antisense regions follows a random pattern across most of the intron span, whereas the sense introns manifest a more biased distribution towards the 5′ start of the introns (Supplementary Figure S2).
Because tandem APA-mediated gene regulation that occurs without a change in the cognate protein sequence (37) is an emerging theme in cancer progression (4,5,38,39), we investigated whether polyadenylated 3′ UTR isoforms are commonly produced. Approximately 30% of genes have multiple tandem polyadenylation sites in their 3′ UTRs in any given tissue (Figure 1E and Supplementary Figure S3A). Most of these genes have precisely two polyadenylation sites. The observed proportion is nearly identical to the previously reported percentage of ∼34%, determined using EST mapping (27). Similarly, in accordance with the EST-mapping study, for those genes with multiple tandem polyadenylation sites, the adjacent polyadenylation sites peak ∼100 nts apart (Figure 1F and Supplementary Figure S3B) and have a median of 368 nts. Thus, the overall genomic characteristics of polyadenylation events captured by both DRS and ESTs are nearly identical.
Polyadenylation patterns of non-coding and novel genes
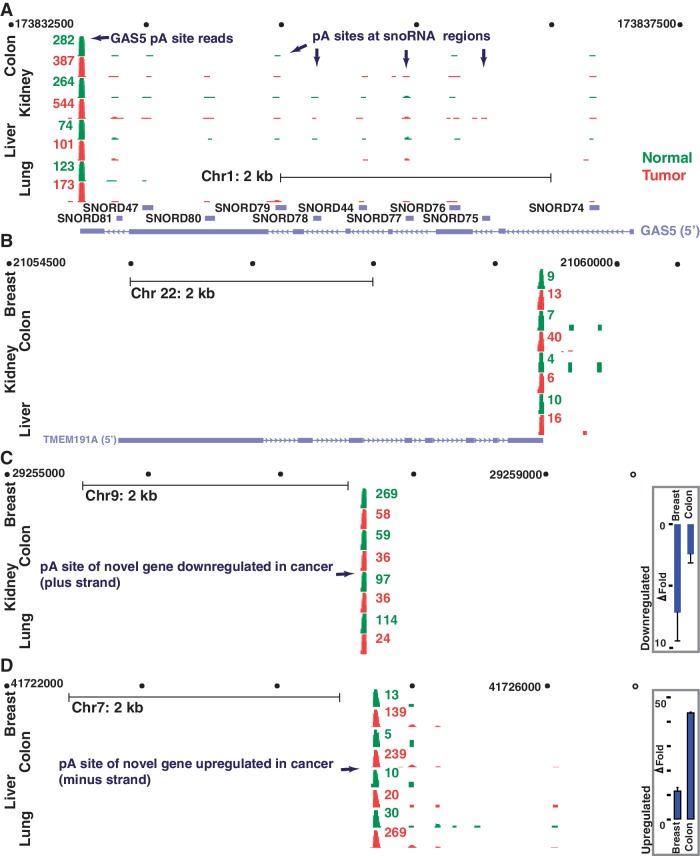
Non-coding RNAs such as miRNAs and lncRNAs (23) are frequently implicated in cancer progression (40) and in some cases are known to undergo polyadenylation (41,42). We analysed whether miRNA and lncRNA regions are polyadenylated and if so whether those polyadenylated forms are aberrantly expressed in cancer. A modest proportion (12.3%) of the 14 403 lncRNAs locations is polyadenylated within 100 nts of their annotated 3′ ends. However, most of these polyadenylated locations are not abundant, as only a small fraction (1.7%; 244 loci) of the lncRNAs generates many (≥10) reads in at least one of the 10 tissue samples tested. Abundantly polyadenylated lncRNAs loci include the well-known breast cancer metastasis-associated HOTAIR (41) that is highly (>12 fold) upregulated in breast tumor (49 versus. 4 reads). Although polyadenylated lncRNAs do not manifest a consistent pattern of differential expression across the majority of the tumors tested, the GAS5 and TMEM191A are two good examples of polyadenylated lncRNA regions that are aberrantly regulated in the majority of tumors tested (Figure 2A and B). The 4 kb long GAS5 that hosts 10 snoRNAs is frequently down-regulated gene in breast cancer (43). In DRS results, GAS5 is also 3-fold down-regulated in breast tumor. Notably, although only 36% of snoRNAs are polyadenylated in at least one of the tissues, all GAS5 snoRNA loci are usually polyadenylated in multiple tissues, generally within ∼5 nts of their annotated 3′ ends. Among all the miRNAs (939 loci) analysed, 25 (2.6%) locations are polyadenylated within 100 nts of the annotated 3′ end of the annotated precursor miRNA (pre-miRNA) sequence, suggesting that polyadenylation among pre-miRNAs is rare. Among these, only three pre-miRNAs (mir-147 b, −886, and −1975), generated at least 10 reads. Two of these loci, miR-886 and miR-1975 represent potentially incorrectly annotated miRNAs (44,45) as they derive from the longer non-coding RNAs, a vault RNA (VTRNA2-1) and a Y-RNA (RNY5). Interestingly, although the genomic location of one of the family members (let-7i) of the well-known let-7 does not contain an annotated neighboring (300 000 nts) gene within its downstream region, a polyadenylation site (∼40 reads in breast) that is within 650 bases downstream of let-7i is readily noticeable in xPAD.
Figure 2.
Genomic view of polyadenylated non-coding RNAs and novel gene locations that are aberrantly expressed in cancer using xPAD. (A–D) All polyadenylation sites detected by DRS reads (green, normal; red, tumor) are indicated for all four gene regions. (A, B) GAS5 (A) and TMEM191A (B) represent lncRNAs that are upregulated in the majority of tumor samples, as indicated. In contrast to the polycistronic GAS5 which hosts multiple polyadenylated snoRNAs, polyadenylation of TMEM191A is limited to its 3′ UTR. (C and D) End locations and the expression levels of two potentially differentially regulated novel genes that are distantly located from known genes. Real-time PCR results also reveal similar expression patterns (fold change, P < 0.001); error bars represent standard deviation (n = 3).
We next investigated the presence of novel polyadenylated genes that may be important for cancer development or progression. To identify such novel RNA transcripts, we probed for novel polyadenylation locations that are at least 1 kb away from any known gene, is abundant (≥10 reads) and is consistently either upregulated or downregulated in the majority of tissues. A total of 9612 potentially novel polyadenylated genes were identified, of which 77 were upregulated and 41 were downregulated in the majority (≥3) of the tumor samples at more than 2-fold levels (DocS2). The DRS results on the differential expression of two of these locations were also confirmed by real-time reverse transcriptase-polymerase chain reaction (Figure 2C and D). These results underscore the notion that many additional RNAs and RNA-isoforms important for cancer and possibly for other diseases exist and that visualization tools for deep sequencing data could be useful in identifying such genes.
Separate measurements of 3′ UTR isoforms reveal candidate APA targets
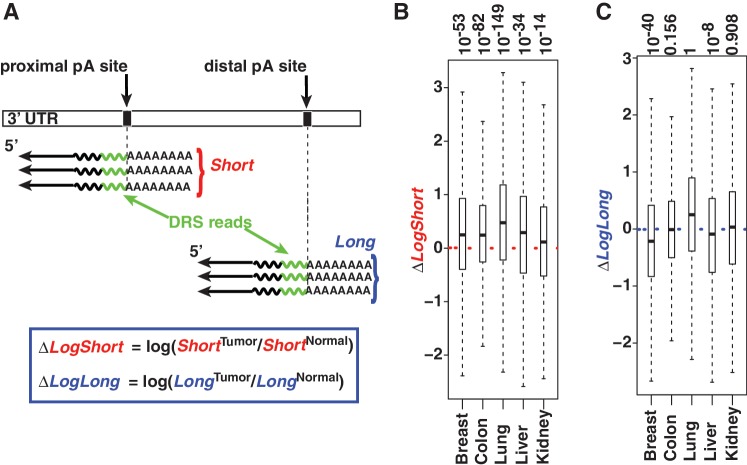
A recent study based on microarrays and a carefully selected set of six genes demonstrated that short isoforms are more upregulated in cancer cells than their long isoforms for at least three genes tested, suggesting that the phenomenon could be widespread (5). Because DRS allows the separation of short and long isoforms readily (Figure 3A), we analysed whether we could gain additional insights into the increased usage of short 3′ UTR isoforms in cancer. We first analysed whether the DRS reads accurately reflect genome-wide gene expression levels. To assess the reliability of the DRS data between two different experiments, the number of reads between each tissue type was compared. Repeatedly (≥10) sequenced reads were highly correlated within each normal-tumor pair (log2 scale; Pearson correlation: breast, 0.86; colon, 0.89; kidney/liver, 0.84; lung, 0.80). As an additional genome-wide validation analysis to assess the overall accuracy of the data, we tested whether the DRS data can replicate previously reported patterns of relative changes between the long and short isoforms of genes expressed in each tissue type. We adopted the index (46) termed Relative Usage of Distal polyadenylation sites (RUD) and used the difference of the index between tumor and normal to monitor each gene’s tendency to have a relatively shortened or lengthened 3′ UTR in tumor cells (DocS1). The analysis indicates that in all tissue types, tumor tissues manifest a lower index than in the corresponding normal tissue (ΔRUD <0, Supplementary Figure S4), which is in full agreement with previous reports based on proliferating cells, cancer cell lines, or cancer tissues (4,5,38). We next made use of the number of reads that are directly assigned to short and long isoforms to evaluate the expression patterns of short and long 3′ UTR isoforms, separately. Analysis of the read-based abundance of short isoforms reveals that short 3′ UTR isoforms tend to manifest upregulation in all tumor tissues tested (Figure 3B). In contrast, the general expression pattern of long isoforms varied across tissues (Figure 3C). These results are consistent with previous reports that the preferential increase of short isoform levels is a general genome-wide phenomenon of proliferating cells (4,5). Thus, the genome-wide data available via xPAD could serve useful in investigating changes at gene isoform-levels in cancer.
Figure 3.
Polyadenylation site usage reveal up-regulation of short isoforms in tumors. (A) Illustration on determining various 3′ UTR isoform-specific quantities (blue box) using DRS. Total number of normalized DRS reads for each isoform, abbreviated as Short (red) and Long (blue), are used to measure various changes between tumor and normal. (B and C) Median (thick line), 75% quantile (upper border), 25% quantile (lower border) and interquartile range (whiskers) are shown for each distribution (P values on top). (B) Short isoforms tend to be up-regulated in all tumor samples as indicated by the median (median >0, P values on top). (C) In contrast to short isoforms that have a consistent pattern, the median expression of long isoforms seems to flip between up/neutral/downregulation.
As DRS reads provided a direct measurement of the levels of each individual 3′ UTR isoforms, we sought to better define the core genetic programs affected by APA. We performed pathway analysis using those short isoforms that are preferentially more up-regulated than their long-isoforms in at least three tissues. Analysis of the 126 genes that are consistently affected by APA yielded top ranking pathways that are generally related to cell–cell (47) and/or cell–ECM contact-initiated signaling (48). Specifically, the top five statistically significant pathways include proteoglycan syndecan-mediated signaling (P = 0.01), the nectin adhesion pathway (P = 0.013), plasma membrane estrogen receptor signaling (P = 0.015) and the integrin signaling pathway (P = 0.017). Consistent with the inferred role of ECM, a more comprehensive network analysis of these genes implicates a large functional network containing two modules that are implicated in tumorigenesis (Supplementary Figure S5): the Rho family small GTPases that receive signals from the ECM and control actin reorganization (49), and the RNA processing machinery that cross-talks with Rho regulators and controls RNA expression, especially upregulation of Pol III transcripts (tRNAs, etc.) in cancer (50). Notable candidate target genes from these two modules include POLR2K, a subunit shared by all three RNA polymerases, which can specifically improve the assembly of the Pol III pre-initiation complex (51), and the neuroepithelial cell transforming 1 gene NET1, which regulates multiple Rho GTPases and controls cell movement and tumor metastasis (52,53). Real-time PCR quantification of both NET1 and POLR2K transcripts across three different tissues also suggests that the short isoforms of these genes are indeed preferentially upregulated with respect to their long isoforms in various tumor samples (Supplementary Figure S6). These observations highlight the notion that separate analysis of gene isoforms could prove useful in defining new genetic signatures of cancers or the underlying pathways.
Polyadenylation sites contain isoform-dependent regulatory marks
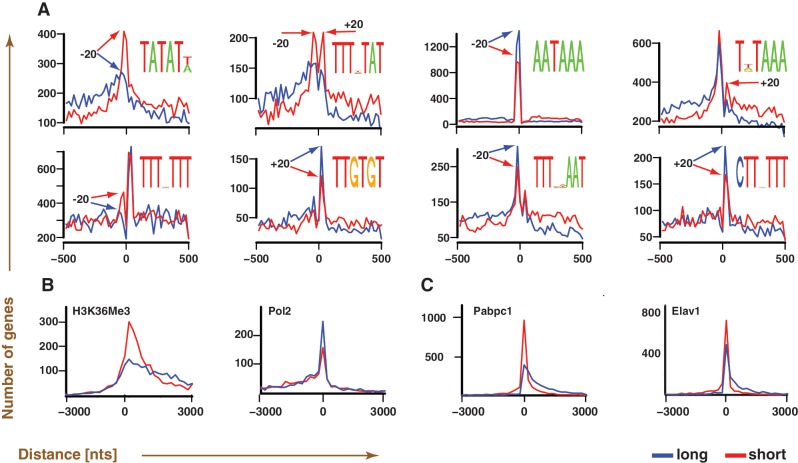
The presence of differentially controlled short and long isoforms raises a fundamental question: how do cells differentiate between the two isoforms of a given gene? As several polyadenylation signals are known to be highly position specific, we tested for the presence of sequence elements that are located in a highly position-specific manner with respect to polyadenylation sites and are differentially used between the long and short isoforms. In particular, an AT-rich motif (TATATW) is highly preferred by short-isoforms and is position specific, occurring dominantly within 20 nts upstream of the polyadenylation sites in short isoforms (Figure 4A and Supplementary Figure S7). Further analysis based on all possible hexamers indicates that its close relative ATATAT is the most short-isoform-specific AT-rich hexamer (Supplementary Figure S8). ATATAT and its close variants correspond to a group of polyadenylation enhancement elements that are located 30–50 nts upstream of the polyadenylation site in yeast (54), and are recognized by Hrp1, a protein involved in mRNA 3′ end formation, surveillance, and export (55,56). Other motifs detected include the canonical polyadenylation signal, AATAAA, which is biased towards the long isoform (9,57). The majority of the motifs are preferentially situated within ±20 bases of the polyadenylation sites, indicating that these motifs are likely involved in positioning cleavage/polyadenylation factors in an isoform-dependent manner.
Figure 4.
Polyadenylation maps enable the identification of isoform-dependent regulatory marks. (A–C) A total of 3270 genes containing both long and short forms that are genomically separated by at least 100 nts at their 3′ ends are used for the analysis. (A) Motifs that are preferentially located near polyadenylation sites (position 0), and are more prevalently used by either short (red) or long (blue) isoform. Readily noticeable locational preferences of the consensus motif, visible as peaks (arrows), are generally within ±20 nts of the polyadenylation sites. (B and C) Chip-Seq/RIP-Seq data comparisons of H3K36Me3 and Pabpc1 to their functional analogs (Pol2 and Elav1) in identical cell lines (B: HepG2, C: GM12878) suggest preferential marking of polyadenylation sites of short isoforms by H3K36Me3 and Pabpc1. The curves correspond to the distance distribution between the location of a given polyadenylation site and the nearest regulatory mark (H3K36Me3, Pol2, Pabpc1 and Elav1), as inferred using Chip-Seq/RIP-Seq.
We next sought to identify proteins that may differentially regulate short and long isoforms. Based on a large number (708) of publically available ENCODE ChIP-Seq or RIP-Seq data sets of transcription factors, histones and RNA binding proteins, we found that the histone mark, H3K36Me3 is consistently more enriched at short isoform polyadenylation site locations than that of long isoforms in all 42 datasets tested. In contrast, Pol2 occupancies at polyadenylation sites of long isoforms in the corresponding cell lines are higher than that of short isoforms (Figure 4B and Supplementary Figure S9). ENCODE RIP-Seq data analysis revealed that polyA-binding protein 1, Pabpc1 (Figure 4C), could be another candidate that preferentially associates with polyadenylation sites of short-isoforms. To ensure that the observed preference is not simply due to the mRNA binding ability of Pabpc1, the Pabpc1 profile was compared with that of a related mRNA binding protein Elav1 (58), performed using identical conditions. The comparisons reveal that the Pabpc1 is more preferentially enriched at short isoform locations, suggesting that Pabpc1 likely has a role in regulating the polyadenylation sites of short isoforms (Figure 4C). In conclusion, multiple regulatory marks that correspond to both transcriptional and post-transcriptional regulatory complexes are present at locations proximal to polyadenylation sites in an isoform-dependent manner, providing a suggestion as to the interplay between these proteins and APA.
DISCUSSION
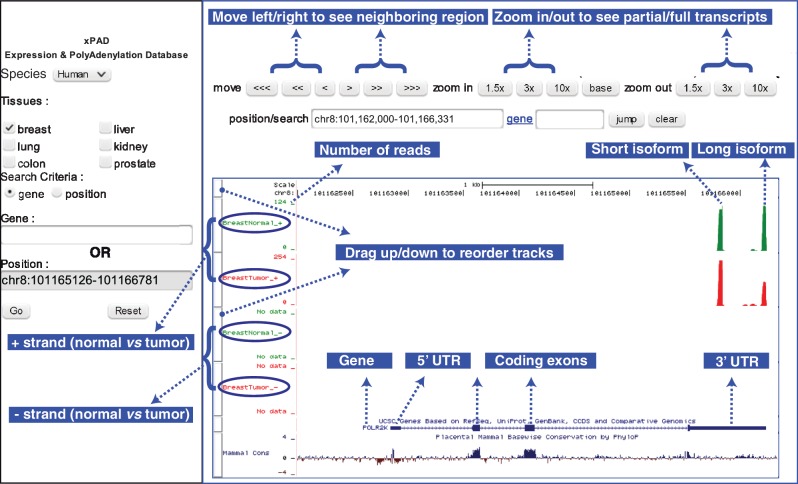
The biogenesis of eukaryotic RNA transcripts is made possible by a myriad of protein complexes that act in concert, orchestrating key tasks such as transcription, splicing, capping, 3′ end cleavage and polyadenylation. Although gene regulation by transcription is a widely studied process, the effects of other terminal processes such as polyadenylation that regulate transcript stability, transport and expression of RNA transcripts are poorly understood. Therefore, to help with the investigations of polyadenylation locations, 3′ UTRs, their different isoforms and usage, we built comprehensive maps of polyadenylation sites and quantified the usage of each polyadenylated gene isoform. The complete polyadenylation landscape is made publicly available through our web-portal, xPAD (Figure 5), which integrates the widely used UCSC genome browser (26) to enable detailed investigation of any genomic region by intuitive queries involving gene name, keywords or gene position.
Figure 5.
Illustration of xPAD. xPAD integrates the UCSC genome browser to provide a web-interface to visualize both the precise polyadenylation locations of different isoforms, as well as their expression levels across tissues of interest. The complete gene structure of POLR2K highlights the utility of DRS; in both normal and tumor tissues, all polyadenylation sites exclusively occur in the 3′ UTR of the gene and within the sense strand, and the 3′ end of the reads (green/red bars) mapping to the long isoform matches within 2 nts of the 3′ UTR polyadenylation site. Normal breast contains 120 reads of long, and 109 reads of short isoforms, whereas breast tumor contains 257 reads of short isoform, which is upregulated, and 134 reads of long isoform that is almost unchanged. For brevity, many additional features such as evolutionary conservation (bottom track) and methylation marks (not shown), which are available via the UCSC browser panel (right) are not illustrated.
Important functional consequences of polyadenylation have emerged recently (14,15,20,59,60). In particular, the disparate usage of various polyadenylation sites within the same 3′ UTR is known to result in reprograming of gene expression in proliferating cells (4), embryonic cells (9,21) and cancer cells (38). The availability of precise polyadenylation sites and their usage provides not only independent confirmation to earlier reports but also generates additional insights into the participation of some functional modules that may be important targets of tandem APA -mediated reprogramming of gene expression (Supplementary Figure S5). Two such modules are noteworthy: the Rho GTPases pathway and the RNA processing machinery. Rho family small GTPases are a class of important cellular signaling molecules with strong implications in cancer (61). Rather than Rho GTPases themselves, their regulators are emerging as the major targets in cancer (61), a theme that recurs in our network analysis, where all four Rho GTPases in the network are not targeted, whereas their regulators and effectors within the network are targeted (Supplementary Figure S5, Rho GTPases module). Rho GTPases are activated by all of these regulators in the network, which leads to actin re-organization, a process important for cancer cell migration and metastasis (49). Another important gene in the RNA processing machinery is POLR2K, the only subunit common to all three polymerases that dose dependently improves the assembly of the Pol III pre-initiation complex (51,62). As Pol III products (tRNAs, 5 S rRNAs and 7SL rRNAs) are required for protein synthesis and tumors have unusually high levels of Pol III activity (63), it is possible that the upregulation of POLR2K could facilitate Pol III assembly and thus contribute to cell proliferation and cancer development.
The different polyadenylated transcript isoforms could be regulated the transcriptional (64,65) as well as posttranscriptional (60) levels, and the role of these two processes in polyadenylation may not be readily separable (66). As most regulatory mechanisms that act in 3′ UTRs are thought to be repressive and not activating (5), it is likely that if the causative factor is a canonical UTR-regulation mechanism such as that of miRNAs, then it must be downregulated in cancer. An alternative explanation is that non-canonical, isoform-dependent, UTR regulatory factors that do not repress, but activate (e.g. polyadenylation/cleavage proteins) short isoform levels are upregulated in cancer. Identification of potential isoform-dependent regulatory roles for Pabpc1, H3K36Me3, and the ATATAT motif that is linked to the yeast heterogeneous nuclear ribonucleoproteins (hnRNP)-like protein Hrp1 highlights the possibility of such isoform-dependent factors. Hrp1 contains two tandem RNA Recognition Motifs domain, an arrangement shared by various human hnRNPs. Although the protein sequence of Hrp1 is similar to all human hnRNPs, the Hrp1 mRNA sequence is most similar to hnRNPA3. Several hnRNP proteins seem upregulated in tumor, including one of the hnRNPA3 isoforms that is upregulated in four of the five tumor samples. Similarly, across all five samples, Pabpc1 reads in tumor samples are higher than that of normal tissues; manifesting greater than 2-fold up-regulation in lung and kidney samples. Given the emerging theme that polyadenylation is interlinked to other key processes such as transcription elongation and posttranscriptional regulation (64,67), some of these regulatory marks such as H3K36Me3 may play a dual role in controlling both transcription and polyadenylation of the isoforms.
In summary, we built a comprehensive polyadenylation and APA map, which is made publically available as a webserver (johnlab.org/xpad) and as a UCSC track hub (http://www.johnlab.org/xpad/Hub/UCSC.txt). We have highlighted potential uses of the resource in studying polyadenylation-mediated gene regulation and generated new insights into the regulation of polyadenylation with an emphasis on gene isoform-dependent signatures in cancer. Although these observations will need to be followed up with more detailed studies, xPAD serves as a unique tool to investigate various questions in biology, relating to polyadenylation, APA-mediated gene regulation, gene expression analysis, and discovery of new genes and gene isoforms. We aim to further expand xPAD with additional samples (tissue-types, cancer-subtypes, and cell lines), with the goal of defining gene-isoform signatures that may be useful for diagnosis or prognosis of specific human diseases, particularly in cancer (39). These studies which provide cell-type specific annotation and usage of 3′ UTRs are expected to also help other genome-wide studies of UTRs such as miRNA target analysis (68,69), which could benefit by incorporating the expression levels of individual 3′ UTR isoforms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–9, Supplementary Methods, Supplementary Dataset and Supplementary References [70,71].
FUNDING
American Cancer Society [RSG-09-054-01 to B.J.]; National Institutes of Health [HG005230 and HG005279 to F.O. and P.M.M., and GM079756 to B.J.]; Magee-Womens foundation [0027467 to B.J.]. Funding for open access charge: American Cancer Society.
Conflict of interest statement. F.O. and P.M.M. are shareholders of Helicos BioSciences Corporation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the laboratories of P.M.M., A.P.M. and B.J. for helpful discussions. They also thank Dr. Gary Bader and Mr. Guanming Wu for advice on pathway analysis, Mr. Frank Doyle for RIP-Seq data discussions, and Dr. Jim Kent and Mr. Tim Dreszer for their help with building a UCSC genome browser track hub for the data.
REFERENCES
- 1.Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 2.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell. Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Hesketh J. 3′-Untranslated regions are important in mRNA localization and translation: lessons from selenium and metallothionein. Biochem. Soc. Trans. 2004;32:990–993. doi: 10.1042/BST0320990. [DOI] [PubMed] [Google Scholar]
- 4.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 13.Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem. Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 14.Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA, Sigurgeirsson B, Thorisdottir K, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, Liu H, Rosenwald A, Muller-Hermelink HK, Ott G, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 17.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 18.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neilson JR, Sandberg R. Heterogeneity in mammalian RNA 3′ end formation. Exp. Cell Res. 2010;316:1357–1364. doi: 10.1016/j.yexcr.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, Woolf B, Shen L, Donahue WF, Tusneem N, Stromberg MP, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc. Natl. Acad. Sci. USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks433. May 18 (doi:10.1093/nar/gks433; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapranov P, St. Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, Thompson JF, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lembo A, Di Cunto F, Provero P. Shortening of 3′UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 41.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 44.Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, Kang GY, Bang JY, Park HS, Leelayuwat C, Lee YS. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meiri E, Levy A, Benjamin H, Ben-David M, Cohen L, Dov A, Dromi N, Elyakim E, Yerushalmi N, Zion O, et al. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res. 2010;38:6234–6246. doi: 10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell. Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- 48.Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011;209:139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White RJ. RNA polymerase III transcription—a battleground for tumour suppressors and oncogenes. Eur. J. Cancer. 2004;40:21–27. doi: 10.1016/j.ejca.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 51.Rubbi L, Labarre-Mariotte S, Chedin S, Thuriaux P. Functional characterization of ABC10alpha, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J. Biol. Chem. 1999;274:31485–31492. doi: 10.1074/jbc.274.44.31485. [DOI] [PubMed] [Google Scholar]
- 52.Alberts AS, Treisman R. Activation of RhoA and SAPK/JNK signalling pathways by the RhoA-specific exchange factor mNET1. EMBO J. 1998;17:4075–4085. doi: 10.1093/emboj/17.14.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murray D, Horgan G, Macmathuna P, Doran P. NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br. J. Cancer. 2008;99:1322–1329. doi: 10.1038/sj.bjc.6604688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Helden J, del Olmo M, Perez-Ortin JE. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jayaseelan S, Doyle F, Currenti S, Tenenbaum SA. RIP: an mRNA localization technique. Methods Mol. Biol. 2011;714:407–422. doi: 10.1007/978-1-61779-005-8_25. [DOI] [PubMed] [Google Scholar]
- 59.Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Z, Kaye DM. Mechanistic insights into the link between a polymorphism of the 3′UTR of the SLC7A1 gene and hypertension. Hum. Mutat. 2009;30:328–333. doi: 10.1002/humu.20891. [DOI] [PubMed] [Google Scholar]
- 61.Ellenbroek SI, Collard JG. Rho GTPases: functions and association with cancer. Clin. Exp. Metastasis. 2007;24:657–672. doi: 10.1007/s10585-007-9119-1. [DOI] [PubMed] [Google Scholar]
- 62.Lefebvre O, Ruth J, Sentenac A. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J. Biol. Chem. 1994;269:23374–23381. [PubMed] [Google Scholar]
- 63.White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 64.Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol. Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, Tian B. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol. Syst. Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 67.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 68.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 69.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.