Abstract
MacroH2A is a histone H2A variant that is typically found in heterochromatic regions of the genome. A positively charged linker that connects the histone-fold with the macro-domain was suggested to have DNA-binding properties, and has been shown to promote oligomerization of chromatin fibers. Here we examine the influence of this basic linker on DNA of mononucleosomes. We find that the macro-linker reduces accessibility to extranucleosomal DNA, and appears to increase compaction of the nucleosome. These properties arise from interactions between the H1-like basic linker region and DNA around the entry/exit site, which increases protection of nucleosomal DNA from exonuclease III digestion by ∼10 bp. By stabilizing the wrapping of DNA around the histone core, this basic linker of macroH2A may alter the distribution of nucleosome-associated factors, and potentially contribute to the more compacted nature of heterochromatin.
INTRODUCTION
By competing with transcription factors and other DNA binding proteins for access to genomic DNA, nucleosomes play a fundamental role in regulating gene expression. This natural ability of histones to restrict access to DNA is in turn regulated by diverse mechanisms, including charge neutralization of basic histone tails by acetylation, active nucleosome repositioning by chromatin remodelers, and stabilization of chromatin fibers and heterochromatic structures by polycomb group proteins and linker histones (1–10). Another means by which chromatin structure can be modulated is through substitution of histone variants, which can alter interactions with other histones, chromatin associated factors and DNA (11,12).
The histone variant macroH2A is emerging as a complex and highly regulated variant linked to heterochromatin. Initially discovered as highly enriched on the inactive X chromosome of female mammals (13), macroH2A is also prevalent on centromeric heterochromatin of X and Y chromosomes during meiosis (14) as well as heterochromatic and transcriptionally repressed regions of autosomes (15–17). In vitro, a central ∼40 residue segment of macroH2A called the macro-linker, which connects the N-terminal histone-fold with the C-terminal macro-domain (Figure 1A), has been shown to stimulate dramatic condensation and oligomerization of chromatin fibers (18). This macro-linker has a highly basic character and is similar in its amino acid composition to the C-terminus of the linker histone H1 (13). Extending from the C-terminus of the H2A-like histone-fold, the macro-linker is positioned adjacent to entry/exit DNA where linker histones are believed to bind to the nucleosome, and thus may serve as a ‘built-in’ linker histone-like tail (18,19). For the linker histone H1, the basic C-terminus plays an important role in nucleosome binding (20–22). Stable binding between linker histones and nucleosomes requires DNA flanking the nucleosome (22,23), and the association of basic C-termini of linker histones with extranucleosomal DNA reduces electrostatic repulsion of DNA, allowing for higher levels of chromatin fiber condensation (21,24).
Figure 1.
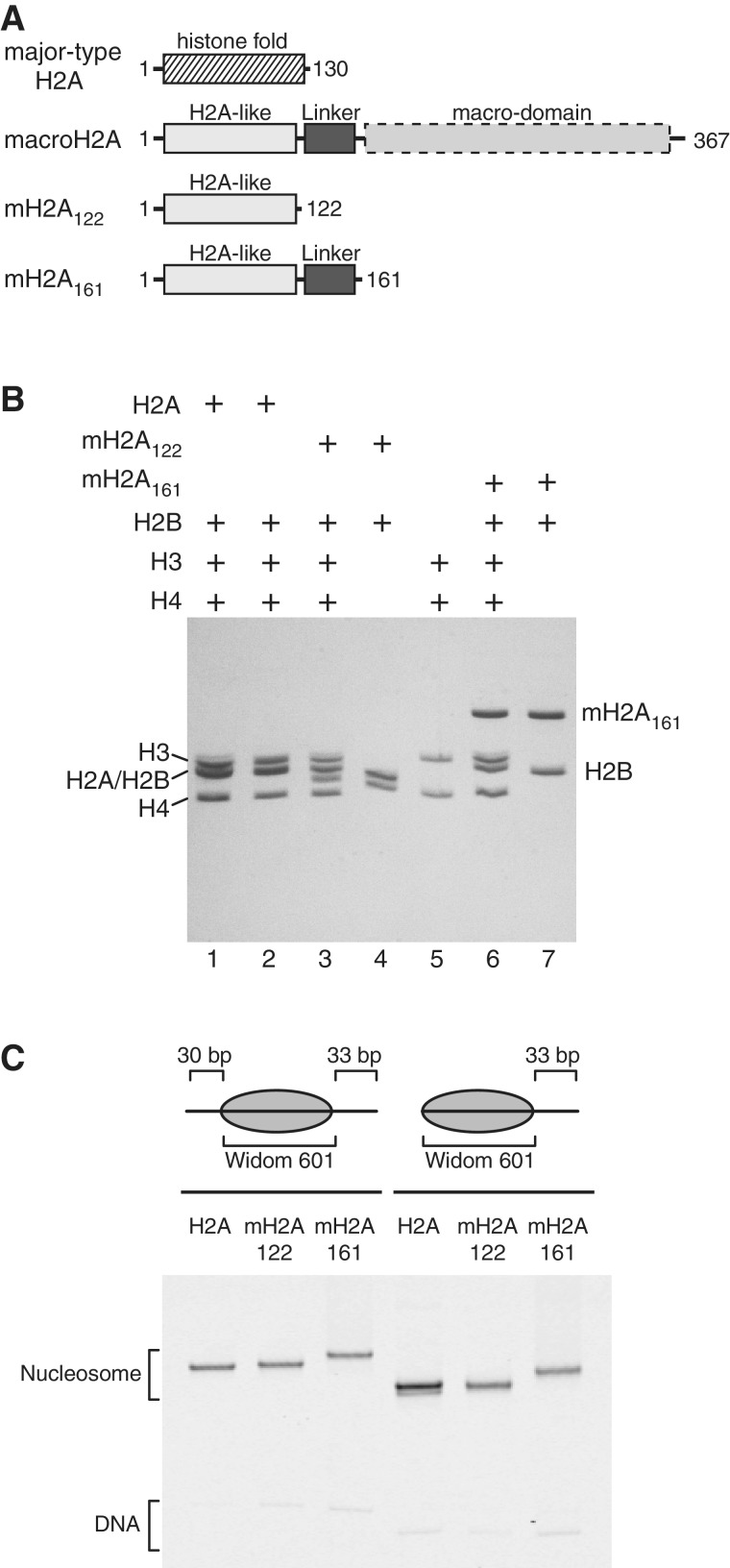
Overview of nucleosomes used in this study. (A) Domain organization of full-length macroH2A, and the truncations used in this study compared with major-type H2A. The truncated macroH2A construct, mH2A122, possesses only the histone-fold portion of macroH2A, and mH2A161 additionally contains the basic macro-linker. (B) Nucleosomes and histone oligomers analyzed by SDS PAGE. To confirm the sizes and ratios of histone proteins in reconstituted nucleosomes, nucleosomes containing major-type H2A (lane 1), mH2A122 (lane 3) and mH2A161 (lane 6) were run alongside the histone components: the histone octamer with major-type H2A (lane 2), mH2A122-H2B dimer (lane 4), (H3-H4)2 tetramer (lane 5) and mH2A161-H2B dimer (lane 7). (C) Nucleosomes analyzed by native PAGE. Fluorescently labeled nucleosomes (centered 30-N-33 and end-positioned 0-N-33) were produced with either major-type H2A, mH2A122 or mH2A161 as indicated.
Localization and participation of macroH2A in chromatin reorganization are regulated in at least two ways. One regulatory mechanism involves phosphorylation of Ser137 of macroH2A, which is located within the basic macro-linker and was found to correlate with exclusion of macroH2A from the inactive X chromosome in female mammals (25). This modification is most prevalent during mitosis, which also correlates with the cell cycle stage when linker histones are hyperphosphorylated and ejected from condensed chromosomes (25,26). Another regulatory mechanism utilizes the C-terminal macro-domain. Although macro-domains found in non-histone proteins have been shown to bind to poly-ADP ribose (PAR) (27), PAR binding appears to be limited to only one splice form of macroH2A, called macroH2A1.1 (28–30). Disruption of PAR binding through mutagenesis of the macro-domain of macroH2A1.1 was found to prevent local chromatin condensation in response to genomic insults such as single-stranded DNA breaks caused by microirradiation (30). In vitro, the macro-domain of macroH2A1.2 blocked the basic macro-linker from condensing and oligomerizing chromatin fibers (18). Thus, the macro-domain appears to provide a switch that allows macroH2A to dynamically reorganize chromatin structure, presumably through the action of the basic macro-linker.
Given its highly charged nature and proximity to the entry/exit DNA on the nucleosome, we were interested in determining whether the macro-linker affected the availability or organization of extranucleosomal DNA for mononucleosomes. Since the manner in which the macro-domain may regulate exposure of the macro-linker is not yet understood, we chose to study the histone-fold and macro-linker portion of macroH2A in the absence of the macro-domain. We find that mononucleosomes possessing the basic macro-linker behave as though more compact and have less accessible extranucleosomal DNA. The apparent increase in compaction was observed by native polyacrylamide electrophoresis, where mononucleosomes that shifted away from DNA ends unexpectedly migrated faster than end-positioned nucleosomes. The presence of the basic macro-linker increased protection of entry/exit DNA from exonuclease III digestion, strengthening a pause site just outside the edge of the nucleosome. These results suggest that the macro-linker interacts with DNA flanking the nucleosome core, and support a functionally similar role to the C-terminal tails of linker histones in stabilizing extranucleosomal DNA.
MATERIALS AND METHODS
Reconstitution of nucleosomes
All histones were recombinantly expressed in Escherichia coli and purified as previously described [(31) and web protocol from the Tsukiyama lab, http://labs.fhcrc.org/tsukiyama/protocols.html]. The macroH2A constructs were derived from macroH2A1.2 from Mus musculus, and major-type H2A and other histones were from Xenopus laevis. According to previously described methods (19,32), major-type H2A was refolded with H2B, H3 and H4 to make octamer, mH2A122 and mH2A161 were each refolded with H2B to make dimers, and H3 and H4 were also independently combined to make (H3-H4)2 tetramer. Dimers, tetramer, and octamer were separately purified by gel-filtration chromatography using a HiPrep Superdex 200 16/60 column (GE Healthcare). Fluorescently labeled DNA was generated using labeled primers and the Widom 601 positioning sequence as template (33) in large-scale (5 ml) polymerase chain reactions. Prior to nucleosome reconstitution, fluorescent DNAs were purified by preparative native gel electrophoresis.
Nucleosomes with major-type H2A were reconstituted by mixing the histone octamer and DNA in a 1:1 molar ratio under buffer conditions optimized previously (10 mM Tris-HCl, pH 7.5, 2 M KCl, 1 mM EDTA and 5 mM β-mercaptoethanol), and dialyzing against a salt gradient from 2 M to zero KCl (32). The same protocol was used to obtain mH2A122 and mH2A161 nucleosomes, except that mH2A122-H2B and mH2A161-H2B dimers were each mixed with (H3-H4)2 tetramer and DNA in a ∼2:1:1 ratio. All nucleosomes were purified with a preparative gel electrophoresis system (Bio-Rad mini prep cell) using a 5.5 cm tall, cylindrical 6% non-denaturing polyacrylamide gel. Nucleosome purity was assessed by 6% native PAGE.
Electrophoretic mobility shift assay
A truncated form of the Saccharomyces cerevisiae Chd1 remodeler (residues 118–1274), which contained the core chromodomains, ATPase motor, and DNA-binding domain (hereafter referred to as Chd1), were expressed and purified as previously described (34). To compare the relative abilities of the Chd1 remodeler to stably associate with different nucleosomes under identical conditions, the Chd1 remodeler was incubated with a mixture of major-type H2A and mH2A122 or mH2A161 nucleosome substrates. Nucleosomes containing major-type H2A were FAM-labeled, and both mH2A122 and mH2A161 nucleosomes were Cy5-labeled. All nucleosomes for the binding experiments were end-positioned 0 -N-33, with the fluorescent label on the zero-end. Chd1 (0, 5, 10, 20, 40, 80 nM) was incubated with nucleosomes (5 nM major-type H2A plus 5 nM mH2A122 or mH2A161) for 1 h on ice and then resolved on 6% native polyacrylamide gels (60:1 acrylamide to bis-acrylamide). Gels were visualized using a Typhoon 9410 variable mode imager (GE Healthcare) and analyzed using ImageJ (http://imagej.nih.gov/ij/).
Nucleosome sliding assay
FAM-labeled nucleosomes containing major-type H2A (50 nM) were mixed with Cy5-labeled nucleosomes containing mH2A122 or mH2A161 histones (also 50 nM) and incubated at room temperature with 20 nM Chd1 in buffer containing 100 mM KCl, 5 mM MgCl2, 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 5% sucrose, 0.1 mg/ml BSA and 5 mM dithiothreitol (DTT). Reactions were stopped at indicated time points by placing on ice after mixing with competitor DNA (pUC18 plasmid with a 12-mer array of the 601 sequence, added to a final concentration of ∼1 mg/ml) and 5 mM EDTA. Reactions were then loaded on 6% native polyacrylamide gels (60:1 acrylamide to bis-acrylamide), visualized using a Typhoon 9410 variable mode imager (GE Healthcare), and analyzed using ImageJ (http://imagej.nih.gov/ij/).
Site-specific histone-DNA contact mapping
The locations of the histone octamer on nucleosomal DNA were mapped at high resolution using previously published methods (35,36). Briefly, nucleosomes reconstituted with H2B having a cysteine substitution at position 53 were modified with p-azidophenacyl bromide (APB) in a 20 µl reaction by incubation in a buffer that contained ∼0.5 µM nucleosomes, 50–100 µM DTT, 2.5 mM APB (from a 50 mM stock in N,N dimethyl formamide), 20 mM Tris-HCl (pH 7.5) and 5% glycerol, for 2–3 h at room temperature and in the dark. Modification reactions were stopped by addition of equal volume of 2X remodeling assay buffer, which contained 10 mM DTT along with 40 mM Tris-HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mg/ml BSA and 10% sucrose. Nucleosomes thus modified were either directly UV cross-linked or further incubated at room temperature with the Chd1 remodeler and 2 mM ATP. Nucleosome sliding reactions contained 50 nM Chd1 and 150 nM nucleosomes and were stopped using competitive DNA (∼1 mg/ml) and 5 mM EDTA. The reaction mixtures were then cross-linked by irradiating with a 312 nm UV transilluminator for ∼1 min, followed by enrichment of the cross-linked species by phenol choloroform extraction as previously described (35). After cleavage of the protein-DNA cross-link with heat and alkali treatment, precipitated DNA samples were resuspended in 5 µl formamide loading dye and applied on 8% polyacrylamide (19:1), 8 M urea sequencing gels run at 65 W for 1 h 20 min. Gels were visualized by using a Typhoon 9410 variable mode imager (GE Healthcare) and analyzed using ImageJ (http://imagej.nih.gov/ij/).
Exonuclease III assay
For exonuclease III mapping experiments, nucleosomes (100 nM) possessed 5′-FAM labels at the zero- or 30-ends. In 10 µl reaction volumes, nucleosomes were incubated at 24°C for 10 min in buffer containing 20 mM HEPES-KOH (pH 7.6), 10 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.1 mg/ml BSA and 5% sucrose. These reactions were then treated with 0–10 units of exonuclease III (New England Biolabs), incubated 5 min longer at 24°C, and then quenched with final concentration of 10 mM EDTA and 1% SDS. Samples were supplemented with carrier agents (5 µg each of E. coli tRNA, oyster glycogen, and linear acrylamide, and 1/10 volume of 3 M sodium acetate, pH 5.2), extracted by phenol:chloroform:isoamyl alcohol (25:24:1) followed by chloroform:isoamyl alcohol (24:1), and then ethanol precipitated. Following a 70% ethanol wash, the samples were dried by speedvac (5 min) and resuspended in 5 µl formamide loading dye. Samples were separated on sequencing gels and visualized as described above for site-specific mapping.
RESULTS
The basic linker of macroH2A (residues 122–161) reduces accessibility of extranucleosomal DNA
In addition to specific DNA interactions provided by the folded portion of the histone octamer, DNA binding energy is also provided by basic residues in the flexible histone tails (37,38). Although histone tails do not appreciably alter the footprint of the core histone octamer on DNA (39,40), interactions between histone tails and nucleosomal DNA can influence association of other proteins. For example, both Gal4-AH and TFIIIA bind more poorly to sites on nucleosomes with intact histone tails, an effect that can be alleviated by reducing positive charges through lysine acetylation or proteolytic removal of the histone tails (41,42). This inhibitory effect of histone tails has been explained as a change in the dynamic unwrapping of nucleosomal DNA from the histone octamer, with a higher number of favorable electrostatic interactions preferentially stabilizing a wrapped and therefore less accessible state of nucleosomal DNA (43).
The linker connecting the histone-fold with the macro-domain of macroH2A is highly basic, with a range of vertebrate species having 1 arginine and 13–14 lysine residues within a ∼40 residue segment (residues 122–161 for mouse macroH2A). This linker extends from the C-terminus of the H2A-like histone-fold, which positions it proximal to the entry/exit DNA of the nucleosome (19). To determine what influence this basic linker may have on extranucleosomal DNA just outside the nucleosome, we generated nucleosomes containing the histone-fold and basic linker portion of macroH2A (residues 1–161, called mH2A161; Figure 1A). To control for biochemical differences arising outside this basic linker, we also generated nucleosomes containing only the histone-fold portion of macroH2A (called mH2A122). Both of these macroH2A-containing nucleosomes were compared with nucleosomes containing the major-type H2A histone.
As previously described, the histone-fold portion of macroH2A in complex with histone H2B remain stably associated with the (H3-H4)2 tetramer in ∼0.5 M KCl, which interferes with the ordered deposition of (H3-H4)2 tetramers and then H2A-H2B dimers onto DNA using gradient salt dialysis (19). Accordingly, (H3-H4)2 tetramers and H2A-H2B dimers were prepared separately. The mH2A122-H2B and mH2A161-H2B dimers each formed 1:1 complexes as expected, and could be combined with (H3-H4)2 tetramers and DNA in a 2:1:1 ratio to form mononucleosomes containing the full complement of histones (Figure 1B). The histone cores were positioned on DNA using the Widom 601 sequence (33). For this study, we designed nucleosomes to be either centrally positioned (denoted 30-N-33) or end-positioned (denoted 0-N-33 or 0-N-80, where N represents the core 145 bp of the Widom 601 sequence (44)). Native polyacrylamide gel electrophoresis (PAGE) was used to evaluate the quality of mononucleosomes, and suggested that each sample had a relatively homogeneous placement of histone octamers on DNA (Figure 1C). The rate of migration in native PAGE is sensitive to the overall size and shape of mononucleosomes, and therefore the presence of multiple bands indicates heterogeneous distribution of histone octamers on the DNA fragment (45,46). It is important to note that the rate of migration with native PAGE also depends on the overall charge of the mononucleosomes. Therefore, the additional basic residues present in mH2A161 nucleosomes may be responsible for the reduced migration speeds compared with major-type H2A and mH2A122 nucleosomes. For a given histone type and length of DNA, nucleosomes typically migrate more rapidly when the histone octamer is end-positioned on DNA than when at more central locations.
Given its highly basic charge and location near the entry/exit site, we were interested to see whether the linker of macroH2A reduced accessibility of extranucleosomal DNA. To evaluate the general accessibility of DNA outside the nucleosome, we used the yeast Chd1 (chromodomain-helicase-DNA-binding factor 1) chromatin remodeler (47). The C-terminal DNA-binding domain of Chd1 is required for stable association with nucleosomes, and binds to DNA duplexes with some sequence preference but is largely sequence-non-specific (48–50). The Chd1 DNA-binding domain interacts poorly with nucleosomal DNA, and a stable complex between Chd1 and nucleosomes requires ∼20 bp of extranucleosomal DNA (49).
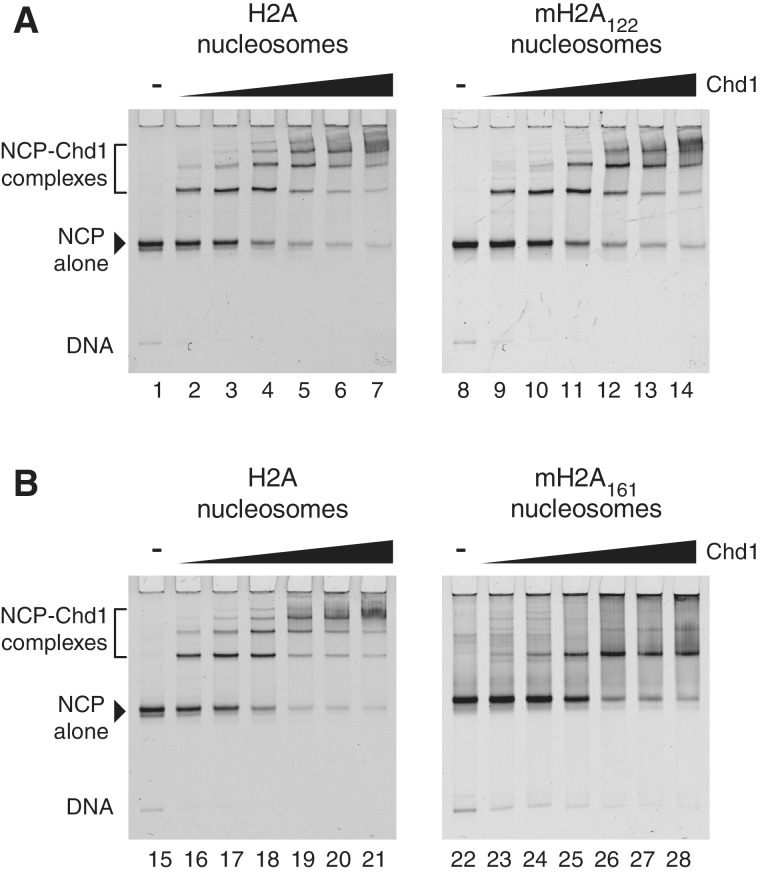
Chd1 was incubated with mixtures of major-type H2A (FAM-labeled) and either mH2A122 or mH2A161 (Cy5-labeled) nucleosomes containing 33 bp of extranucleosomal DNA at one entry/exit site, and monitored by native PAGE (Figure 2). For both major-type H2A and mH2A122 nucleosomes, super-shifted bands were apparent at all Chd1 concentrations, with ladders produced at higher Chd1 concentrations indicative of nucleosomes associated with multiple Chd1 proteins (lanes 1–14). In contrast, nucleosomes containing the macro-linker (mH2A161) failed to yield super-shifted bands at the lowest protein concentrations used, and at higher concentrations primarily produced a single super-shifted band rather than a ladder (lanes 22–28). This reduction in both apparent affinity and the number of super-shifted bands suggests that the basic macro-linker effectively reduced the availability of extranucleosomal DNA for Chd1 binding.
Figure 2.
The basic macro-linker reduces the availability of extranucleosomal DNA for Chd1 binding. (A) Mixtures nucleosomes containing major-type H2A and mH2A122 (5 nM each) were incubated with increasing amounts of the chromatin remodeler Chd1 (5, 10, 20, 40, 60 and 80 nM) for 1 h on ice and resolved on a 6% non-denaturing gel. Left image is an FAM-scan showing the locations of major-type H2A nucleosomes, and right image is a Cy5-scan showing the mH2A122 nucleosomes. (B) Similar experiment as shown in (A), except that Cy5-labeled nucleosomes contained mH2A161. These gels are representative of more than three experiments carried out under similar conditions.
The basic linker of macroH2A appears to increase compaction of mononucleosomes
In the context of chromatin fibers, nucleosomes containing full-length macroH2A behaved similarly to those with major-type H2A, yet the presence of macro-linker without the C-terminal macro-domain dramatically altered fiber properties (18). We wondered whether the macro-linker would also change properties of mononucleosomes in a way that might affect nucleosome sliding. Previous studies demonstrated that nucleosomes containing either the histone-fold portion only or the full-length macroH2A could be readily shifted by Swi/Snf, RSC (Swi/Snf-type) and ACF (Iswi-type) chromatin remodelers (51). To test whether the macro-linker alone could change the outcome of a nucleosome sliding reaction, we incubated 0-N-33 nucleosomes with the Chd1 remodeler and monitored the reaction by native PAGE (Figure 3A). As observed for other remodelers, movement of mH2A122 nucleosomes by Chd1 paralleled those of major-type H2A nucleosomes. Both nucleosomes showed slower migration over time by native PAGE, suggestive of histone octamers shifting to more central locations on the DNA (lanes 1–20). The apparent rates at which both nucleosomes shifted from end to more centered locations were comparable, indicating that nucleosome sliding by Chd1 is not affected by substitution of major-type H2A with the histone-fold portion of macroH2A.
Figure 3.
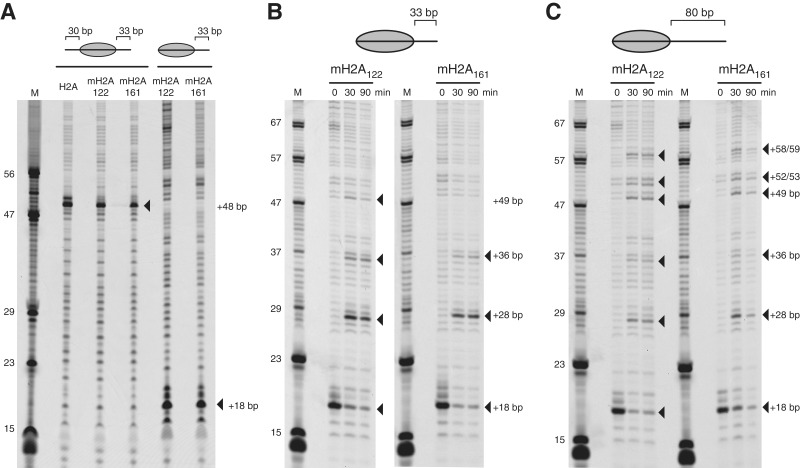
End-positioned nucleosomes containing mH2A161 migrate aberrantly in native gels when shifted by the Chd1 remodeler. (A) A representative nucleosome sliding experiment of end-positioned 0-N-33 nucleosomes that consisted of a mixture of major-type H2A nucleosomes (FAM-labeled) and either mH2A122 or mH2A161 nucleosomes (Cy5-labeled). A total of 100 nM nucleosome (50 nM each) was incubated with 20 nM Chd1 in a buffer containing 50 mM KCl, 5 mM MgCl2, 20 mM Tris.HCl (pH 7.5), 0.1 mM EDTA, 0.1 mg/ml BSA, 5 mM EDTA, 5% sucrose and 2 mM ATP. (B) A similar nucleosome sliding experiment as shown in (A), except using 0-N-80 end-positioned nucleosomes, and with sliding buffer containing 100 mM KCl. For all nucleosome sliding reactions, the same pattern of shifted nucleosomes migrating faster than the starting material was observed for mH2A161, whereas both major-type H2A and mH2A122 nucleosomes showed the expected slower migration indicative of nucleosome centering.
In contrast, sliding of mH2A161 nucleosomes produced an unexpected result: instead of showing a reduced migration in the native gel, as expected for movement toward a more central location, we observed more rapid migration of nucleosomes in the presence of Chd1 and ATP (lanes 21–30). The lower, faster migrating band appeared rapidly, and at later time points a slower band, similar in position to the starting material, began to increase in intensity. This downward movement of the starting mH2A161 nucleosome band was observed consistently in an ATP-dependent fashion. Nucleosomes were also generated with longer extranucleosomal DNA (0-N-80), which allows for more intermediate nucleosome positions to be resolved between end and more central locations (Figure 3B). Similar to the 0-N-33 nucleosomes, the 0-N-80 mH2A161 nucleosomes also displayed a striking downward shift of the primary nucleosome band relative to the starting material (lanes 61–70). At later time points, the mH2A161 nucleosome bands also shifted upward, and appeared to migrate at a location similar to the starting material.
Two possible explanations for this aberrant migration of mH2A161 nucleosomes are that the histone octamer was initially mis-positioned relative to the 601 positioning sequence, and/or that the macro-linker altered the preferred direction of sliding by Chd1. To determine whether the macro-linker altered the expected location of the histone octamer on the 601 sequence, we employed high-resolution nucleosome mapping using the photoactivatable cross-linker p-azidophenacyl bromide (APB) (35,36). The site of APB modification was residue 53 of histone H2B (mutated Ser→Cys), which has been shown to cross-link to one strand of nucleosomal DNA approximately two helical turns from the entry/exit site (35,36). Dimers of H2A/H2B containing this H2B Cys-53 mapping histone were generated for both mH2A122 and mH2A161, and incorporated into nucleosomes. As we were concerned from the migration profile that the mH2A161 octamer may have been mis-positioned from being close to one end of the DNA duplex, we also designed a centrally located 30-N-33 nucleosome. For both 30-N-33 and 0-N-33 nucleosomes, mapping revealed cross-links corresponding to the expected positions of the histone octamer on the 601 sequence for both mH2A122 and mH2A161 nucleosomes (Figure 4A). Therefore, the presence of the macro-linker did not alter the placement of the histone octamer on the 601 positioning sequence, and the aberrant migration in the native gel must arise from another property of these nucleosomes.
Figure 4.
The presence of the macro-linker does not affect the position of the histone octamer on the 601 sequence in the presence or absence of the Chd1 remodeler. Histone octamer positions were mapped by photochemical cross-linking of APB-modified histone H2B as previously described (35,36). (A) Centrally positioned (30-N-33) and end-positioned (0-N-33) nucleosomes were mapped containing major-type H2A, mH2A122 and mH2A161 as indicated. For all nucleosomes, the predominant sites of cross-linking were found ∼2 turns of DNA from entry/exit sites expected from positioning by the 601 sequence. (B) End-positioned 0-N-33 nucleosomes containing either mH2A122 or mH2A161 were mapped before and after sliding with Chd1. After 30 or 90 min incubations with Chd1, most new cross-linking sites were observed for both mH2A122 and mH2A161 nucleosomes (filled triangles). (C) Similar nucleosome sliding experiments as shown in (B), except using 0-N-80 nucleosomes. For the nucleosome sliding experiments in (B) and (C), the close match of new cross-linking sites for mH2A122 and mH2A161 nucleosomes show that the macro-linker does not impair the direction of sliding by Chd1, nor alter the preferential locations of repositioned histone octamers in the context of the 601 positioning sequence.
To further investigate the possible causes of the aberrant migration of mH2A161 nucleosomes, we used nucleosome mapping to determine whether the direction of nucleosome sliding was altered by the macro-linker. Given the similar patterns of mH2A122 and major-type H2A nucleosomes upon treatment with Chd1, we mapped mH2A122 in parallel with mH2A161 nucleosomes. For both mH2A122 and mH2A161 nucleosomes, similar patterns of histone cross-linking were observed (Figure 4B and C). For the 0-N-33 nucleosomes, cross-linking shifted from the initial position at +18 bp to +28 bp and +36 bp, corresponding to movements of the histone octamer of 10 bp and 18 bp along the DNA, respectively. An additional cross-link at +49 bp was evident for mH2A122 but not mH2A161 nucleosomes, which corresponded to a 31 bp shift of the histone octamer. The failure to observe this +49 cross-link for mH2A161 may have been due to weaker binding of Chd1 to centered nucleosomes with short extranucleosomal DNA (e.g. to 18-N-15). A similar but more extended pattern of cross-linking was also observed with 0-N-80 nucleosomes (Figure 4C). In agreement with their poorer binding to Chd1, mH2A161 nucleosomes were mobilized more slowly than mH2A122 nucleosomes (Supplementary Figure S1). However, given sufficient time and Chd1 protein, both nucleosomes produced similar patterns (Figure 4C), indicating that the macro-linker did not interfere with either the direction of nucleosome sliding, nor the final preferred distributions of histone octamer on the DNA.
These mapping experiments demonstrated that the aberrant migration of mH2A161 nucleosomes on native gels (Figure 3) did not arise from initial mis-positioning of the mH2A161 octamer on the 601 sequence, nor from a different direction of sliding by the Chd1 remodeler. Instead, this aberrant migration suggested that as mH2A161 nucleosomes shifted away from DNA ends, the molecular shape changed in a way that differed from nucleosomes lacking the basic macro-linker. One explanation for the increased rates of migration is that the macro-linker promoted a more compact organization of the nucleosome. Since dynamic unwrapping of DNA from the nucleosome would increase the effective size of the particle, a more compact organization may be achieved if the macro-linker stabilized the interactions of entry/exit site DNA with the histone core.
The linker domain of macroH2A stabilizes DNA at the entry/exit site of the nucleosome
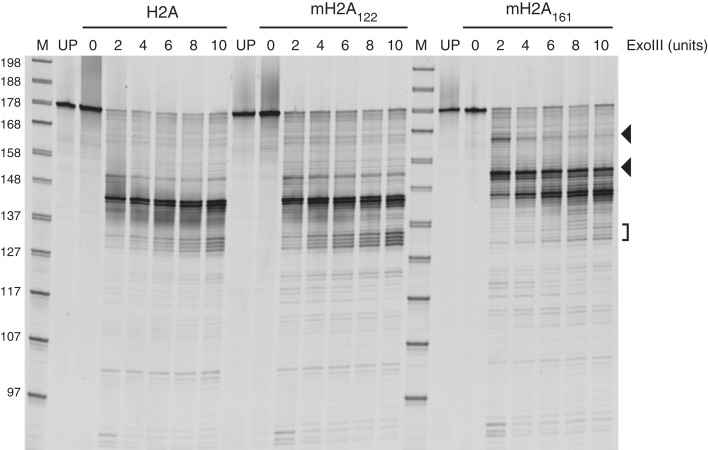
To further probe the organization of DNA at the nucleosome entry/exit site, we treated nucleosomes with limiting amounts of exonuclease III. End-positioned 0-N-33 nucleosomes containing major-type H2A, mH2A122 or mH2A161 were used, all of which possessed 5′-FAM labels at the zero-length side of the nucleosome. Since exonuclease III digests DNA 3′–5′, the FAM label allowed digestion through the 33 bp extranucleosomal DNA to be visualized. For major-type H2A and mH2A122 nucleosomes, the major products corresponded to the nucleosome edge, ∼144 bp from the FAM label. Two minor sites flanked this position, one of which was ∼10 bp more internal on the nucleosome, whereas another was ∼10 bp external, at ∼154 bp. Exonuclease III digestion of mH2A161 nucleosomes produced related, but noticeably distinct profiles. The major digestion products around 144 bp were still apparent with higher concentrations of exonuclease III, but were accompanied by substantial increases in pausing at the ∼154 bp site. At the lowest amount of exonuclease III used, an additional site of protection was observed farther away from the nucleosome edge, giving rise to a digested product ∼165 bp in length. These digestion patterns were highly reproducible, and similar relative locations of exonuclease III pausing were also observed for 30-N-33 nucleosomes (Supplementary Figure S2). The pause sites at the entry/exit site with 33 bp of extranucleosomal DNA were therefore not dependent on the presence or absence of extranucleosomal DNA at the other entry/exit site.
These exonuclease experiments show that the basic macro-linker increased the amount of DNA protected by the histone core. Given the highly charged and presumed flexible nature of the macro-linker, a defined site of protection due to direct interactions with the macro-linker was not expected. Since the two major sites of protection for mH2A161 were also observed for H2A and mH2A122 nucleosomes, the macro-linker appears to strengthen the minor pause site just outside the nucleosome edge. This behavior is consistent with a decreased propensity for unwrapping at the entry/exit site compared with major-type H2A nucleosomes, and agrees with the more compact nature of mH2A161 nucleosomes suggested by native PAGE.
DISCUSSION
Histone variants possess chemically and structurally unique features at strategic locations on the nucleosome, which provide a rich medium for maintaining epigenetic information. These distinct surfaces of histone variants are targets for post-translational modifications, but also can dramatically alter the biophysical properties of both mononucleosomes and chromatin fibers. The variant H2A.Bbd, for example, promotes decondensation of chromatin fibers, having both a less negatively charged acidic patch, and a reduced ability to stabilize DNA wrapping at entry/exit sites of the nucleosome (52–54). In contrast, macroH2A has a highly basic linker extending from the histone-fold domain that promotes oligomerization and condensation of chromatin fibers (18). These chromatin compaction properties were only observed in the absence of the C-terminal macro-domain, demonstrating that the macro-domain can functionally mask the basic macro-linker. In this study, we focused on how the macro-linker, independent of the macro-domain, influenced properties of mononucleosomes.
We report here that the basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. The presence of the basic macro-linker, but not the macroH2A histone-fold, increased the mobility of partially shifted nucleosomes in native polyacrylamide gels (Figure 3). Such behavior may be explained by increased interactions with entry/exit DNA. The shape of a mononucleosome largely depends on the lengths and directions of DNA protruding from the histone core. Although crystal structures show converging trajectories of DNA exiting both side of the nucleosome (55,56), DNA unwrapping, which has been shown to occur prevalently in solution (57,58), would result in DNA extending from the nucleosome with divergent trajectories. Sliding histones away from DNA ends, which shortens the flanking DNA from one side while exposing previously protected DNA on the other, typically slows migration in native gels (45,46). However, if the newly protruding short segment from the ‘zero’ side of the nucleosome were stabilized against the histone core as the opposite side were shortened, the resulting nucleosome shape may provide a net increase in mobility. Further sliding away from DNA ends, however, would eventually allow DNA from the short side to protrude significantly enough to reduce migration. Thus, interactions of entry/exit DNA with the basic macro-linker may be responsible for the more rapid and then slowed nucleosome migration as the histone octamer shifts away from one end of DNA to a more central location.
In agreement with stabilization of DNA around entry/exit sites, we found that the macroH2A linker increased protection from exonuclease III digestion (Figure 5). In the presence of the macro-linker, the two major pause sites for exonuclease were shifted by ∼10 bp toward extranucleosomal DNA, and for 0-N-33 nucleosomes a new minor pause site was observed an additional ∼10 bp further away. Although changing the distribution of paused sites, this shift appeared to maintain the pattern of exonuclease pausing observed for nucleosomes lacking the macro-linker, and corresponded with phasing of DNA on the nucleosome. Since the macro-linker is not expected to have specific interactions with DNA due to its sequence and predicted flexibility, the shift in exonuclease pausing likely reflects stabilization of DNA wrapping of the nucleosome.
Figure 5.
The macro-linker extends protection of DNA flanking the nucleosome from exonuclease III digestion. End-positioned (0-N-33) nucleosomes containing major-type H2A, mH2A122 or mH2A161 were digested with increasing amounts of exonuclease III. DNA fragments contained the 5′-FAM label on the zero-end of the nucleosome. Increased digestion was apparent for mH2A161 at ∼154 bp and ∼165 bp, corresponding to ∼81 bp and ∼92 bp from the nucleosome dyad (filled triangles). DNA appeared more protected at 132–135 bp, corresponding to ∼59–62 bp from the dyad (bracket), likely due to increased exonuclease III pausing at the nucleosome edge. M = marker, UP = unprocessed.
A similar increase in native gel mobility has been previously described for mononucleosomes in the presence of linker histone H5 (9). In that study, the changes in mononucleosome migration were dependent on the basic C-terminus of the linker histone. Consistent with faster migration, the presence of the histone H5 C-terminus was also shown by electron microscopy to bridge the two segments of entry/exit DNA on either side of the nucleosome into a stem-like structure (9,59). This notion of a stem-like organization of extranucleosomal DNA stabilized by basic polypeptide segments has been supported by subsequent FRET studies, where non-acetylated histone tails and histone H1 were shown to reduce separation of DNA segments extending from the nucleosome core (60,61). Consistent with reduced repulsion of extranucleosomal DNA segments, we also observed that size-exclusion chromatography elution profiles of 30-N-33 nucleosomes were retarded when containing mH2A161 compared with mH2A122 and major-type H2A histones, also suggestive of a more compact nucleosome shape (data not shown).
Strengthening of histone-DNA interactions around the entry/exit site of the nucleosome has been observed for nucleosome-associated factors such as linker histones and HMG family proteins. Linker histones have long been known to extend the DNA protection pattern beyond the nucleosome core, a characteristic of the so-called chromatosome (62). Although the ∼80 amino acid globular domain of linker histone H1 is essential for nucleosome binding and chromatosome formation (63), the highly basic and unstructured ∼100 amino acid C-terminal domain, which is largely responsible for H1 binding to chromatin in vivo, influences the length of extranucleosomal linker DNA organized at the entry/exit sites (22,64,65).
The nucleosome sliding experiments presented here suggest that while the linker region of macroH2A can slow the rate of nucleosome remodeling, it does not noticeably alter the relative translational positions of the histone core along the DNA for remodeling intermediates (Figure 4). An important difference from previous nucleosome sliding experiments with macroH2A (51) is that our nucleosomes lacked the macro-domain. Interactions between the macro-linker and DNA around the entry/exit site that stabilize histone-DNA interactions may explain the reduced affinity and activity of the yeast Chd1 remodeler. Based on these experiments, we expect that other remodelers sensitive to the presence and/or position of extranucleosomal DNA would also have reduced activity on nucleosomes where the basic macro-linker is exposed.
Neither ACF nor Swi/Snf-type remodelers were significantly challenged for mobilizing nucleosomes with full-length macroH2A (51), and we speculate that neither of these remodelers would be sensitive to nucleosomes with exposed macro-linkers. Although the Iswi-type remodelers such as ACF have several biochemical properties in common with Chd1 remodelers, such as nucleosome assembly, spacing, and centering (66,67), these two enzymes differ in their ability to mobilize nucleosomes associated with linker histones. ACF but not Chd1 was found to both incorporate and actively mobilize chromatin in the presence of histone H1 (66,68). Thus, despite the sensitivity toward flanking DNA that would be expected for a remodeler that produces evenly spaced nucleosome arrays, the ability of ACF to mobilize nucleosomes in the presence of linker histones makes it unlikely to be sensitive to the presence of an exposed macro-linker. Swi/Snf-type remodelers are known for sliding nucleosomes independently of available extranucleosomal DNA (69), and therefore would also likely not be blocked by the macro-linker. However, Swi/Snf remodelers are often recruited to chromatin by sequence-specific binding proteins (70,71), and thus while not directly blocked, the reduced accessibility of extranucleosomal DNA due to macroH2A may reduce affinity of targeting factors, thereby indirectly reducing Swi/Snf remodeling activity.
Reduced binding affinity due to the macro-linker could affect other chromatin-specific factors as well. Both HMG family proteins and linker histones require extranucleosomal DNA for stable binding to nucleosomes (72), and associations between the macro-linker and flanking DNA may make DNA less available for these factors. In agreement with this notion, chromatin containing macroH2A has been shown to be devoid of histone H1, perhaps as a consequence of the basic macro-linker effectively competing away the C-terminus of linker histones (73). We look forward to future studies that describe how the macro-linker activity is regulated by the macro-domain, and how altering DNA organization at the mononucleosome level may contribute to physical properties of heterochromatin stabilized by macroH2A.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–2.
FUNDING
National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) [R01 GM084192-5, R01 GM084192-S1 to G.D.B.]. Funding for open access charge: NIH/NIGMS [R01 GM084192-5].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Karolin Luger for the histone expression plasmids, Li Ma and Guy Montelione (Rutgers University) for chaperone co-expression plasmids for Chd1 production, Blaine Bartholomew for discussions regarding histone-DNA mapping, and members of the Bowman lab for comments and suggestions.
REFERENCES
- 1.Wolffe AP, Guschin D. Review: chromatin structural features and targets that regulate transcription. J. Struct. Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 2.Ura K, Hayes JJ, Wolffe AP. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 1995;14:3752–3765. doi: 10.1002/j.1460-2075.1995.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vettese-Dadey M, Grant PA, Hebbes TR, Crane- Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 4.Nightingale KP, Wellinger RE, Sogo JM, Becker PB. Histone acetylation facilitates RNA polymerase II transcription of the drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe AP. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas G, Leibovitch BA, Elgin SC. Chromatin organization and transcriptional control of gene expression in drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- 7.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 8.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 9.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem. Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 12.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev. Cell. 2010;19:662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- 14.Turner JM, Burgoyne PS, Singh PB. M31 and macroH2A1.2 colocalise at the pseudoautosomal region during mouse meiosis. J. Cell. Sci. 2001;114:3367–3375. doi: 10.1242/jcs.114.18.3367. [DOI] [PubMed] [Google Scholar]
- 15.Choo JH, Kim JD, Chung JH, Stubbs L, Kim J. Allele-specific deposition of macroH2A1 in imprinting control regions. Hum. Mol. Genet. 2006;15:717–724. doi: 10.1093/hmg/ddi485. [DOI] [PubMed] [Google Scholar]
- 16.Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 2009;16:1074–1079. doi: 10.1038/nsmb.1665. [DOI] [PubMed] [Google Scholar]
- 17.Changolkar LN, Singh G, Cui K, Berletch JB, Zhao K, Disteche CM, Pehrson JR. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol. Cell. Biol. 2010;30:5473–5483. doi: 10.1128/MCB.00518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muthurajan UM, McBryant SJ, Lu X, Hansen JC, Luger K. The linker region of macroH2A promotes self-association of nucleosomal arrays. J. Biol. Chem. 2011;286:23852–23864. doi: 10.1074/jbc.M111.244871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 21.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 22.Caterino TL, Fang H, Hayes JJ. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol. Cell. Biol. 2011;31:2341–2348. doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes JJ, Wolffe AP. Preferential and asymmetric interaction of linker histones with 5S DNA in the nucleosome. Proc. Natl Acad. Sci. USA. 1993;90:6415–6419. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arya G, Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein E, Muratore-Schroeder TL, Diaz RL, Chow JC, Changolkar LN, Shabanowitz J, Heard E, Pehrson JR, Hunt DF, Allis CD. A phosphorylated subpopulation of the histone variant macroH2A1 is excluded from the inactive X chromosome and enriched during mitosis. Proc. Natl Acad. Sci. USA. 2008;105:1533–1538. doi: 10.1073/pnas.0711632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth SY, Allis CD. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 1992;17:93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- 27.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pehrson JR, Costanzi C, Dharia C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 29.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 30.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 31.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 32.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 33.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 34.Patel A, McKnight JN, Genzor P, Bowman GD. Identification of residues in chromo-helicase-DNA-binding protein 1 (Chd1) required for coupling ATP hydrolysis to nucleosome sliding. J. Biol. Chem. 2011;286:43984–43993. doi: 10.1074/jbc.M111.282970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassabov SR, Bartholomew B. Site-directed histone-DNA contact mapping for analysis of nucleosome dynamics. Meth. Enzymol. 2004;375:193–210. doi: 10.1016/s0076-6879(03)75013-7. [DOI] [PubMed] [Google Scholar]
- 36.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cary PD, Crane-Robinson C, Bradbury EM, Dixon GH. Effect of acetylation on the binding of N-terminal peptides of histone H4 to DNA. Eur. J. Biochem. 1982;127:137–143. doi: 10.1111/j.1432-1033.1982.tb06847.x. [DOI] [PubMed] [Google Scholar]
- 38.Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J. Biol. Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 39.Hayes JJ, Clark DJ, Wolffe AP. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl Acad. Sci. USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer WR, Hayes JJ, White JH, Wolffe AP. Nucleosome structural changes due to acetylation. J. Mol. Biol. 1994;236:685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 41.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 42.Vettese-Dadey M, Walter P, Chen H, Juan LJ, Workman JL. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol. Cell. Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 44.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meersseman G, Pennings S, Bradbury EM. Mobile nucleosomes–a general behavior. EMBO J. 1992;11:2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Längst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 47.Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl Acad. Sci. USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stokes DG, Perry RP. DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 2011;30:2596–2609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma A, Jenkins KR, Héroux A, Bowman GD. Crystal structure of the chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA. J. Biol. Chem. 2011;286:42099–42104. doi: 10.1074/jbc.C111.294462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang EY, Ferreira H, Somers J, Nusinow DA, Owen-Hughes T, Narlikar GJ. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry. 2008;47:13726–13732. doi: 10.1021/bi8016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao Y, Konesky K, Park YJ, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K. Nucleosomes containing the histone variant H2A.bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyen CM, Montel F, Gautier T, Menoni H, Claudet C, Delacour-Larose M, Angelov D, Hamiche A, Bednar J, Faivre-Moskalenko C, et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 55.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 56.Tan S, Davey CA. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 58.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 59.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J. Mol. Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 60.Tóth K, Brun N, Langowski J. Trajectory of nucleosomal linker DNA studied by fluorescence resonance energy transfer. Biochemistry. 2001;40:6921–6928. doi: 10.1021/bi002695m. [DOI] [PubMed] [Google Scholar]
- 61.Tóth K, Brun N, Langowski J. Chromatin compaction at the mononucleosome level. Biochemistry. 2006;45:1591–1598. doi: 10.1021/bi052110u. [DOI] [PubMed] [Google Scholar]
- 62.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 63.Hayes JJ, Pruss D, Wolffe AP. Contacts of the globular domain of histone H5 and core histones with DNA in a "chromatosome". Proc. Natl Acad. Sci. USA. 1994;91:7817–7821. doi: 10.1073/pnas.91.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang H, Clark DJ, Hayes JJ. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 2012;40:1475–1484. doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hendzel MJ, Lever MA, Crawford E, Th'ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 66.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 67.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maier VK, Chioda M, Rhodes D, Becker PB. ACF catalyses chromatosome movements in chromatin fibres. EMBO J. 2008;27:817–826. doi: 10.1038/sj.emboj.7601902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 70.Yudkovsky N, Logie C, Hahn S, Peterson CL. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nightingale K, Dimitrov S, Reeves R, Wolffe AP. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 73.Abbott DW, Chadwick BP, Thambirajah AA, Ausió J. Beyond the Xi: macroH2A chromatin distribution and post-translational modification in an avian system. J. Biol. Chem. 2005;280:16437–16445. doi: 10.1074/jbc.M500170200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.