Abstract
Reactive oxygen species constantly generated as by-products of cellular metabolism readily attack genomic DNA creating mutagenic lesions such as 7,8-dihydro-8-oxo-guanine (8-oxo-G) that promote aging. 8-oxo-G:A mispairs arising during DNA replication are eliminated by base excision repair initiated by the MutY DNA glycosylase homologue (MUTYH). Here, by using formaldehyde crosslinking in mammalian cell extracts, we demonstrate that the WRN helicase/exonuclease defective in the premature aging disorder Werner syndrome (WS) is recruited to DNA duplex containing an 8-oxo-G:A mispair in a manner dependent on DNA polymerase λ (Polλ) that catalyzes accurate DNA synthesis over 8-oxo-G. Similarly, by immunofluorescence, we show that Polλ is required for accumulation of WRN at sites of 8-oxo-G lesions in human cells. Moreover, we show that nuclear focus formation of WRN and Polλ induced by oxidative stress is dependent on ongoing DNA replication and on the presence of MUTYH. Cell viability assays reveal that depletion of MUTYH suppresses the hypersensitivity of cells lacking WRN and/or Polλ to oxidative stress. Biochemical studies demonstrate that WRN binds to the catalytic domain of Polλ and specifically stimulates DNA gap filling by Polλ over 8-oxo-G followed by strand displacement synthesis. Our results suggest that WRN promotes long-patch DNA repair synthesis by Polλ during MUTYH-initiated repair of 8-oxo-G:A mispairs.
INTRODUCTION
Reactive oxygen species constantly produced in living organisms as byproducts of normal cellular metabolism or as a consequence of environmental exposure to various physical and chemical agents can generate a variety of oxidized DNA bases that are highly mutagenic and hence compromise genomic stability, promoting aging and carcinogenesis (1–4). One of the most frequent oxidative lesions is 7,8-dihydro-8-oxo-guanine (8-oxo-G) with a steady-state level of about 103 lesions per cell in normal tissue (5). Replication of genomic DNA containing 8-oxo-G lesions frequently leads to the formation of 8-oxo-G:A mispairs, giving rise to a G:C to T:A transversion mutations (6). Interestingly, these transversions are among the predominant somatic mutations found in lung, breast, ovarian, gastric and colorectal cancers, suggesting that a failure to eliminate 8-oxo-G lesions can initiate tumorigenesis and drive tumor progression (7).
Oxidized base lesions are primarily eliminated by the base excision repair (BER) system (8). In mammalian cells, the repair of 8-oxo-G:A mispairs is achieved via two BER events that occur sequentially on the two DNA strands (9). The first event is initiated by excision of the mispaired A residue by the MutY glycosylase homologue (MUTYH) in a reaction coordinated by proliferating cell nuclear antigen (PCNA) (10–12). This is followed by cleavage of the apurinic site (AP) by the AP endonuclease 1 (APE1), creating a DNA gap with a 3′-OH moiety (12,13). PCNA and replication protein A (RPA) then govern the bypass of the 8-oxo-G lesion by the DNA polymerase λ (Polλ), which in the presence of these two auxiliary factors preferentially incorporates dCTP opposite the lesion (12,14,15). Following lesion bypass, RPA dissociates and PCNA recruits flap endonuclease 1 (FEN1) to remove the 5′-single-stranded DNA (ssDNA) flap resulting from the limited strand displacement synthesis by Polλ (12). Finally, DNA ligase I interacts with PCNA loaded on the nick arising from FEN1 cleavage and seals it, creating the substrate for a second BER event, which leads to the elimination of the 8-oxo-G lesion (12). 8-oxo-G paired with C is predominantly excised by the OGG1 glycosylase in a short patch BER reaction in which Polβ fills the DNA gap and the DNA ligase III/XRCC1 complex restores the continuity of the damaged DNA strand (8).
Werner syndrome (WS) is an autosomal recessive disorder characterized by premature aging, cancer predisposition and genomic instability (16). It is caused by mutations in the WRN gene which encodes a multifunctional protein (WRN) possessing 3′–5′ DNA helicase and 3′–5′ exonuclease activities (16). Interestingly, WRN-deficient cells accumulate 8-oxo-G lesions at a much higher rate than WRN-proficient cells (17,18). However, the molecular basis of this phenomenon is not known. Here we present several lines of evidence suggesting that WRN cooperates with Polλ to carry out long-patch DNA repair synthesis during MUTYH-initiated repair of 8-oxo-G:A mispairs. Loss of such an activity might explain many cellular phenotypes associated with WS including accumulation of oxidative DNA lesions, accelerated telomere attrition and genomic instability.
MATERIALS AND METHODS
Antibodies and purified proteins
All primary antibodies used for immunofluorescence staining and immunoblotting are described in Supplementary Materials and Methods. Recombinant human Polλ protein was expressed and purified as previously described (19). His-tagged recombinant human Polλ fragments were purified on Ni-NTA agarose (Invitrogen) as recommended by the manufacturer. Recombinant human WRN protein and its mutants were produced and purified as previously described (20). These protein preparations had a purity of >95% (Supplementary Figure S1A) and did not contain any contaminating DNA polymerase activity (Supplementary Figure S1C). Purified wild-type WRN could unwind efficiently a forked DNA duplex (Supplementary Figure S1B). RECQ5 and BLM proteins were purified as previously described (21,22).
Cell culture experiments
All cell lines (HeLa, U2OS, HEK293, MEFs, MRC5) used in this study were maintained in DMEM (Gibco) supplemented with 10% fetal calf serum (Gibco) and streptomycin/penicillin (100 U/ml). Where required, H2O2 (Sigma) was added to cell cultures to a final concentration as indicated. Hydroxyurea (Sigma) was used at a concentration of 2 mM to synchronize cells at G1/S boundary. To arrest DNA replication, cells were treated with aphidicolin (Sigma) at a concentration of 1 µg/ml. Transfection of siRNA oligonucleotides was carried out using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s instructions. Cells were analyzed 72 h after siRNA transfection. The sequences of the siRNA oligonucleotides used in this study are indicated in Supplementary Materials and Methods.
Crosslinking assay
Formaldehyde crosslinking assays with cell extracts and hairpin oligonucleotide substrates attached to streptavidin magnetic beads (Invitrogen) were performed as described previously (12).
Chromatin-binding assay
Chromatin fractionation was done as previously described (23). Briefly, to obtain cytoplasmic fraction, HEK293T cells were resuspended in buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 1 mg/ml digitonin, Roche complete protease inhibitor cocktail) and incubated on ice for 5 min. Nuclei were collected by centrifugation at 1500g for 4 min, washed once with buffer A, resuspended in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, Roche complete protease inhibitor cocktail) and incubated on ice for 15 min. Chromatin was separated from nucleoplasmic fraction by low speed centrifugation at 2000g for 4 min, washed twice in buffer B, resuspended in SDS loading buffer and sonicated. Bound proteins were analyzed by western blotting.
Immunofluorescence assays
Cells cultured on glass coverslips were fixed with 3.7% formaldehyde for 10 min at room temperature (RT) and subsequently permeabilized by soaking in 0.2% (v/v) Triton X-100 for 5 min at RT. After blocking in PBS containing 5 mg/ml BSA for 30 min at RT, the fixed cells were incubated overnight at 4°C with appropriate primary antibodies. The slides were washed with PBS and incubated for 1.5 h at RT with secondary antibodies diluted in blocking solution: fluorescein isothiocyanate- (FITC) conjugated sheep anti-rabbit IgG (Sigma; 1:700) and Texas Red-conjugated donkey anti-mouse IgG (Jackson Immunoresearch, 1:200). After washing with PBS, coverslips were mounted on Vectashield (Vector Laboratories) and images were captured on an Olympus IX81 fluorescence microscope. At least 100 nuclei were analyzed in each of three independent experiments. For simultaneous detection of WRN and 8-oxo-G, cells were fixed and sequentially incubated with rabbit polyclonal anti-WRN antibody and anti-rabbit FITC-conjugated secondary antibody. Stained cells were then fixed with 100% cold methanol for 30 min at −20°C and immersed in 100% cold acetone for 30 s. After washing, the fixed cells were treated with 2M HCl for 30 min to denature the DNA and then neutralized with 0.1 M borate buffer (pH 8.5). After washing and blocking, cells were stained with mouse monoclonal anti-8-oxo-G (IgM) antibody (1:100) followed by Texas Red-conjugated donkey anti-mouse IgM secondary antibody (Jackson Immunoresearch, 1:150). After washing, coverslips were mounted and analyzed as described above. The same procedure was used for simultaneous detection of Polλ and 8-oxo-G lesion.
Immunoprecipitation assays
Total cell extract preparation and immunoprecipitation (IP) were carried out as described previously (24). For IP of purified recombinant proteins, a mixture of equal amounts of WRN (300 ng) and Polλ (300 ng) was incubated for 2 h at 4°C and then added to Protein A/G-agarose beads (20 µl) coated with rabbit polyclonal anti-WRN IgGs (2 µg). After incubation for 2 h at 4°C, the beads were washed and subjected to western blot analysis. All the IP reactions were conducted in the presence of DNase I (Roche) to exclude the possibility of protein–protein interaction mediated through DNA.
Gap-filling assay
Annealing of a 72-mer containing 8-oxo-G lesion (or a normal G) with the 5′-[32P]-labeled 39-mer primer created a primer/template substrate with the lesion (or a normal G) at the +1 position relative to single-strand/double-strand junction. Annealing of a 32-mer oligonucleotide to the region 3′ of the lesion site in this structure yielded a duplex containing a one-nucleotide (1-nt) gap opposite 8-oxo-G (or normal G). The 32-mer oligonucleotide was phosphorylated by T4 polynucleotide kinase (NEB) prior to annealing. The reaction mixtures (10 µl) contained 50 mM Tris–HCl (pH 7.5), 2 mM DTT, 0.25 mg/ml BSA, 10 µM dNTP, 1 mM MgCl2 and 10 fmol of the 5′-[32P]-labeled DNA substrate. Concentrations of Polλ and WRN are indicated in figures and figure legends. Reactions were carried out at 37°C for 10 min. Reaction mixtures were separated on a denaturing urea-polyacrylamide gel and radiolabeled DNA species were visualized by phosphorimaging on a Typhoon 9400. Gel images were quantified using ImageQuant software.
Biotin pull-down assay
Pull-down assays using biotinylated gapped DNA duplexes with or without 8-oxo-G lesion were performed as described previously (15).
Cell viability assay
Cell viability assays were done as previously described (25). Briefly, HeLa cells were seeded at a confluency of 25% in cell culture plates containing complete medium (DMEM, 10% FCS and penicillin/streptomycin). After 24 h, cells were transfected with the appropriate siRNAs. 2 days after transfection, siRNA-treated cells were harvested and seeded in a 96-well plate at a density of 2500 cells/well in a volume of 100 µl of complete medium. After 24 h, cells were treated with different concentrations of H2O2 (Sigma) ranging from 0 to 640 µM. Experiments were carried out in hexaplicates for each H2O2 concentration. 2 h after H2O2 addition, cells were washed twice with PBS, and allowed to grow in complete medium. After 2 days, a mixture of resazurin and complete medium (ratio of 1:10; 100 µl) was added to individual wells. Cell viability was measured after 4 h of incubation using a SpectraMax reader M5 (Molecular Devices). The percentage of viable cells was calculated relative to mock treated cells and plotted using GraphPad Prism as mean ± SD. Data from at least two independent experiments were plotted.
RESULTS
WRN is involved in the processing of 8-oxo-G:A mispairs in mammalian cell extracts
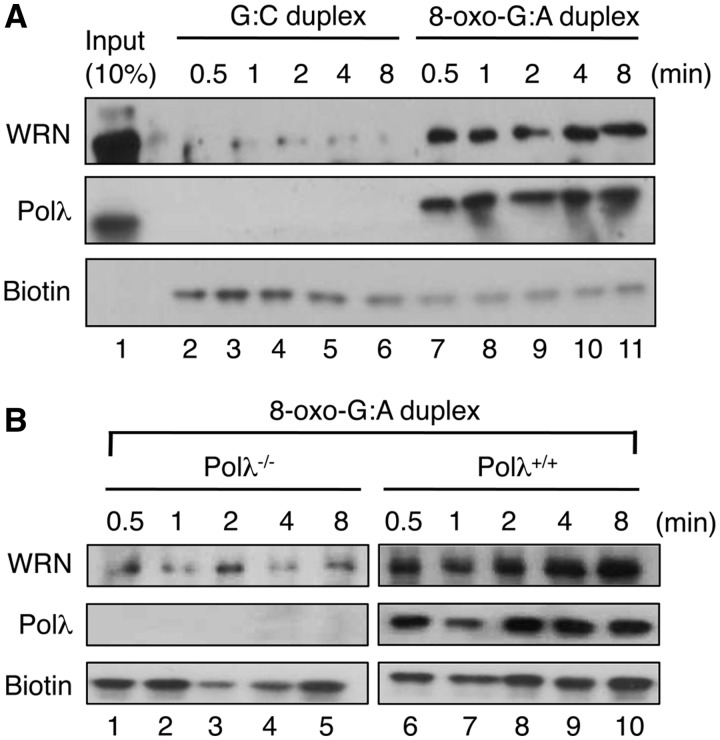
To investigate whether WRN is involved in the repair of 8-oxo-G:A mispairs, we employed a previously established assay using formaldehyde crosslinking in human cell extracts to monitor the recruitment of BER proteins to damaged DNA (26). A 3′-biotinylated hairpin loop oligoduplex (27 bp) containing a single 8-oxo-G:A mispair was incubated with HeLa whole-cell extract in the presence of Mg2+ to initiate repair (12). As a control reaction, the corresponding lesion-free substrate was also tested. At different time points, the reactions were stopped by the addition of formaldehyde and cross-linked DNA–protein complexes, isolated using streptavidin magnetic beads, were analyzed by western blotting. We observed a rapid, damage-specific recruitment of WRN to the DNA substrate (Figure 1A). A robust damage-specific recruitment of Polλ was also detected as previously reported (12). To address whether the recruitment of WRN to the hairpin duplex containing 8-oxo-G:A mispair was dependent on Polλ, whole-cell extracts from Polλ−/− and Polλ+/+ mouse embryonic fibroblasts (MEFs) were prepared and subjected to formaldehyde crosslinking assay. We found that in the absence of Polλ, WRN was not cross-linked to the hairpin substrate, suggesting that Polλ mediates the recruitment of WRN to the sites of repair of 8-oxo-G:A mispairs (Figure 1B). This is further supported by the finding that complementation of Polλ−/− MEFs with human Polλ cDNA lead to a restoration of WRN recruitment to the hairpin duplex in formaldehyde crosslinking assay (Supplementary Figure S2). These results provide a strong evidence for the involvement of WRN in the repair of 8-oxo-G:A mispairs.
Figure 1.
Polλ-dependent recruitment of WRN to sites of repair of 8-oxo-G:A mispairs in cell extracts. (A) Formaldehyde crosslinking assay using G:C (lanes 2–6) or 8-oxo-G:A (lanes 7–11) biotinylated hairpin DNA substrate and HeLa whole-cell extract. (B) Formaldehyde crosslinking assay using biotinylated 8-oxo-G:A hairpin substrate and whole-cell extracts from Polλ−/− (left panel, lanes 1–5) or Polλ+/+ (right panel, lanes 6–10) MEFs. The experiments were performed under the conditions specified in the ‘Materials and Methods’ section. Blots were probed with antibodies against WRN and Polλ. Biotin was detected using HRP-conjugated Streptavidin. The indicated time points refer to the length of incubation of extracts with DNA before addition of the crosslinking agent. Note that western blot analysis of extracts from Polλ+/+ and Polλ−/− MEFs is shown in Figure 4B (right panel).
WRN accumulates at sites of 8-oxo-G lesions during S-phase in a manner dependent on DNA replication and the presence of Polλ
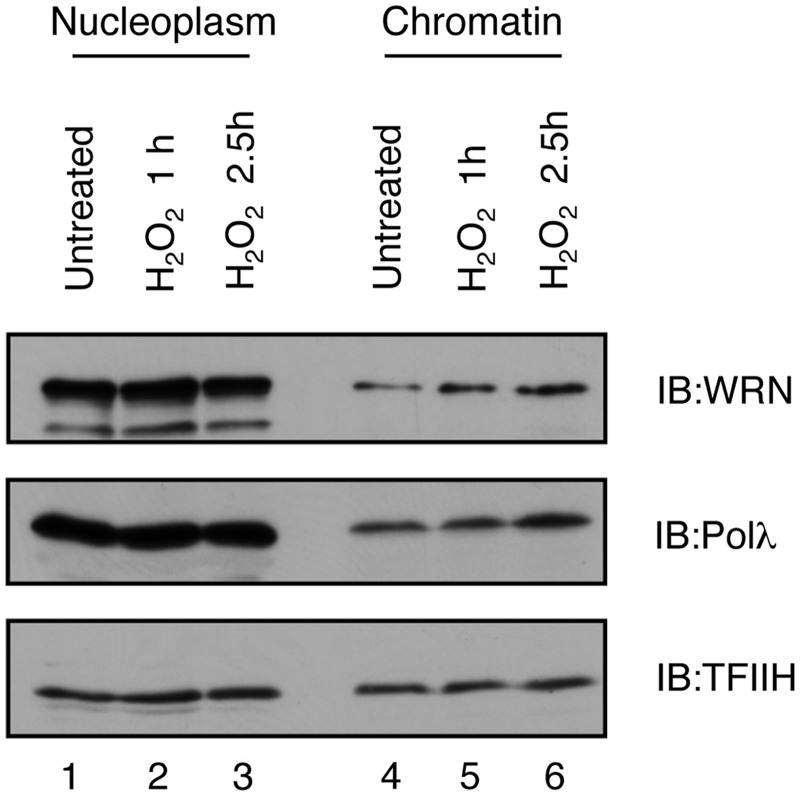
To investigate whether WRN is recruited to sites of oxidative DNA damage in vivo, we first performed sub-cellular fractionation to analyze chromatin binding of WRN in HEK293T cells prior to and after treatment with H2O2. We found that at 2.5 h after H2O2 addition, the amount of WRN bound to chromatin substantially increased as compared to non-treated cells (Figure 2). Likewise, we observed that H2O2 treatment promoted chromatin binding of Polλ (Figure 2).
Figure 2.
Accumulation of WRN and Polλ on chromatin after oxidative stress. HEK293 cells were treated with 500 µM H2O2 and fractionated under conditions specified in the ‘Materials and Methods’ section. Nucleoplasmic and chromatin fractions were analyzed by western blotting. Blots were probed with antibodies against WRN, Polλ and TFIIH.
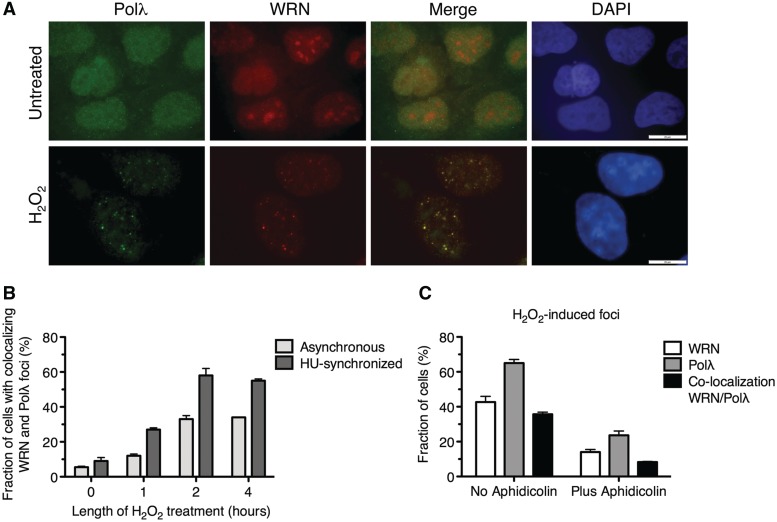
Next, we used indirect immunofluorescence technique to monitor the spatial distribution of WRN and Polλ in U2OS cells after H2O2 treatment. In agreement with previous reports, WRN was observed to localize to the nucleolus in the majority of non-treated cells, whereas Polλ showed a dispersed nuclear staining (Figure 3A, top row). Upon treatment of cells with 500 µM H2O2 for 2 h, WRN and Polλ each formed ≥10 distinct foci per nucleus in 45 and 65% of cells, respectively. Approximately in 35% of these cells, more than 75% of WRN foci co-localized with Polλ foci (Figure 3A, bottom row, and Supplementary Table S1). Notably, the frequency of nuclei with co-localizing foci of WRN and Polλ increased substantially when H2O2 was added to cells that had been released from G1/S blockade, suggesting that the re-localization of these proteins induced by oxidative stress is specific to S-phase cells (Figure 3B, Supplementary Figure S3). In accordance with this assumption, we found that after H2O2 treatment, WRN and Polλ formed foci only in cells expressing cyclin A (Supplementary Figure S4). To substantiate these findings, cells were treated with aphidicolin for 20 min prior to addition of H2O2 to inhibit DNA replication. We found that aphidicolin impaired the formation of WRN and Polλ foci after oxidative stress, suggesting that WRN and Polλ are recruited to site of oxidative DNA damage in a manner dependent on DNA replication that can give rise to 8-oxo-G:A mispairs (Figure 3C).
Figure 3.
DNA replication-dependent co-localization of WRN and Polλ at nuclear foci in response to oxidative stress. (A) WRN co-localizes with Polλ after exposure of U2OS cells to oxidative stress. Cells grown on glass coverslips were either left untreated or treated with 500 µM H2O2 for 2 h. After treatment, cells were fixed and immunostained using antibodies against Polλ (green) and WRN (red). DAPI staining of the nucleus is shown in blue. Scale bar, 20 µm. (B) Frequency of the formation of co-localizing foci of WRN and Polλ in synchronized and non-synchronized populations of U2OS cells following H2O2 treatment. Prior to addition of H2O2, cells were synchronized at G1/S boundary by treatment with hydroxyurea (HU) for 16 h and then released to S phase by adding fresh medium without HU. (C) Effect of aphidicolin on the nuclear focus formation of WRN and Polλ in U2OS cells after H2O2 treatment. Aphidicolin was added to cells at a concentration of 1 µg/ml at 20 min prior to addition of H2O2 to arrest cellular DNA replication. The data points in (B) and (C) represent the mean of three independent experiments with at least 100 nuclei scored in each experiment.
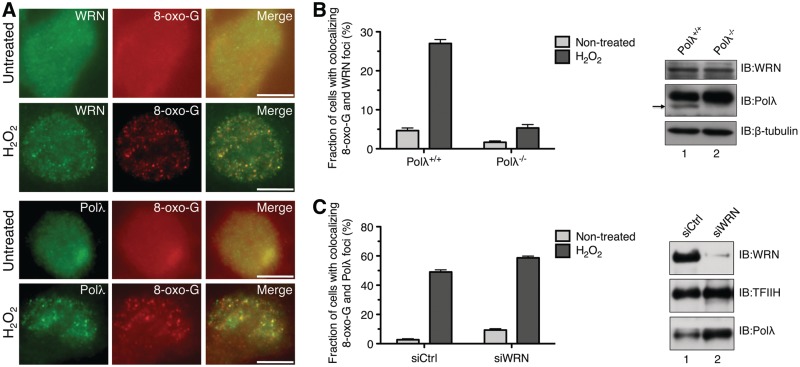
To prove that WRN and Polλ co-localized at sites of oxidative DNA damage, untreated and H2O2-treated cells were co-immunostained either for visualization of WRN and 8-oxo-G or for visualization of Polλ and 8-oxo-G. The results indicated that WRN and Polλ co-localized with sites of 8-oxo-G lesions in ∼40 and ∼50% of cells, respectively (Figure 4A, Supplementary Tables S2 and S3). To explore the mechanism underlying the recruitment of WRN to sites of 8-oxo-G lesions in vivo, we analyzed by immunofluorescence the spatial distribution of WRN in Polλ+/+ and Polλ−/− MEFs prior to and after treatment with H2O2. The results demonstrated that genetic ablation of Polλ impaired the re-localization of WRN to sites of 8-oxo-G lesions (Figure 4B and Supplementary Figure S5), which is consistent with the data from the crosslinking experiments described above (Figure 1B). We also tested the effect of WRN depletion on the accumulation of Polλ at sites of 8-oxo-G lesions in U2OS cells. We found that lack of WRN did not abolish Polλ focus formation after oxidative stress (Figure 4C). Instead, a small increase in the frequency of cells with co-localizing foci of 8-oxo-G and Polλ was observed in WRN-deficient cells relative to WRN-proficient cells both in the presence and in the absence of H2O2 (Figure 4C). This is in agreement with the previous finding of elevated levels of spontaneous oxidative DNA damage in WRN-deficient cells (17).
Figure 4.
WRN localizes to sites of 8-oxo-G lesions in vivo in a manner dependent on Polλ. (A) WRN and Polλ accumulate at sites of 8-oxo-G lesions. U2OS cells were left either non-treated (top panels) or treated with 500 µM H2O2 for 2 h (lower panels), fixed and co-stained either with antibodies against WRN (green) and anti-8-oxo-G (red) or with antibodies against Polλ (green) and 8-oxo-G (red) under conditions described in the ‘Materials and Methods’ section. Images were captured on an Olympus IX81 fluorescence microscope. Scale bar, 20 µm. (B) Graph showing the proportion of Polλ+/+ and Polλ−/− MEFs positive for co-localization of WRN and 8-oxo-G foci after mock- or H2O2-treatment, respectively. H2O2 was present at a concentration of 100 µM for 2 h. After treatment, cells were fixed and stained for WRN and 8-oxo-G. (Right panel) Western blot analysis of extracts from Polλ+/+ and Polλ−/− MEFs. Blots were probed with antibodies against WRN, Polλ and β-tubulin (loading control). The arrowhead indicates the band corresponding to mouse Polλ. (C) Graph showing the proportion of siCtrl- and siWRN-treated cells positive for co-localization between Polλ and 8-oxo-G foci after mock- or H2O2-treatment. Cells were treated with 500 µM H2O2 (or mock-treated) for 2 h, fixed and immunostained to visualize Polλ and 8-oxo-G. Treatment was carried out 72 h after siRNA transfection (right panel). Western blot analysis of extracts of U2OS cells transfected with WRN siRNA (siWRN) and control siRNA (siCtrl), respectively. Cells were harvested 72 h post-transfection. Blots were probed with antibodies against WRN, Polλ and TFIIH (loading control). The data points in (B) and (C) represent the mean of three independent experiments with at least 100 nuclei counted in each experiment.
Collectively, the results described above provide strong evidence that WRN is recruited to sites of 8-oxo-G lesions in a DNA replication- and Polλ-dependent manner.
WRN is required for repair of 8-oxo-G:A mispairs in human cells
Earlier studies revealed that Polλ−/− MEFs were more sensitive to oxidative stress than normal MEFs (27). To prove that WRN functions in repair of oxidative DNA damage in conjunction with Polλ, we employed resazurin-based cell viability assay to test the effect of single and combined depletions of WRN and Polλ on sensitivity of HeLa cells to H2O2 treatment. In agreement with the previous report, we found that Polλ-depleted cells displayed a higher sensitivity to H2O2 compared to mock-depleted cells (Figure 5A and E). WRN-depleted cells exhibited a similar degree of sensitivity to H2O2 as Polλ-depleted cells (Figure 5A and E). Co-depletion of WRN and Polλ did not result in a further decrease in H2O2 sensitivity compared to singly-depleted cells, suggesting that WRN and Polλ act in the same pathway of oxidative DNA damage repair (Figure 5A and E).
Figure 5.
Depletion of MUTYH confers oxidative stress tolerance to WRN- and Polλ-deficient cells and impairs the recruitment of WRN and Polλ to sites of oxidative DNA damage. (A–D) Graphs showing sensitivity of siRNA-depleted HeLa cells to indicated concentrations of H2O2. siCtrl, siMUTYH, siWRN and siPolλ represent siRNA against luciferase, MUTYH, WRN and Polλ, respectively. Cell viability assays were performed as described in the ‘Materials and Methods’ section. Each data point represents mean ± SD (n = 12). (E) Western blot analysis of extracts of HeLa cells transfected with different siRNAs as indicated. W, P and M stand for WRN, Polλ and MUTYH, respectively. Cells were harvested 72 hours after transfection. Blots were probed with antibodies against MUTYH, WRN, Polλ and TFIIH (loading control). IB, immunoblotting. (F) Effect of MUTYH depletion on nuclear focus formation of WRN and Polλ in U2OS cells after oxidative stress. Cells were treated with 500 µM H2O2 for 2 h, fixed and immunostained as in Figure 3A. H2O2-treatment was carried out 72 h after siRNA transfection. The data points represent the mean of three independent experiments with at least 100 nuclei scored in each experiment. (Right panel) Western blot analysis of extracts of U2OS cells transfected with siCtrl and siMUTYH, respectively. Cells were harvested 72 h post-transfection. Blots were probed with antibodies against WRN, Polλ, MUTYH and TFIIH (loading control).
To substantiate the above findings, we investigated whether depletion of WRN and Polλ caused cell death in response to oxidative stress. Using Annexin V-binding assay, we found that lack of either protein leads to a significant increase in the frequency of dead cells after H2O2 treatment as compared to mock-depleted cells (Supplementary Figure S6). Again, no additive effects on cell death were observed upon co-depletion of WRN and Polλ, confirming that these proteins act epistatically to protect cells against oxidative DNA damage (Supplementary Figure S6).
We reasoned that the hypersensitivity of cells deficient for WRN and Polλ to H2O2 was a consequence of accumulation of ssDNA breaks resulting from MUTYH-mediated processing of 8-oxo-G:A mispairs. Therefore, we tested whether the defect conferred by WRN/Polλ deficiency could be rescued by the elimination of MUTYH. Indeed, we found that depletion of MUTYH from HeLa cells lacking WRN and/or Polλ restored the cellular sensitivity to H2O2 to control levels (Figure 5B–E and Supplementary Figure S6). Depletion of MUTYH alone did not significantly alter H2O2 sensitivity of cells proficient for WRN and Polλ (Figure 5, compare panels B–D with panel A). Together, these data provide strong evidence that WRN and Polλ are required for repair of 8-oxo-G:A mispairs initiated by the MUTYH DNA glycosylase.
To obtain further evidence for the above statement, we tested the effect of MUTYH depletion on the re-localization of WRN and Polλ to nuclear foci in response to oxidative stress. We found that lack of MUTYH dramatically impaired the nuclear focus formation of both WRN and Polλ in U2OS cells after H2O2 treatment, suggesting that these foci represent sites of repair of 8-oxo-G:A mispairs (Figure 5F).
WRN physically interacts with Polλ
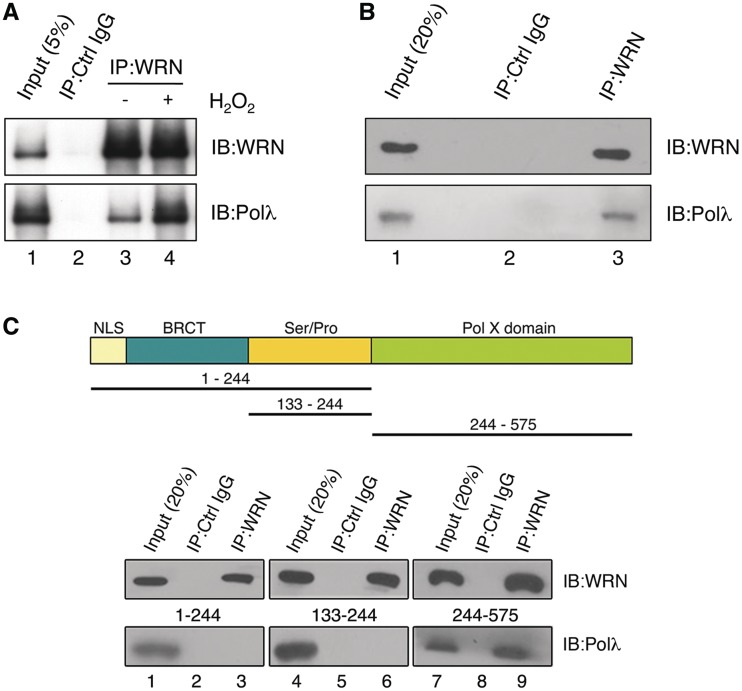
To investigate whether WRN and Polλ interact physically, an extract of HEK293 cells was subjected to IP with anti-WRN antibody. We found that Polλ co-immunoprecipitated with WRN, while it was not present in an immunoprecipitate obtained with control IgG, suggesting that these proteins formed a stable complex in vivo (Figure 6A). Interestingly, the level of WRN–Polλ complex in the extract from cells treated with H2O2 was notably higher compared to that in the extract from non-treated cells, suggesting that the formation of WRN–Polλ complex in cells might be stimulated by oxidative DNA damage (Figure 6A, compare lanes 3 and 4). We also performed co-IP experiments with a mixture of purified recombinant proteins and found that WRN and Polλ co-precipitated with anti-WRN antibody, but not with control IgG, indicating that WRN and Polλ interact directly (Figure 6B).
Figure 6.
Physical interaction between WRN and Polλ. (A) Co-immunoprecipitation of Polλ with WRN from extracts of non-treated and H2O2-treated HEK293 cells. Cells were treated with 500 µM H2O2 for 2 h or mock-treated. WRN was immunoprecipitated using rabbit polyclonal anti-WRN antibody as described under the ‘Materials and Methods’ section. Blots were probed with anti-Polλ and anti-WRN antibodies. Lane 2, control immunoprecipitate obtained with preimmune rabbit IgGs. (B) Co-immunoprecipitation of Polλ with WRN from a mixture of purified proteins (300 ng of each protein) pre-incubated on ice for 2 h. Blots were probed as in (A). (C) Mapping of WRN-interacting domain on Polλ. (Top panel) Domain organization of Polλ. NLS, nuclear localization sequence; BRCT, BRCA1 C-terminal domain; Ser/Pro, serine/proline rich domain; PolX domain, catalytic domain conserved in the X family DNA polymerases. Black lines indicate fragments used for mapping. (Bottom panel) Co-immunoprecipitation assay. Purified (His)6-tagged Polλ fragments were incubated with full-length WRN followed by immunoprecipitation with rabbit polyclonal antibody against WRN or with control (Ctrl) IgG. Immunoprecipitates were analyzed by western blotting as in (A).
To map the WRN-binding site on Polλ, co-IP experiments were performed with different deletion variants of Polλ (Figure 6C). A stable interaction of WRN with Polλ244–575, but not with Polλ1–244 and Polλ133–244 was observed, suggesting that WRN binds to the catalytic core domain of Polλ (Figure 6C). We also used GST pull-down assay to map the region of WRN that binds to Polλ. We found that Polλ bound well to GST–WRN949–1432 and GST–WRN500–946, but not to GST–WRN51–499 or to GST alone (Supplementary Figure S7). These results indicated that Polλ interacted with the helicase domain of WRN (amino acids 500–946) as well as with the C-terminal region of WRN, which contains winged-helix domain, a binding site of a number of other proteins shown to interact with WRN (28).
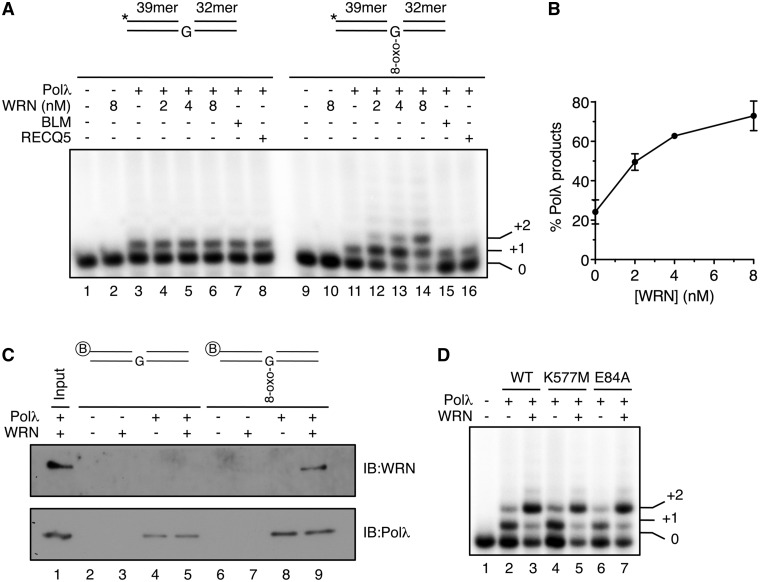
WRN specifically stimulates the bypass of 8-oxo-G by Polλ on gapped DNA duplex
In order to gain insight into the role of WRN in the repair of 8-oxo-G:A mispairs, we tested its effect on the gap-filling activity of Polλ using a synthetic DNA duplex containing a centrally located 1-nt gap opposite to 8-oxo-G (Figure 7A). We found that WRN significantly stimulated Polλ to fill the gap opposite the 8-oxo-G lesion and to carry out strand displacement synthesis (Figure 7A, lanes 11–14). Quantification of gel images from these experiments indicated that WRN increased the extent of Polλ reaction about 3-fold (Figure 7B). In contrast, WRN did not stimulate the gap-filling activity of Polλ on DNA substrate containing a normal G opposite the gap (Figure 7A, lanes 3–6). Moreover, stimulation of Polλ was not observed with other RecQ helicases such as BLM and RECQ5, and WRN did not enhance 8-oxo-G bypass by Polβ on gapped DNA duplex (Figure 7A, lanes 11, 15 and 16, and Supplementary Figure S8). These data indicate that WRN stimulates specifically Polλ-mediated bypass of 8-oxo-G on gapped DNA duplex.
Figure 7.
WRN promotes bypass of 8-oxo-G by Polλ on gapped DNA duplex. (A) Effect of WRN, BLM and RECQ5 on primer extension by Polλ across a 1-nt DNA gap opposite either a normal G or an 8-oxo-G. Reaction mixtures contained 1 nM DNA substrate, 5 nM Polλ, 10 µM dNTPs and indicated concentrations of WRN. BLM and RECQ5 were present at concentrations of 20 nM and 80 nM, respectively. Schemes of the DNA substrate are shown above the gels. Asterisks indicate radioactive label on the 5′-end of the primer. Reactions were analyzed as specified under the ‘Materials and Methods’ section. (B) Quantification of Polλ activity on gapped DNA duplex containing 8-oxo-G in the presence of increasing amounts of WRN as shown in (A). Relative concentration of Polλ products is plotted. Data points represent the mean of values from three independent experiments. (C) Binding of WRN and Polλ to 1-nt gap DNA duplexes with or without 8-oxo-G lesion. 20 nM Polλ and 20 nM WRN were incubated alone or in combination with 100 nM 5′-biotinylated gapped DNA duplexes in the presence of 1 µM dCTP. Bound proteins were isolated using streptavidin beads and analyzed by immunoblotting (IB). Lane 1 contains 100% of input material. (D) Incorporation of dCTP by Polλ on 8-oxo-G-containing gapped DNA substrate in the presence of helicase-deficient (K557M) and exonuclease-deficient (E84A) mutants of WRN. Reactions were carried out as in (A). Wild type (WT) and mutant forms of WRN were present at a concentration of 8 nM.
We also tested the effect of WRN on 8-oxo-G bypass by Polλ following MUTYH/APE1-mediated processing of an 8-oxo-G:A mispair in a 100-bp DNA duplex (12). We found that WRN significantly enhanced the trans-lesion synthesis mediated by Polλ in this system (∼1.7-fold), providing evidence that WRN promotes lesion bypass by Polλ during MUTYH-initiated repair of 8-oxo-G:A mispairs (Supplementary Figure S9).
To substantiate these findings, we tested binding of WRN to the 8-oxo-G-containing and lesion-free gapped DNA substrates in the presence and in the absence of Polλ. Reactions also contained dCTP to allow Polλ to incorporate 2 nt. We found that Polλ was bound to both DNA substrates irrespective of the presence or absence of WRN, showing a higher affinity for the 8-oxo-G substrate (Figure 7C, compare lanes 4, 5, 8 and 9). Importantly, WRN did not bind to the gapped duplex containing normal G opposite the gap even in the presence of Polλ, while it did bind efficiently to the 8-oxo-G-containing gapped duplex providing that Polλ was present (Figure 7C, lanes 3, 5, 7 and 9).
To address whether the enzymatic activities of WRN were required for its stimulatory effect on DNA synthesis by Polλ, we used previously established mutants of WRN defective either in the exonuclease (E84A) or in the helicase (K577M) activity (29,30). We found that both mutants stimulated translesion synthesis by Polλ on the 8-oxo-G-containing gapped duplex to a similar degree as wild-type WRN (Figure 7D). In agreement with this result, we found that expression of either WRN mutant in WS fibroblasts restored oxidative stress tolerance to the wild-type level, suggesting that WRN might act non-catalytically in repair of oxidative DNA damage in vivo (Supplementary Figure S10).
RPA and PCNA were shown to promote accurate bypass of 8-oxo-G by Polλ, by repressing the formation of 8-oxo-G:A mispair and stimulating the formation of 8-oxo-G:C base pair (12). Therefore, we finally investigated the effect of WRN on incorporation by Polλ of individual nucleotides opposite 8-oxo-G. We found that WRN stimulated the incorporation of both dCTP and dATP opposite 8-oxo-G on gapped DNA duplex, indicating that WRN does not improve the fidelity of 8-oxo-G bypass by Polλ per se (Supplementary Figure S11).
DISCUSSION
The WRN protein has been implicated in a number of cellular processes including telomere maintenance, homologous recombination, replication fork recovery and repair of alkylation DNA damage (31–35). There are also reports suggesting a role for WRN in repair of oxidative DNA damage, but the underlying mechanism is not clear (17,18). Here we provide several lines of evidence suggesting that WRN is required for the correction of 8-oxo-G:A mispairs, which is mediated by the long patch BER pathway initiated by the MUTYH DNA glycosylase (12). Firstly, we found that WRN accumulated at sites of 8-oxo-G lesions in a manner dependent on Polλ, an X-family DNA polymerase that mediates translesion synthesis during MUTYH-initiated repair of 8-oxo-G:A mispairs (12). Nuclear focus formation of WRN and Polλ after exposure of cells to oxidative stress was found to be dependent on DNA replication and on the presence of MUTYH, strongly suggesting that WRN and Polλ accumulated at sites of 8-oxo-G:A mispairs.
Secondly, we found that depletion of WRN from human cells increased their sensitivity to H2O2 to a level similar to that displayed by cells lacking Polλ. Together with the finding that co-depletion of WRN and Polλ did not have an additive effect on the cellular sensitivity to oxidative stress, these data demonstrate that WRN and Polλ act in the same DNA repair pathway. Most importantly, the hypersensitivity of WRN- and Polλ-deficient cells to H2O2 was completely suppressed by depletion of MUTYH, strongly suggesting that WRN and Polλ are involved in the repair of 8-oxo-G:A mispairs initiated by the MUTYH DNA glycosylase. One can imagine a scenario where in absence of Polλ and/or WRN, ssDNA breaks resulting from the processing of 8-oxo-G:A mispairs by MUTYH and APE1 are converted to DNA double-strand breaks in the next round of DNA replication, leading to cell cycle arrest followed either by DNA repair or by cell death (Supplementary Figure S12).
Finally, we show that WRN forms a stable complex with Polλ in vivo and in vitro and stimulates specifically the bypass of 8-oxo-G by Polλ on a gapped DNA duplex resembling the DNA intermediate generated during MUTYH-initiated repair of 8-oxo-G:A mispairs. Moreover, we show that WRN stimulates 8-oxo-G bypass by Polλ following MUTYH/APEI-mediated processing of 8-oxo-G:A mispairs in vitro. WRN stimulated not only DNA gap filling by Polλ opposite 8-oxo-G, but it also enhanced the subsequent strand displacement synthesis by Polλ. Therefore, it appears that the role of WRN in MUTYH-initiated repair of 8-oxo-G:A mispair is to promote this pathway in direction of long-patch BER. It is known that DNA ligase I is not able to efficiently ligate a DNA substrate containing a nick in the vicinity of 8-oxo-G:C pair, but is able to seal a nick located 1 nt downstream of the correctly incorporated C (12). Thus stimulation of Polλ-mediated strand displacement synthesis would enable more efficient completion of accurate MUTYH-initiated BER. Interestingly, previous studies have shown that WRN physically interacts with FEN1 and strongly stimulates FEN1-catalyzed cleavage of 5′-flap substrates (36). Such an activity is essential to generate a ligateable 5′-P end following strand displacement synthesis by Polλ during the MUTYH-initiated repair of 8-oxo-G:A mispairs. Thus, it is possible that, in addition to stimulation of Polλ-mediated long-patch DNA repair synthesis, WRN promotes the endonucleolytic cleavage by FEN1 in this BER pathway.
Our study has shown that the helicase and exonuclease activities of WRN are dispensable for Polλ stimulation by WRN. Similarly, the functional interaction of WRN with FEN1 was independent of the WRN catalytic functions (36). These findings suggested that the stimulatory effect of WRN on these enzymes stems from direct protein–protein interaction. It is possible that physical interaction between WRN and Polλ triggers a conformational change in the polymerase, which alters its catalytic properties. Of note, WRN was shown to stimulate DNA synthesis by the Y-family translesion polymerases η, ι and κ by increasing the reaction rate (37).
We have also obtained evidence suggesting that the catalytic activities of WRN are not essential for repair of oxidative DNA damage in the cell. This notion is consistent with the fact that there are no case reports of WS patients carrying loss-of-function mutations in the exonuclease or in the helicase domains of WRN. All WS-causing mutations reported thus far result either in the elimination of the nuclear localization signal located at the C-terminus of WRN or lead to an unstable protein (38).
Importantly, WRN stimulated DNA gap filling and strand displacement synthesis by Polλ only if the DNA substrate contained an 8-oxo-G lesion opposite the gap. Accordingly, although Polλ could bind to both lesion-free and 8-oxo-G-containing substrates, WRN was bound only to the latter substrate in a manner dependent on Polλ. Thus, it is possible that binding of Polλ to the lesion generates a particular structure either on the DNA or in Polλ itself, allowing Polλ-WRN interaction. Our finding of direct interaction of WRN with the catalytic domain of Polλ suggest that a lesion-induced structural change in Polλ leading to the exposure of the WRN interaction site is the more likely scenario. In the complex of Polλ with lesion-free gapped duplex, the WRN interaction site on Polλ might be hindered by DNA. Future structural studies will be needed to shed more light on this issue.
Cells derived from individuals suffering from WS show premature senescence and accelerated telomere shortening, which explains the early onset of aging manifested in these patients and suggest that WRN plays a role in telomere metabolism (16). Evidence indicates that genomic instability in WS cells depends directly on telomere dysfunction, suggesting that the primary cause of high cancer incidence in WS is a breakdown in telomere integrity (39). It is well known that oxidative stress accelerates telomere shortening and shortens replicative lifespan of cultured human primary fibroblasts (40). A recent study has revealed that disruption of the mouse Ogg1 gene is associated with accumulation of oxidative guanine lesions in telomeres and increased telomere attrition upon oxidative stress, suggesting that oxidative stress compromises telomere integrity through induction of oxidative base damage (41). Thus, based on the earlier finding that WRN deficiency leads to accumulation of 8-oxo-G lesions in the genomic DNA (17,18) and our data implicating WRN in the repair of 8-oxo-G:A mispairs, it is tempting to speculate that WS phenotypes arise from a defect in repair of oxidative DNA base damage in telomeres. CO-FISH analysis of telomeric repeats revealed that the oxidative stress-induced telomere shortening in Ogg1−/− primary MEFs most frequently arises from single-strand breaks in the G-strand, which compromise telomere lagging strand synthesis (41). Interestingly, preferential telomere lagging strand loss was also demonstrated in cells lacking WRN or FEN1 (31,42). Thus, it is possible that the accelerated telomere shortening observed in these cells stems from persistence of 8-oxo-G lesions within the telomeric G-strand due to defective repair of 8-oxo-G:A mispairs. Therefore, it will be interesting to evaluate the role of the other enzymes acting in this BER pathway, such as MUTYH or Polλ, in promoting telomere integrity upon oxidative stress. Such studies will shed more light on how oxidative stress contributes to aging and cancer development.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–12, Supplementary Materials and Methods and Supplementary References [12,22,24,43–45].
FUNDING
Swiss National Science Foundation [31003A-129747/1 to P.J.]; [3100-109312/2 to U.H., P.P. and B.V.L.]; UBS AG to P.J. and R.K.; Oncosuisse [KLS-02344-02-2009 to P.J.]; Stiftung zur Krebsbekämpfung (to P.J.); University of Zurich (to U.H., B.V.L. and P.J.). National Institutes of Health Intramural Program of the National Institute on Aging [Z01-AG000726-17, in part]. Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank S.H. Wilson for Polλ+/+ and Polλ−/− MEFs, L. Blanco for antibody against mouse Polλ, K. Surendranath for help with Annexin V-binding assay and A. Franchitto for WS fibroblasts stably expressing WRN-E84A and WRN-K577M, respectively. We are also grateful to J. Jiricny and G. Maga for fruitful discussions.
REFERENCES
- 1.Valko M, Izakovic M, Mazur M, Rhodes CJ, Telser J. Role of oxygen radicals in DNA damage and cancer incidence. Mol. Cell. Biochem. 2004;266:37–56. doi: 10.1023/b:mcbi.0000049134.69131.89. [DOI] [PubMed] [Google Scholar]
- 2.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 3.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative stress and oxidative damage in carcinogenesis. Toxicol. Pathol. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 4.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, DC: ASM Press; 2006. p. 16. [Google Scholar]
- 6.Avkin S, Livneh Z. Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat. Res. 2002;510:81–90. doi: 10.1016/s0027-5107(02)00254-3. [DOI] [PubMed] [Google Scholar]
- 7.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst.) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst.) 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DzNA glycosylase. Nucleic Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi H, Tominaga Y, Hirano S, McKenna AE, Nakabeppu Y, Matsumoto Y. Replication-associated repair of adenine:8-oxoguanine mispairs by MYH. Curr. Biol. 2002;12:335–339. doi: 10.1016/s0960-9822(02)00686-3. [DOI] [PubMed] [Google Scholar]
- 12.van Loon B, Hubscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc. Natl Acad. Sci. USA. 2009;106:18201–18206. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29:743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 15.Maga G, Crespan E, Wimmer U, van Loon B, Amoroso A, Mondello C, Belgiovine C, Ferrari E, Locatelli G, Villani G, et al. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc. Natl Acad. Sci. USA. 2008;105:20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst.) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Kobbe C, May A, Grandori C, Bohr VA. Werner syndrome cells escape hydrogen peroxide-induced cell proliferation arrest. FASEB J. 2004;18:1970–1972. doi: 10.1096/fj.04-1895fje. [DOI] [PubMed] [Google Scholar]
- 18.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J. Biol. Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 19.Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hubscher U. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 20.Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 2003;278:8795–8803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 24.Saydam N, Kanagaraj R, Dietschy T, Garcia PL, Pena-Diaz J, Shevelev I, Stagljar I, Janscak P. Physical and functional interactions between Werner syndrome helicase and mismatch repair initiation factors. Nucleic Acids Res. 2007;35:5706–5716. doi: 10.1093/nar/gkm500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanagaraj R, Huehn D, Mackellar A, Menigatti M, Zheng L, Urban V, Shevelev I, Greenleaf AL, Janscak P. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 2010;38:8131–8140. doi: 10.1093/nar/gkq697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons JL, Dianov GL. Monitoring base excision repair proteins on damaged DNA using human cell extracts. Biochem. Soc. Trans. 2004;32:962–963. doi: 10.1042/BST0320962. [DOI] [PubMed] [Google Scholar]
- 27.Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD, Wilson SH. DNA polymerases beta and lambda mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS One. 2010;5:e12229. doi: 10.1371/journal.pone.0012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JW, Harrigan J, Opresko PL, Bohr VA. Pathways and functions of the Werner syndrome protein. Mech. Ageing Dev. 2005;126:79–86. doi: 10.1016/j.mad.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′–>5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 31.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 32.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in Werner syndrome. Mol. Cell. Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29:3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrigan JA, Wilson DM, III, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc. Natl Acad. Sci. USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, et al. The spectrum of WRN mutations in Werner syndrome patients. Hum. Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proc. Natl Acad. Sci. USA. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, de Souza-Pinto NC, Liu Y. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6:e1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 44.Wimmer U, Ferrari E, Hunziker P, Hubscher U. Control of DNA polymerase lambda stability by phosphorylation and ubiquitination during the cell cycle. EMBO Rep. 2008;9:1027–1033. doi: 10.1038/embor.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.