Abstract
Transsynaptic circuit tracing using genetically modified Rabies virus (RV) is an emerging technology for identifying synaptic connections between neurons. Complementing this methodology, it has become possible to assay the basic molecular and cellular properties of neuronal lineages derived from embryonic stem (ES) cells in vitro, and these properties are under intense investigation towards devising cell replacement therapies. Here, we report the generation of a novel mouse ES cell (mESC) line that harbors the genetic elements to allow RV-mediated transsynaptic circuit tracing in ES cell-derived neurons and their synaptic networks. To facilitate transsynaptic tracing, we have engineered a new reporter allele by introducing cDNA encoding tdTomato, the Rabies-G glycoprotein, and the avian TVA receptor into the ROSA26 locus by gene targeting. We demonstrate high-efficiency differentiation of these novel mESCs into functional neurons, show their capacity to functionally connect with primary neuronal cultures as evidenced by immunohistochemistry and electrophysiological recordings, and show their ability to act as source cells for presynaptic tracing of neuronal networks in vitro and in vivo. Together, our data highlight the potential for using genetically engineered stem cells to investigate fundamental mechanisms of synapse and circuit formation with unambiguous identification of presynaptic inputs onto neuronal populations of interest.
Keywords: monosynaptic, ES cells, retrograde, TVA, Rabies-G, circuit, synapse
Introduction
The mammalian nervous system is built upon an elaborate collection of cells that form precise patterns of synaptic connectivity. Due to the complexity of neuronal tissue, cracking the code of brain wiring has remained a significant challenge. Limitations that have obscured our understanding of circuit formation stem from a lack of precise technology for marking, manipulating, and mapping patterns of connectivity between and among targeted neuronal subsets. To better understand how the mammalian brain forms and functions, we need to devise new approaches to elucidate patterns of synaptic connectivity. Genetically engineered ES cells harboring components that allow for direct visualization of connectivity between cells would aid this goal.
Historically, light microscopy has been the standard approach to dissect complex neuroanatomy. With advances in recombinant technologies, stem cell biology, in vitro culturing methods, fluorescent imaging, and electrophysiological analyses, it is now becoming feasible to investigate the molecular mechanisms that underlie synaptogenesis, circuit formation, neuronal plasticity, and cell survival with impressive precision1–3. Additionally, manipulating the genome in ES cells now provides us with the ability to address fundamental aspects of gene or neuron function both in vitro and in vivo. Advances in stem cell technology raise the notion that transplantation of stem cell lineages into the intact brain could perhaps provide therapeutic avenues for treatment of some neurological disorders and/or neurodegenerative disease4–9. Several groups have reported the successful transplantation of ES cells into the rodent nervous system10–12. However, implantation of undifferentiated stem cells has led to teratoma formation11–15. Consequently, significant effort has been placed on establishing in vitro differentiation methods of stem cells14,16–27. It is now possible to obtain high numbers of cultured ES cell-derived neurons in vitro following routine neuronal differentiation and isolation protocols. Following transplantation of neurons obtained from ES cells, teratomas form with significantly lower incidence compared to undifferentiated ES cells11,28,29. However, it remains unknown whether transplanted neurons form appropriate synaptic connections with established circuits in vivo. Not only is ectopic circuit formation a major concern, but knowledge regarding the propensity for transplanted neurons to form proper connectivity in the intact brain is also lacking.
In an effort to elucidate patterns of synaptic connectivity between neurons, various transsynaptic viral tracing methods have been developed30–34. The recently described method of monosynaptic circuit tracing using genetically engineered RV provides a means to genetically target discrete neuronal subsets for transsynaptic circuit labeling1,31,33–35. RV is a negative-strand RNA virus that shows neurotropism and is propagated transsynaptically in a retrograde manner33–36. RV preferentially spreads to neighboring cells via chemical synapses, thus RV infection occurs primarily between functionally connected neurons32,33. Once infected with genetically engineered RV, neurons can survive for more than 10 days before any cytopathic changes are visualized in vivo or in vitro 33. The RV glycoprotein G mediates binding of wildtype (wt) virus to neurons and is responsible for the transsynaptic spread. Deleting G from the RV genome and replacing it with an EGFP reporter (ΔG-EGFP) renders the mutant RV replication incompetent35,36. Pseudotyping the ΔG-EGFP RV with the foreign coat protein EnvA makes it infectious only to cells that express the TVA receptor34–37. If cells targeted for pseudotyped ΔG-EGFP RV infection harbor a cDNA encoding wt G, the virus can utilize available G protein to package “live” particles capable of retrograde transsynaptic transfer. Once transferred to presynaptic cells that lack G, the ΔG-EGFP RV can no longer spread. To differentiate between the source cell and presynaptic inputs, a red fluorescent protein such as tdTomato can be included. Consequently, the source cell would be labeled both with tdTomato and GFP, while monosynaptic inputs will remain GFP positive only. This technology allows for unambiguous identification of presynaptic inputs onto neuronal populations of interest.
Here we report the generation of a novel mESC line expressing the genetic components necessary to perform monosynaptic tracing. We further show high-efficiency differentiation of these ES cells into functional neurons, demonstrate their capacity to form functional synaptic connections with primary neurons and brain slices in culture, and reveal their ability to act as source cells for presynaptic tracing of neuronal networks in vitro and in vivo. Finally, we provide evidence that genetically engineered stem cells can be used to investigate fundamental mechanisms of synapse and circuit formation both in vitro and in vivo, which may ultimately lead to valuable insight towards designing new methods for cell replacement therapy.
Materials and Methods
Expression Plasmid Construction
To generate the monosynaptic tracing allele, cDNA encoding Rabies G was excised from pHCMV-RabiesG38 and cloned into a custom expression construct harboring an Ef1α promoter, tdTomato, a 3′ WPRE element, and an hGH polyA sequence. A P2A peptide cleavage sequence39 was introduced between tdTomato and Rabies G by annealed oligos. For positive selection of ES cell clones, bi-cistronic expression of a Neomycin resistance gene (pIRES-Neo3, Clontech, Mountain View, CA) was inserted downstream of Rabies G. For tri-cistronic expression of the TVA receptor, the TVA cDNA from pCMMP-TVA80040 was PCR amplified and cloned downstream of an IRES2 element (Clontech, Mountain View, CA) for insertion 3′ to NeoR, generating pEf1α-tdTomato-Rabies G-IRES-Neo-IRES-TVA (pTomRITVA).
Generation of ROSA26-tomRITVA mESCs
pTomRITVA was shuttled into the pROSA-acceptor targeting plasmid using PacI and AscI41 to generate the ROSA26-tomRITVA targeting vector. This targeting construct was linearized and electroporated into 129/SvJ ES cells. Following G418 selection, clones were picked, propagated, DNA isolated, and PCR genotyping was performed across the targeting vector junction.
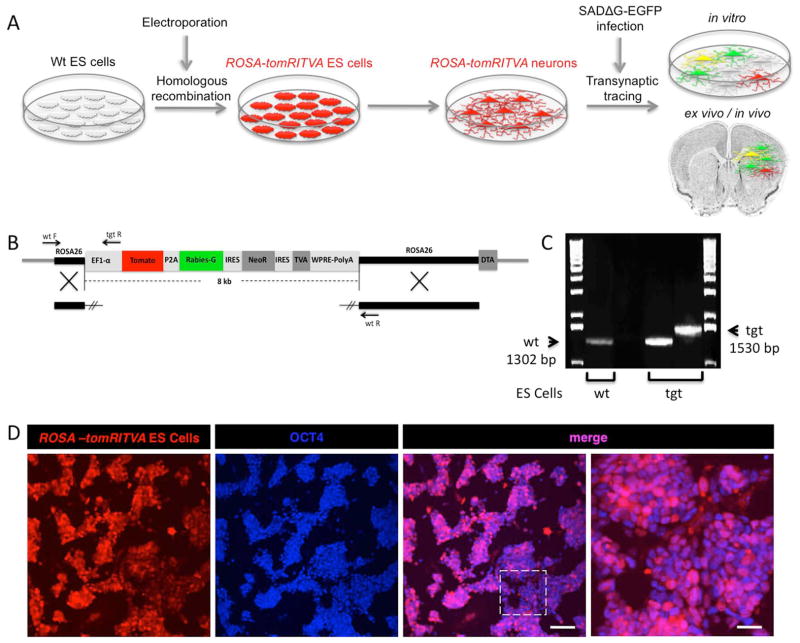
Genotyping was performed using Herculase II Fusion DNA polymerase (Agilent Technologies, Santa Clara, CA) in 50 μl reactions. Primer sequences were as follows: wt/tgt forward primer -GCCTAAAGAAGAGGCTGTGC (112 bp upstream of 5′ ROSA homologous arm), wt reverse primer -GGAAGTCTTGTCCCTCCAAT, tgt reverse primer -ACCTGTGTTCTGGCGG (318 bp downstream of the EF1α promoter start site). Wt transcript size: 1302 bp; tgt size: 1530 bp. Two positively screened clones and the parental cell line were karyotyped via chromosomal metaphase spread G-banding to identify potential aneuploidies.
mESC Culture
Mouse ES cells were grown on gelatinized 10 cm tissue culture plates with Leukemia-Inhibitory Factor (LIF)-containing GMEM medium (Lonza Group Ltd, Switzerland) supplemented with 10% FBS, 1% 10 mM nonessential amino acids, 1% 100 mM sodium pyruvate, 1% 100X Penicillin/Streptomycin, 560 μl β-Mercaptoethanol (55 mM in PBS, Sigma), 1% 200 mM L-glutamine) in the absence of feeder layers.
Neuronal Differentiation of Targeted mESCs
The ES cell differentiation assay was modified from Bibel et al.24. Briefly, ES cells were grown on 10 cm gelatinized tissue culture plates in ES cell medium containing LIF. Cells were trypsinized with 0.05% trypsin/EDTA for 5 min, and 5 ml ES cell medium without LIF was added to inactivate the trypsin. Cells were pelleted and resuspended in 15 ml of ES cell medium without LIF and transferred to 10 cm bacteriological petri dishes (Greiner #633102) for neurosphere culture in suspension. After two days, neurospheres were transferred to 50 ml Falcon tubes, allowed to settle, and the supernatant was replaced with 15 ml ES cell medium without LIF. Next, the cell suspension was added to a new bacteriological petri dish. After 4 days, culture medium was changed as above with the addition of retinoic acid (Sigma, 1:1000 from 5 mM stock solution in DMSO). Cell aggregates were allowed to grow for 6 days, and the medium was changed once more to ES cell medium containing retinoic acid, but lacking LIF. Two days later, cell aggregates were washed twice with PBS and trypsinized for 5 min in 0.05% trypsin/EDTA. 10 ml of ES cell medium without LIF was added to inactivate the trypsin and the cell suspension was pelleted and resuspended in NB/B27 medium (Neurobasal medium (GIBCO), 2% B27, 5% fetal bovine serum, 1% 200 mM L-glutamine, 1 μg/ml Gentamycin). ~ 5×105 cells were plated on Poly-D-Lysine (PDL) (Sigma) coated glass coverslips in 12 well plates in the presence of 2 ml NB/B27 medium. Within 5 to 7 days, elaborate neuronal morphology was observed.
Confocal Imaging and Immunohistochemistry
ES cell-derived neurons grown on glass coverslips were rinsed with PBS, fixed in 4% PFA/PBS for 15 min at RT, rinsed in PBS, then incubated in blocking solution (10% normal goat serum, 0.3% Triton X-100 in PBS, pH 7.35) at 4°C for 2 hours. The following primary antibodies were used: anti-NeuN (1:700, Millipore MAB377), anti-β-III-Tubulin (1:500, Chemicon MAB1637), anti-TUJ1 (1:500, Covance MMS-435P), anti-Synapsin (1:50, Hybridoma Bank), anti-Gephyrin (1:2000, Synaptic Systems 147011), anti-Bassoon (1:2000, Synaptic Systems 141004). Antibodies were diluted in blocking solution and applied overnight at 4° C. The next day, coverslips were washed 4 × 10 min in PBS with 0.1% Triton X-100. Secondary Alexa-488 anti-mouse IgG, and Alexa-488 anti-rabbit (Invitrogen, Carlsbad, CA) were then added to a final dilution of 1:300 and incubated for 1 hour at room temperature. Coverslips were washed 4 × 15 min each and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Microscopy was performed using a Leica TCS SPE confocal microscope under a 10x or 20x objective. Neuronal differentiation efficiency was quantified per culture per 400 by 400 μm field of view and is reported as percentage ± SEM. Statistical significance of neuronal differentiation efficiency between parental and ROSA-tomRITVA ES cells was determined by a Student’s t-test and one-way analysis of variance (ANOVA) using SPSS analysis software.
Primary Neuronal and Slice Co-Culture
Primary neurons were obtained from E18 C57Bl/6 wt embryos. Cortical tissue was dissected into cold DM (HBSS (Invitrogen), 10 mM HEPES, 0.3% BSA, 12 mM MgSO4, pH 7.4), centrifuged 2 min at 1100 rpm, supernatant was aspirated, and 8 ml prewarmed digestion solution was added (4.2 mM HaHCO3, 25 mM HEPES, 137 mM NaCl, 5 mM KCl, 7 mM Na2HPO4, pH 7.4, 4 ml 2.5% trypsin, 2 mg DNAse (Sigma)). The solution was incubated 8 min at 37°C, centrifuged as above, supernatant removed, and 3 ml trypsin inhibitor solution (70 mg in 3 ml PBS, Sigma) added and incubated for 2 min at 37°C. The tissue was again centrifuged, supernatant removed, the pellet was washed twice with ice cold DM and resuspended in fresh DM. Cells were counted and seeded a density of 2×105 primary neurons onto PDL coated coverslips containing 0.5% ROSA-tomRITVA expressing neurons in 2 ml NB/B27 medium. Cells were grown for 5 to 7 days before infection with low titer SADΔG-EGFP-RV. SADΔG-EGFP-RV was produced as previously described35. Infected cells were allowed to grow for 2, 4, or 6 days post infection, and imaged for presynaptic jumping of SADΔG-EGFP-RV using confocal microscopy.
Cortical slice cultures were obtained from P4 C57Bl/6 wt pups. Brain tissue was sliced using a Compresstome (Precisionary Instruments, NC) at 300 μm thickness, and collected in modified Gey’s Balanced Salt Solution (GBSS (Sigma), 0.5% D-glucose, 1% 3M KCl). Slices were placed onto 30 mm Millicell-CM culture membranes (Millipore), transferred to 6 well plates filled with 1.2 ml culture medium (25% horse serum (Gibco), 25% HBSS, 0.5% D-glucose in Opti-MEM (Gibco)), and allowed to recover for 1 d. Medium was changed to 1.2 ml NB/B27 before seeding with 1×104 ROSA-tomRITVA ES cell-derived neurons. Slices were infected 3 to 5 days later with SADΔG-EGFP-RV. Images were taken 2, 4, and 6 days post infection.
Electrophysiological Recordings
ES cell-derived neurons and primary neuronal co-cultures were placed in a RT chamber mounted on an Olympus upright microscope (BX50WI) and perfused with buffered (5% CO2, 95% O2) ACSF (in mM: 122 NaCl, 3 KCl, 1.2 NaH2PO4, 26 NaHCO3, 20 glucose, 2 CaCl2, 1 MgCl2, 305–310 mOsm, pH 7.3). Cells and slices were visualized under differential interference contrast (DIC) imaging. Recording pipettes were pulled with tip resistance between 4–7 MΩ. Data were obtained via a Multiclamp 700B amplifier, low-pass Bessel filtered at 4 kHz, and digitized on computer disk (Clampex, Axon Instruments). Whole-cell patch-clamp recordings were obtained from visually targeted GFP+/tdTomato+ cells or GFP+/tdTomato− cells using pipettes filled with internal solution (in mM: 120 K-gluconate, 5 KCl, 2 MgCl2, 0.05 EGTA, 10 HEPES, 2 Mg-ATP, 0.4 Mg-GTP, 10 creatine phosphate, 290–300 mOsm, pH 7.3). For paired cell recordings, 40 pA current was injected into the GFP+/tdTomato− cell (presynaptic) for 1s to initiate action potentials, and postsynaptic recordings (GFP+/tdTomato+ cell) were performed in voltage clamp mode at −80 mV. Blockage of fast synaptic transmission was performed via bath application of 20 μM CNQX, 20 μM APV and 50 μM Bicuculline. Presynaptic neurons were stimulated and recordings were made from the postsynaptic neuron before drug application, 5 min after drug application, and 10 min after wash out. The traces were averaged over 15 sweeps from the same neuron.
ES cell and ES cell-derived neuron transplantation
For postnatal ES cell transplants, P1 C57Bl/6 wt pups were injected into the lateral ventricle with 1×105 cells with 0.01% Fast green (Sigma) using a 30g Hamilton syringe. Thirty days later, animals were anesthetized with a ketamine/dormatore mixture, placed in a stereotaxic injection frame, and 10 × 50 nl of SADΔG-EGFP RV (6 X 103 particles/μl) was injected at a rate of 23 nl/s using a Drummond Nanoject II at 20 s intervals into the core of the olfactory bulb, 800 μm from the dorsal surface. Tissues were harvested for transsynaptic analysis 7 days post injection (dpi). For adult ES cell-derived neuronal transplants, 3–6 week old C57Bl/6 mice were anesthetized as above, placed in a stereotaxic frame, and 1×105 cells were injected into different parts of the brain using a Drummond Nanoject II (coordinates in mm from bregma and dorsal surface: cortex ML 1.12 mm, AP 2.46, DV 1.7, striatum ML 1.45, AP 1.1, DV 3.31, SVZ ML 0.65, AP 1.1, DV 3.69). Thirty days later, SADΔG-EGFP RV was injected into the original injection site as described above.
Results
A new ES cell line for transsynaptic viral tracing in vitro and in vivo
Homologous recombination and ES cell technology affords the routine ability to generate genetically engineered pluripotent cell lines that can be investigated at the cellular level in vitro, in the context of intact tissue, or in living mouse models. Understanding gene, cell, and tissue function in the mammalian nervous system where delineating precise patterns of synaptic connectivity is required to understand how the brain forms and functions benefits from multifaceted experimental manipulations. Towards this goal, we have generated a novel ES cell line that harbors the genetic elements necessary to perform transsynaptic viral circuit tracing using the deletion-mutant Rabies virus strain SADΔG-EGFP RV36. Through gene targeting42,43, we generated ROSA-tomRITVA ES cells, differentiated these cells into neurons, and performed in vitro, ex vivo, and in vivo transsynaptic circuit tracing (Fig. 1A). The construct targeted to the ROSA26 locus included an EF1α promoter for high levels of tdTomato reporter expression, followed by a P2A element fused to the Rabies-G sequence. Neomycin resistance and TVA expression were linked using internal ribosomal entry sites (IRES). We included a Diphtheria toxin element outside vector homology to allow for negative selection (Fig. 1B). After linearizing the targeting vector, the construct was electroporated into wt 129SvJ ES cells by standard methods. Following positive and negative selection in vitro, we first visually identified potential clones using tdTomato expression and infectivity by SADΔG-EGFP RV. Out of 96 fluorescent clones, we picked 20 with the brightest tdTomato reporter and GFP expression following infection with pseudotyped virus (data not shown). We further screened the reporter-positive clones by PCR genotyping to validate correct targeting at the ROSA26 locus (Fig. 1B and C). As a control, we used wt 129SvJ ES cell DNA and demonstrated the absence of the mutant allele. The correctly targeted ES cell clones showed two bands, one at 1302 bp (wt) and the other at 1530bp (tgt) (Fig. 1C). Following identification of positive ES cell clones, visual inspection showed high efficiency tdTomato reporter expression as well as the ES cell marker Oct4 (Fig. 1D). To verify that the newly generated ROSA-tomRITVA ES cells did not show chromosomal abnormalities, G-banding was performed both on the parental cell line and two ROSA-tomRITVA ES cell clones. Cytogenetic evaluation of the parental wild type mouse ES cells and one of the ROSA-tomRITVA ES cell clones revealed a normal male diploid karyotype in all analyzed metaphases (Fig S1). The other ROSA-tomRITVA ES cell clone showed normal male diploid karyotype in 17/20 examined metaphases. We proceeded with the 100% normal ROSA-tomRITVA ES cell clone for all experimentation. Thus, we have successfully generated and identified a new ES cell line harboring the monosynaptic tracing components genetically targeted to the ROSA26 locus.
Figure 1.
Transsynaptic tracing in vitro and in vivo using ROSA-tomRITVA ES cells. (A): Scheme for generating ROSA-tomRITVA ES cells for transsynaptic tracing. (B): ROSA-tomRITVA targeting vector, highlighting primer pairs for genotyping. (C): PCR genotyping of ROSA-tomRITVA ES cells. The wildtype allele (wt) is 1302 bp, whereas the ROSA-tomRITVA knock-in allele (tgt) is 1530 bp. (D): Undifferentiated ROSA-tomRITVA ES cells immunostained for OCT4. Far right panel represents inset shown in panel to the left. Scale bars left to right: 100 μm and 30 μm.
Genetically engineered ES cells show neuronal differentiation in vitro
Transient transfection, overexpression, and knockdown experiments in cultured cell lines have provided significant insight into neurodevelopmental pathways. Direct modification of the genome, however, provides stable and reproducible molecular expression and/or cellular phenotypes. Genetic manipulations of ES cells now bridge these two approaches; ES cells are readily manipulated genetically, and can routinely be differentiated into desired cell and tissue types in vitro.
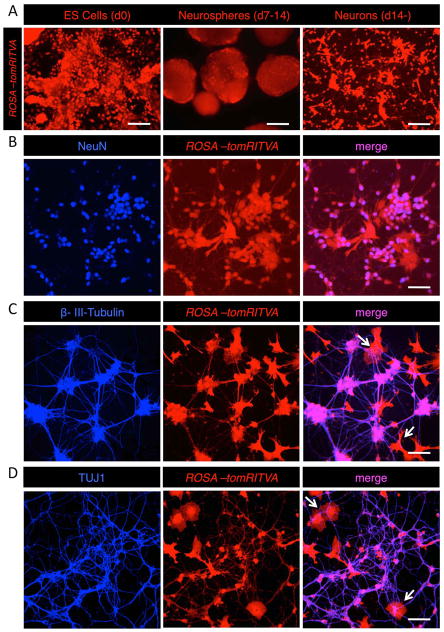
Following generation of ROSA-tomRITVA ES cells, we differentiated them in vitro into neurons (see Materials and Methods). Post in vitro plating, ROSA-tomRITVA ES cell-derived neurons showed neuronal morphology with extensive neurite outgrowth (Fig. 2). To verify neuronal differentiation, we assayed ES cell lineages for neuronal marker expression via immunohistochemical analysis. Fourteen days post plating, ROSA-tomRITVA ES cell-derived neurons expressed molecular markers of neuronal differentiation, including NeuN (Fig. 2B), β-III-Tubulin (Fig. 2C), and TUJ1 (Fig. 2D). In addition to general markers of neuronal differentiation, ES cell-derived neurons also showed colocalization and punctate expression of synaptic proteins (Fig. S2). Notably, we did not observe statistically significant differences in the differentiation efficiency between the parental and genetically targeted ES cell lines using Student’s t-test and one-way analysis of variance (ANOVA). 81% ± 1.6% of differentiated wt ES cells expressed NeuN, whereas NeuN was detected in 82.3% ± 1% in ROSA-tomRITVA ES cell derived neurons (n = 4 coverslips each: p > 0.05 for all groups). Additionally, we also noted the presence of cells with glial morphologies (arrows, Figs. 2C and D). Cells that did not express NeuN were GFAP positive, suggesting differentiation of a minor fraction of the differentiated cells into an astrocyte lineage (21.5% ± 0.5%; n = 6 coverslips). Further, no delay in maturation was observed as evidenced by quantification of β-III-Tubulin and TUJ1 positive cells (β-III-Tubulin: 77.1% ± 1.1% wt vs. 78.2% ± 1.2% ROSA-tomRITVA, n = 5 coverslips each; p > 0.05; TUJ1: 86.2% ± 0.6% wt vs. 78.7% ± 1.5% ROSA-tomRITVA, n = 5 coverslips each; p > 0.05). Worth noting, the longer we allowed the mESC-derived neurons to grow in culture, neuronal marker and synaptic protein expression became further enriched (data not shown), demonstrating that ES cell-derived neurons mature with increased time in culture. Thus, ES cells harboring ROSA-tomRITVA show equal differentiation efficiency and maturation as the parental wt ES cell line. Together, these data show that this new line of ROSA-tomRITVA ES cells can be efficiently differentiated into neurons in vitro by established protocols, and with maturation show robust expression of molecular markers of neuronal differentiation and synapse formation.
Figure 2.
Neuronal differentiation of ROSA-tomRITVA ES cells in vitro. (A): ROSA-tomRITVA ES cells are grown as neurospheres with retinoic acid to induce neuronal differentiation in vitro (scale bars left to right: 60 and 100 μm). (B): Neuronal differentiated ES cells show expression of NeuN (scale bar: 60 μm), (C): β-III-Tubulin (scale bar: 60 μm), and (D): Tuj1 (scale bar: 60 μm). Arrows in C and D point to glial cells.
Genetically engineered ES cell-derived neurons show synaptic coupling and transsynaptic viral spread in vitro
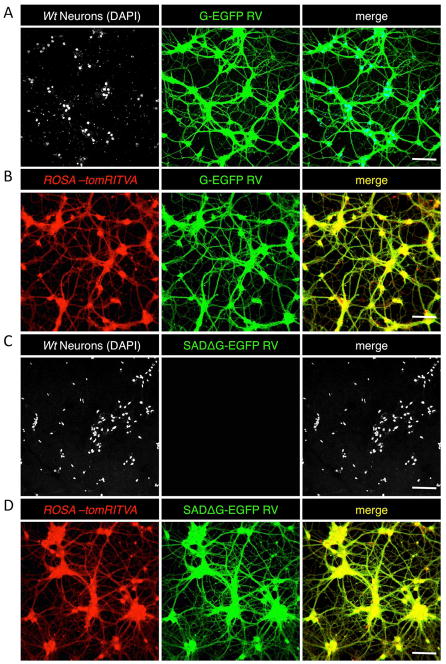
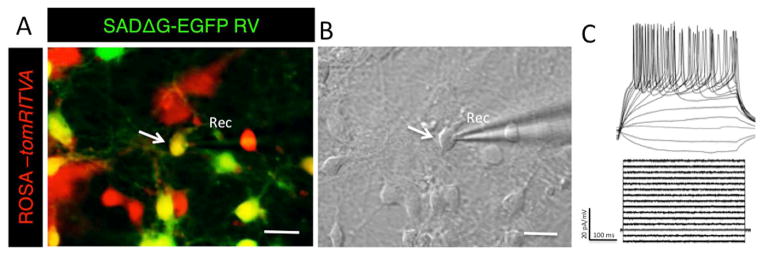
Upon demonstrating that ROSA-tomRITVA ES cells can be efficiently differentiated into neurons in vitro, we next sought to determine if the mESC-derived neurons appropriately expressed the necessary elements required for transsynaptic tracing, wire up in vitro as functional synaptic networks, and show properties of mature neurons. Towards this, we targeted ROSA-tomRITVA ES cell-derived neurons for transsynaptic viral circuit tracing using an EnvA pseudotyped deletion mutant strain of Rabies Virus SADΔG-EGFP RV, which does not infect wild type neurons35,36. We infected both wild type and ROSA-tomRITVA ES cell-derived neuronal cultures that underwent identical differentiation protocols both with a normal G-typed (non-pseudotyped) and pseudotyped SADΔG-EGFP RV 5 to 7 days post plating, which is sufficient time for extensive neurite formation and axonal outgrowth in vitro. Three days post RV infection, nearly all wild type and ROSA-tomRITVA ES cell-derived neurons became infected with the G-typed virus and robustly expressed GFP, demonstrating nonselective neuronal binding and uptake of the normal G-typed virus (Fig. 3A and B). Concurrently, we performed the same experiment using EnvA pseudotyped SADΔG-EGFP RV to determine the propensity for differential infectivity of ROSA-tomRITVA ES cell-derived neurons. Using EnvA pseudotyped SADΔG-EGFP RV, we observed highly selective and targeted infection to ROSA-tomRITVA clones, but not to wild type ES cell-derived neurons (Fig. 3C and D). We next aimed to determine if the mESC-derived neurons showed membrane and ion channel properties of mature neurons, and thus were capable of firing action potentials. The mESC-derived neurons were infected with a low titer SADΔG-EGFP RV (100 μl of 2.4 × 104 infectious particles/ml) for sub-saturating levels of infection, and whole cell patch clamp recordings were performed on tdTomato and GFP double positive neurons (Fig. 4A and B). In current clamp mode, ES cell-derived neurons fired action potentials at high frequency to controlled current injections. Neurons reproducibly fired action potentials upon reaching threshold (−40 mV) following a series of stepwise current injections (Fig. 4C). These data show that neuronally differentiated ROSA-tomRITVA ES cells express the components necessary for transsynaptic tracing, are highly susceptible to EnvA SADΔG-EGFP RV infection, and that reporter expression does not interfere with in vitro neuronal maturation as shown by targeted whole cell recordings.
Figure 3.
ROSA-tomRITVA neurons are selectively infected by SADΔG-EGFP RV. (A) and (B): Both wt neurons (marked by DAPI) and ROSA-tomRITVA neurons show infectivity by G-EGFP RV, (C): WT neurons are not infected by the pseudotyped SADΔG-EGFP RV. (D): ROSA-tomRITVA neurons infected with SADΔG-EGFP RV (scalebars 70 μm).
Figure 4.
ROSA-tomRITVA ES cell-derived neurons show neuronal properties in vitro. (A): Fluorescent merge of recorded ROSA-tomRITVA neuron infected with GFP-expressing Rabies Virus (RV) (scale bar: 20 μm). (B): DIC image of recorded cell, arrow (scale bar: 20 μm) and (C): representative responses to ramped current injections.
Genetically engineered ES cell-derived neurons form functional synapses with primary neurons and slice explants in vitro
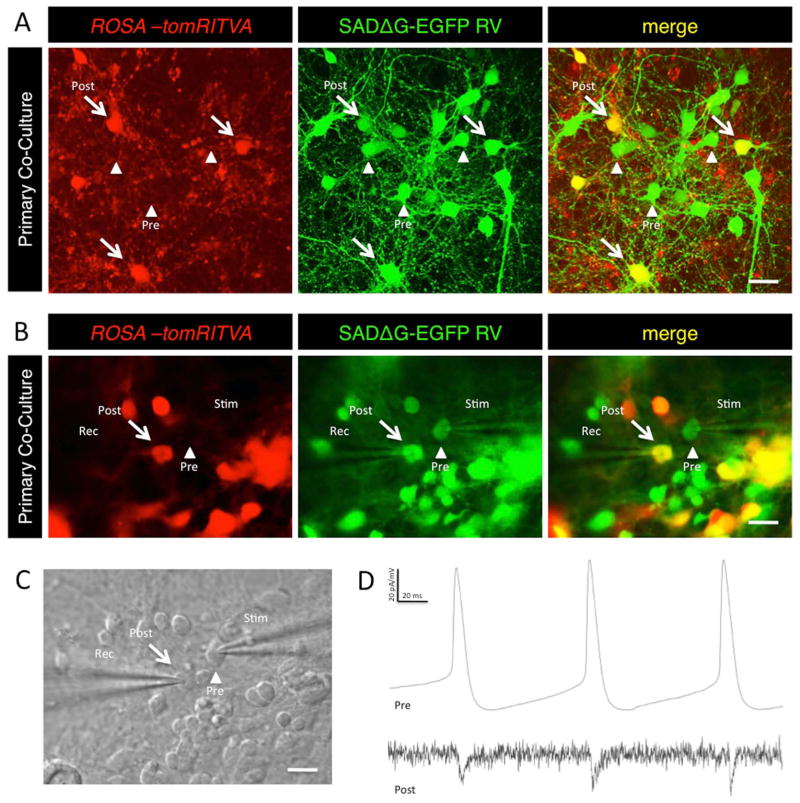
A hallmark of neuronal differentiation is the formation of functional synapses. We next sought to determine if the mESC-derived neurons functionally connect with primary neurons co-cultured in vitro, and if these artificially generated neuronal networks are capable of transsynaptic circuit tracing and synaptic transmission. To test this, we co-cultured ROSA-tomRITVA neurons at low density (0.5% of total cells) with excess wt primary cortical neurons, and waited 7 days before infection with SADΔG-EGFP RV. Three days post-infection, transsynaptic tracing was evident by the presence of few doubly labeled source cells and high numbers of GFP+ presynaptic cells (Fig. 5A). Next, to determine if transsynaptically labeled neurons showed functional synaptic transmission, we performed paired whole cell recordings of infected source cells (tdTomato+ and GFP+ ROSA-tomRITVA neurons) and their presynaptic partners (GFP+ only neurons) 3 days post RV infection (Fig. 5B and 5C). This time point is ideal for our studies since transsynaptic labeling is the primary means for RV transfer, whereas non-specific particle shedding into the neuronal culture and/or RV-mediated labeling from cell death or glial-mediated synaptic remodeling processes in vitro is minimized. Upon establishing whole cell configuration in pre- and postsynaptic pairs, current was injected into the presynaptic (GFP+) neurons to initiate depolarizing events and drive action potentials (Fig. 5B). Postsynaptic responses were simultaneously measured in double positive (tdTomato+/GFP+) source cells. In multiple pairs (n = 7), we found that cells were synaptically coupled (Fig. 5D), and that the GFP+ cell was indeed presynaptic to our source cell (Fig. 5B-4D). To rule out the possibility of other non-synaptic electrical connections or field effects, we established paired whole cell recordings and bath-applied both GABA and glutamate receptor blockers (CNQX, APV, Bicuculline) to interfere with fast synaptic transmission. Drug application blocked electrophysiological responses between RV labeled pairs, consistent with transsynaptic transfer of RV particles and synaptic connectivity between recorded cell pairs. Synaptic responses were recovered following washout (Fig. S3).
Figure 5.
ROSA-tomRITVA neurons synaptically couple with primary neurons in vitro. (A): Primary neuronal co-culture shows transsynaptic spread of SAD-ΔG-EGFP RV. Arrows, postsynaptic neurons; arrowheads, presynaptic neurons (scale bar: 20 μm). (B): Fluorescent images of synaptic pairs targeted for paired whole cell recordings (scale bar: 15 μm). (C): DIC image of (B) (scale bar: 15 μm). (D): Representative trace showing synaptic coupling. Top, changes in membrane potential with presynaptic current injection; bottom, postsynaptic current responses.
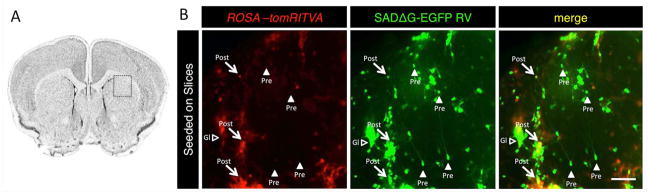
To determine if ROSA-tomRITVA ES cell-derived neurons could form functional connections with neurons in more intact nervous tissue, we next seeded ROSA-tomRITVA neurons onto cortical slice explants that were made from wild type postnatal day 4 (P4) pups. Three to five days after seeding, the co-culture explants were infected with SADΔG-EGFP RV. Three days post infection we observed extensive GFP reporter expression in presynaptic neurons as well as a minor fraction of glial cells (Fig 6A and 6B). Together, our data demonstrate that neurons derived from ROSA-tomRITVA ES cells form functional synaptic connections with neuronal cell types in vitro. These connections can be revealed by transsynaptic reporter expression, and electrophysiological recordings validate synaptic connectivity between RV labeled neuronal pairs.
Figure 6.
ROSA-tomRITVA ES cell-derived neurons form synapses with brain slice explants. (A): Slices seeded with ROSA-tomRITVA neurons and infected with SADΔG-EGFP RV. The square shows the region of the seeded brain slice that was imaged. (B): Transplanted neurons show transsynaptic viral spread in intact brain tissue. Arrows, postsynaptic ROSA-tomRITVA neurons; arrowheads, presynaptic input neurons; hollow arrowheads, labeled glial cells (scale bar: 150 μm).
Genetically engineered ES cells allow transsynaptic circuit tracing in vivo
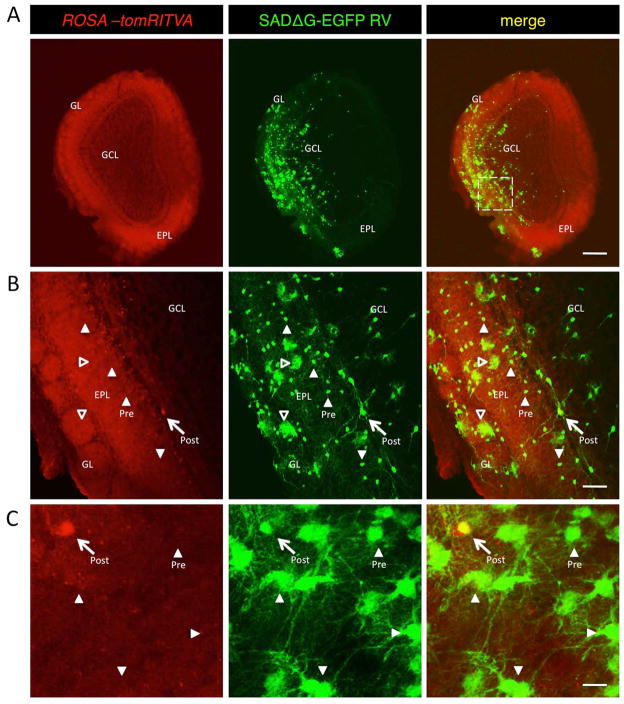
Numerous studies have reported the introduction, differentiation, and survival of ES cells into living rodent nervous tissues11–13,28,44. Although, evidence of circuit integration has been substantiated by the expression of fluorescent reporters44–46, recovery of pathological conditions11,45,48–51, detection by magnetic resonance imaging (MRI) or Positron Emission Tomography (PET) signals45,47,52, and behavior17,49,51,53, direct visualization of synaptic connectivity of transplanted cells in these preparations has been lacking. Having shown that the ROSA-tomRITVA mESC-derived neurons form functional synaptic connections with primary neurons and slice explants in vitro (Figs. 5 and 6), we next wanted to determine if these cells could be used for transsynaptic tracing in vivo. Exploiting the feature of continued neurogenesis in the murine olfactory system54–60, we set out to determine if this cell line would appropriately differentiate, migrate, and integrate into olfactory bulb circuits in vivo. For this, we transplanted ROSA-tomRITVA ES cells (2 × 104 cells) into the lateral ventricles of postnatal C57Bl/6 mice, followed by a 30 day period of maturation, and subsequent introduction of SADΔG-GFP RV into the olfactory bulb. Points of infection and transsynaptic labeling were analyzed 7 days later (Fig. 7A). We observed numerous presynaptic partners as evidenced by extensive SADΔG-GFP RV spread throughout the olfactory bulb. Moreover, we determined that transplanted ES cells differentiated, became infected by SADΔG-GFP RV, and subsequently formed synaptic connections with local olfactory bulb cell types previously described to form connections with newborn neurons31 (Fig. 7B). Interestingly however, we also observed some GFP labeling in cells with glial morphology. Glial cell labeling via RV transfer has been documented31, but it remains to be determined if this occurs through glial cell-mediated synaptic remodeling, or actual synaptic connectivity between neurons and glial subtypes. Together these data show that ROSA-tomRITVA ES cells are capable of neuronal differentiation and synapse formation following in vivo transplantation.
Figure 7.
ROSA-tomRITVA ES cells show transsynaptic circuit tracing in vivo. (A): ES cell-derived neurons migrate to the olfactory bulb and show transsynaptic tracing of host circuits. GL, glomerular layer; GCL, granule cell layer; EPL, external plexiform layer. Scale bar: 500 μm. Square is the magnified region shown in B. (B): High magnification of highlighted region shown in (A) (scale bar: 100 μm). (C): ROSA-tomRITVA neurons transplanted into prefrontal cortex of adult mice, followed by infection with SADΔG-EGFP RV. Arrows in B and C point to postsynaptic source cell, arrowheads point to presynaptic host neurons, hollow arrowheads point to labeled glial cells (scale bar: 20 μm).
In agreement with previous reports11–14,28, one major limitation we observed following transplantation of undifferentiated ES cells was teratoma formation. Injection of undifferentiated ES cells resulted in a 20% (4 of 20 transplanted animals) incidence of teratomas. To decrease the incidence of teratoma formation, and to increase the chances of cell integration into other parts of the adult brain, we next performed transplantations of in vitro differentiated ROSA-tomRITVA neurons into the brain parenchyma. After differentiation, we transplanted ROSA-tomRITVA neurons at low density (2 × 104 cells) into adult mouse somatosensory cortex. Following transplantation, we waited 30 days and injected SADΔG-EGFP RV into the transplantation sites. Seven days later, we observed successful integration of ROSA-tomRITVA expressing cells into the pre-existing host brain circuits. 17 of 20 transplanted animals contained ROSA-tomRITVA cells one month later. Transsynaptic tracing was evident in cortical regions where numerous cells were identified as being presynaptic to our source cells (Fig. 7C). Interestingly, when implanted into the SVZ, differentiated ROSA-tomRITVA neurons incorporated into the walls of the lateral ventricle and SVZ but did not give rise to known adult-born SVZ-derived neuronal lineages. This may be due to the postmitotic state of ES cell-derived neurons, and therefore integration into the olfactory bulb was rarely observed following SVZ transplantation. Taken together, we were able to show that ROSA-tomRITVA ES cell-derived neurons functionally integrate into pre-existing host circuits in vivo and that these circuits are susceptible to transsynaptic viral circuit tracing using genetically-modified RV.
Discussion
Delineating precise patterns of synaptic connectivity in the mammalian nervous system is essential if we are to ultimately gain mechanistic insight into normal neural development, neurological disease, or potential avenues for brain repair. Numerous experimental approaches ranging from imaging and electrophysiology, to genetic engineering and stem cell biology, have been broadly implemented towards our working knowledge in these areas. Unfortunately, progress in this field has been somewhat tempered due to the innate complexity of brain tissue. Thus, new experimental approaches and methodologies for investigating patterns of synaptic connectivity are both useful and necessary.
We have generated a novel ES cell line with all the necessary elements for transsynaptic viral tracing in nervous tissue using genetically modified RV. Through immunohistochemistry, transsynaptic fluorescent reporter analysis, and electrophysiological recordings, we have demonstrated the efficient differentiation of these cells into functional neurons in vitro, and have shown that that they can wire up effectively with host neuronal circuits in vitro and in vivo. Transsynaptic tracing technology using genetically-engineered RV is a powerful tool given its general utility to genetically identify neurons that are connected via chemical synapses32–37. In contrast to most lipophilic dye-based synaptic tracers, lectin-based conjugates, and other viral technology, RV spread occurs exclusively in a retrograde manner, is not thought to cross gap junctions, and requires the presence of intact presynaptic structures for neuronal infection and transsynaptic transfer31–36. However, it is important to consider other mechanisms of viral labeling and spread that can occur in vitro. Unlike the specificity of synapse formation within intact nervous tissue, in vitro preparations facilitate numerous “unnatural” cellular interactions. For example, differentiating cells and neurons in culture can form transient synaptic connections through time. This poses the potential for non-specific viral transfer and thus labeling of cell types that do not show long-lived functional synaptic connectivity. Moreover, in vitro culture environments do not foster the longevity of cells in vivo, and thus cultured neuronal cells can die off earlier than in vivo. In this scenario, infected neurons might shed RV into the culture medium upon death, and these viral particles have the capacity to be taken up by nearby neurons without synaptic connectivity. Importantly, these non-specific labeling phenomena can be minimized when in vitro RV infection times are kept as short as possible. For this reason, we performed all tracing experiments within 3–7 days after infection. During this time, infected cells remain alive, healthy, and function as expected for in vitro growth33.
Neuronal differentiation of modified ES cells allows the unique opportunity to investigate basic molecular and cell biological mechanisms that underlie synapse and circuit formation in vitro, but also affords the ability to efficiently cross over to in vivo experimentation via cell transplantation or generation of mutant mice. A beneficial facet of our approach that complements preexisting tracing methods is that circuit integration can be studied at any stage of neuronal differentiation and in any part of the nervous system. Having a genetically targeted ES cell line that harbors the elements for transsynaptic viral tracing allows for differentiation of these cells into virtually any cell type, and facilitates the identification of the various types of presynaptic inputs in diverse regions of the nervous system, both healthy and diseased. Experimentation becomes feasible in which ROSA-tomRITVA ES cells are differentiated into specific neuronal subtypes and subsequently combined with either neurons or tissues harboring known genetic lesions to address cell non-autonomous mechanisms of synapse formation, or alternatively, introduce compound mutations or shRNA expression into ROSA-tomRITVA cells to address cell autonomous gene function. These approaches could also be extended to in vivo applications, where transplanted ES cells could interact with intact brain tissue. Alternatively, the generation of novel mouse lines from these ES cells could be powerful in delineating patterns of connectivity in the context of normal development or disease. One application that could prove to be extremely useful would be to generate mosaic mice through morula aggregation, in which select subsets of neurons could be targeted for transsynaptic tracing in a semi-stochastic manner. This could also be combined with additional genetic alterations to tease out mechanisms of synapse wiring.
Of course we must also consider the limitations. ES cell technology is in its infancy. Although we have learned a great deal about driving neural differentiation in vitro, we still have much to learn about the generation and maintenance of certain neuronal subtypes. It will be imperative for future work to more precisely characterize and describe methodology to enrich and isolate neuronal subtypes of given neurotransmitter or receptor expression properties. Additionally, current applications for ES-derived neuronal transplants remain prominent in rodents, and in particular inbred strains of mice. Teratoma formation and immune responses have stifled applications in other mammalian systems. Induced pluripotent stem (iPS) cells offer promise in this area. It is conceivable that similar genetic expression systems for transsynaptic analysis could be introduced into tailored iPS cell lines, and that these could subsequently be used to investigate synaptic mechanisms in other tissues or model systems.
Considering the widespread use of ES cells to generate mouse models of brain development and dysfunction, the broad array of available alleles expressed in the nervous system, and advancements in iPS cell technology, in vitro experimentation that directly transfers from the culture dish to in vivo application is thus becoming much more feasible. Permutations of the experimentation we performed here using the described ROSA-tomRITVA ES cells hold promise to provide future insight into synaptic wiring mechanisms at work in the mammalian brain.
Conclusions
With the ultimate goal of elucidating the mechanisms that underlie synaptogenesis, circuit formation, circuit integration and neuronal survival, we have developed a novel ES cell line harboring all the necessary elements for transsynaptic circuit tracing using genetically modified RV. We have shown that these cells efficiently undergo differentiation into functional neurons and can be used for transsynaptic tracing both in vitro and in vivo. With this novel technology, it will be feasible to identify precise presynaptic inputs in various healthy and diseased brain regions. This knowledge will allow us to better understand why certain neurons form correct synapses while others fail to properly integrate. In the long term, such knowledge could potentially lead to novel therapeutic methods in neurodegenerative diseases.
Supplementary Material
Supplementary Figure S1. Wt and ROSA-tomRITVA ES cells show normal chromosomal consistency. (A) and (B): Karyotype of wt 129SvJ parental and ROSA-tomRITVA ES cells.
Supplementary Figure S2. ROSA-tomRITVA ES cell-derived neurons show expression of pre- and postsynaptic protein markers. (A): Neurons express Synapsin, (B): Gephyrin, and (C): Bassoon. Scale bars: 60 μm.
Supplementary Figure S3. ROSA-tomRITVA ES cell-derived neurons form synapses with primary cortical neurons. (A) DIC image of synaptic pairs targeted for paired whole cell recordings (scale bar: 10 μm). Presynaptic cell is ROSA-tomRITVA ES cell-derived SADΔG-EGFP infected neuron. Postsynaptic is primary cortical neuron (B): Representative traces of synaptic coupling. Top, presynaptic changes in membrane potential with current injection; bottom, postsynaptic current responses. Responses are blocked via bath application of CNQX, APV, and Bicuculline. Washout rescues synaptic responses (traces are averaged over 15 sweeps of the same neuron).
Acknowledgments
This work was supported by the McNair Medical Institute and NINDS grants R00NS064171 and R01NS078294 to B.R.A. We would like to thank Michael Ehlers and Irina Lebedeva for input and assistance with previous versions of this experimentation, and Mirjana Maletic-Savatic, Ben Deneen, and Fan Wang for critical review of this manuscript.
Footnotes
Author Contributions:
Isabela Garcia: Experimental design, data collection, and manuscript writing.
Longwen Huang: Data collection and data analysis.
Kevin Ung: Data collection.
Benjamin Arenkiel: Experimental conception and design, manuscript writing, and financial support.
References
- 1.Arenkiel BR, Ehlers MD. Molecular genetics and imaging technologies for circuit-based neuroanatomy. Nature. 2009;461:900–907. doi: 10.1038/nature08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Connor DH, Huber D, Svoboda K. Reverse engineering the mouse brain. Nature. 2009;461:923–929. doi: 10.1038/nature08539. [DOI] [PubMed] [Google Scholar]
- 3.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez Dolado M, Broccoli V. GABAergic neuronal precursor grafting: implications in brain regeneration and plasticity. Neural Plats. 2011;2011:384216. doi: 10.1155/2011/384216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson SC, Barker RA. Cell-based therapies for Parkinson’s disease. Expert Rev Neurother. 2011;11:831–844. doi: 10.1586/ern.11.33. [DOI] [PubMed] [Google Scholar]
- 6.Glavaski-Joksimovic A, Virag T, Chang QA, et al. Reversal of dopaminergic degeneration in a parkinsonian rat following micrografting of human bone marrow-derived neural progenitors. Cell Transplant. 2009;18:801–814. doi: 10.3727/096368909X470801. [DOI] [PubMed] [Google Scholar]
- 7.Hattiangady B, Shetty AK. Neural stem cell grafting in an animal model of chronic temporal lobe epilepsy. Curr Protoc Stem Cell Biol. 2011;Chapter 2(Unit2D):7. doi: 10.1002/9780470151808.sc02d07s18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hattiangady B, Shuai B, Cai J, et al. Increased dentate neurogenesis after grafting of glial restricted progenitors or neural stem cells in the aging hippocampus. Stem Cells. 2007;25:2104–2117. doi: 10.1634/stemcells.2006-0726. [DOI] [PubMed] [Google Scholar]
- 9.Nicoleau C, Viegas P, Peschanski M, et al. Human Pluripotent Stem Cell Therapy for Huntington’s Disease: Technical, Immunological, and Safety Challenges. Neurotherapeutics. 2011 doi: 10.1007/s13311-011-0079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deacon T, Dinsmore J, Costantini LC, et al. Blastula-stage stem cells can differentiate into dopaminergic and serotonergic neurons after transplantation. Exp Neurol. 1998;149:28–41. doi: 10.1006/exnr.1997.6674. [DOI] [PubMed] [Google Scholar]
- 11.Bjorklund LM, Sanchez-Pernaute R, Chung S, et al. Embryonic stem cells develop into functional dopaminergic neurons after transplantation in a Parkinson rat model. Proc Natl Acad Sci U S A. 2002;99:2344–2349. doi: 10.1073/pnas.022438099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdo F, Buhrle C, Blunk J, et al. Host-dependent tumorigenesis of embryonic stem cell transplantation in experimental stroke. J Cereb Blood Flow Metab. 2003;23:780–785. doi: 10.1097/01.WCB.0000071886.63724.FB. [DOI] [PubMed] [Google Scholar]
- 13.Seminatore C, Polentes J, Ellman D, et al. The postischemic environment differentially impacts teratoma or tumor formation after transplantation of human embryonic stem cell-derived neural progenitors. Stroke. 2010;41:153–159. doi: 10.1161/STROKEAHA.109.563015. [DOI] [PubMed] [Google Scholar]
- 14.Barberi T, Klivenyi P, Calingasan NY, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 15.Erdo F, Trapp T, Buhrle C, et al. Embryonic stem cell therapy in experimental stroke: host-dependent malignant transformation. Orv Hetil. 2004;145:1307–1313. [PubMed] [Google Scholar]
- 16.Caiazzo M, Dell’Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 17.Chung S, Moon JI, Leung A, et al. ES cell-derived renewable and functional midbrain dopaminergic progenitors. Proc Natl Acad Sci U S A. 2011;108:9703–9708. doi: 10.1073/pnas.1016443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleidi M, Cooper O, Hargus G, et al. Oct4-induced reprogramming is required for adult brain neural stem cell differentiation into midbrain dopaminergic neurons. PLoS One. 2011;6:e19926. doi: 10.1371/journal.pone.0019926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emdad L, D’Souza SL, Kothari HP, et al. Efficient Differentiation of Human Embryonic and Induced Pluripotent Stem Cells into Functional Astrocytes. Stem Cells Dev. 2011 doi: 10.1089/scd.2010.0560. [DOI] [PubMed]
- 20.Jing Y, Machon O, Hampl A, et al. In vitro differentiation of mouse embryonic stem cells into neurons of the dorsal forebrain. Cell Mol Neurobiol. 2011;31:715–727. doi: 10.1007/s10571-011-9669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juliandi B, Abematsu M, Sanosaka T, et al. Induction of superficial cortical layer neurons from mouse embryonic stem cells by valproic acid. Neurosci Res. 2011 doi: 10.1016/j.neures.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Maroof AM, Brown K, Shi SH, et al. Prospective isolation of cortical interneuron precursors from mouse embryonic stem cells. J Neurosci. 2010;30:4667–4675. doi: 10.1523/JNEUROSCI.4255-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jing XT, Wu HT, Wu Y, et al. DIXDC1 promotes retinoic acid-induced neuronal differentiation and inhibits gliogenesis in P19 cells. Cell Mol Neurobiol. 2009;29:55–67. doi: 10.1007/s10571-008-9295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bibel M, Richter J, Lacroix E, et al. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 25.Lee G, Kim H, Elkabetz Y, et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- 26.Hubner K, Fuhrmann G, Christenson LK, et al. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- 27.Reubinoff BE, Itsykson P, Turetsky T, et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 28.Bieberich E, Silva J, Wang G, et al. Selective apoptosis of pluripotent mouse and human stem cells by novel ceramide analogues prevents teratoma formation and enriches for neural precursors in ES cell-derived neural transplants. J Cell Biol. 2004;167:723–734. doi: 10.1083/jcb.200405144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjorklund LM, Isacson O. Regulation of dopamine cell type and transmitter function in fetal and stem cell transplantation for Parkinson’s disease. Prog Brain Res. 2002;138:411–420. doi: 10.1016/S0079-6123(02)38090-7. [DOI] [PubMed] [Google Scholar]
- 30.Arenkiel BR. Genetic approaches to reveal the connectivity of adult-born neurons. Front Neurosci. 2011;5:48. doi: 10.3389/fnins.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arenkiel BR, Hasegawa H, Yi JJ, et al. Activity-Induced Remodeling of Olfactory Bulb Microcircuits Revealed by Monosynaptic Tracing. PLoS One. 2011 doi: 10.1371/journal.pone.0029423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ugolini G. Rabies virus as a transneuronal tracer of neuronal connections. Adv Virus Res. 2011;79:165–202. doi: 10.1016/B978-0-12-387040-7.00010-X. [DOI] [PubMed] [Google Scholar]
- 33.Ugolini G. Advances in viral transneuronal tracing. J Neurosci Meth. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickersham IR, Finke S, Conzelmann KK, et al. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickersham IR, Lyon DC, Barnard RJ, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnard RJ, Elleder D, Young JA. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology. 2006;344:25–29. doi: 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Sena-Esteves M, Tebbets JC, Steffens S, et al. Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Tang W, Ehrlich I, Wolff SB, et al. Faithful expression of multiple proteins via 2A-peptide self-processing: a versatile and reliable method for manipulating brain circuits. J Neurosci. 2009;29:8621–8629. doi: 10.1523/JNEUROSCI.0359-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayan S, Barnard RJ, Young JA. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J Virol. 2003;77:1977–1983. doi: 10.1128/JVI.77.3.1977-1983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 43.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 44.Steinbeck JA, Koch P, Derouiche A, et al. Human embryonic stem cell-derived neurons establish region-specific, long-range projections in the adult brain. Cell Mol Life Sci. 2011 doi: 10.1007/s00018-011-0759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang YL, Chen SJ, Kao CL, et al. Docosahexaenoic Acid Promotes Dopaminergic Differentiation in Induced Pluripotent Stem Cells and Inhibits Teratoma Formation in Rats with Parkinson-like Pathology. Cell Transplant. 2011 doi: 10.3727/096368911X580572. [DOI] [PubMed] [Google Scholar]
- 46.Yang JR, Liao CH, Pang CY, et al. Directed differentiation into neural lineages and therapeutic potential of porcine embryonic stem cells in rat Parkinson’s disease model. Cell Reprogram. 2010;12:447–461. doi: 10.1089/cell.2009.0078. [DOI] [PubMed] [Google Scholar]
- 47.Daadi MM, Li Z, Arac A, et al. Molecular and magnetic resonance imaging of human embryonic stem cell-derived neural stem cell grafts in ischemic rat brain. Mol Ther. 2009;17:1282–1291. doi: 10.1038/mt.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui YF, Xu JC, Hargus G, et al. Embryonic stem cell-derived L1 overexpressing neural aggregates enhance recovery after spinal cord injury in mice. PLoS One. 2011;6:e17126. doi: 10.1371/journal.pone.0017126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu JM, Zhao YY, Chen SD, et al. Functional recovery after transplantation of neural stem cells modified by brain-derived neurotrophic factor in rats with cerebral ischaemia. J Int Med Res. 2011;39:488–498. doi: 10.1177/147323001103900216. [DOI] [PubMed] [Google Scholar]
- 50.Nagai N, Kawao N, Okada K, et al. Systemic transplantation of embryonic stem cells accelerates brain lesion decrease and angiogenesis. Neuroreport. 2010;21:575–579. doi: 10.1097/WNR.0b013e32833a7d2c. [DOI] [PubMed] [Google Scholar]
- 51.Yang D, Zhang ZJ, Oldenburg M, et al. Human embryonic stem cell-derived dopaminergic neurons reverse functional deficit in parkinsonian rats. Stem Cells. 2008;26:55–63. doi: 10.1634/stemcells.2007-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang HL, Sun HP, Wu X, et al. Detection of neural stem cells function in rats with traumatic brain injury by manganese-enhanced magnetic resonance imaging. Chin Med J (Engl) 2011;124:1848–1853. [PubMed] [Google Scholar]
- 53.Redmond DE, Jr, Bjugstad KB, Teng YD, et al. Behavioral improvement in a primate Parkinson’s model is associated with multiple homeostatic effects of human neural stem cells. Proc Natl Acad Sci U S A. 2007;104:12175–12180. doi: 10.1073/pnas.0704091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitman MC, Greer CA. Adult neurogenesis and the olfactory system. Prog Neurobiol. 2009;89:162–175. doi: 10.1016/j.pneurobio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizrahi A. Dendritic development and plasticity of adult-born neurons in the mouse olfactory bulb. Nat Neurosci. 2007;10:444–452. doi: 10.1038/nn1875. [DOI] [PubMed] [Google Scholar]
- 56.Lledo PM, Saghatelyan A. Integrating new neurons into the adult olfactory bulb: joining the network, life-death decisions, and the effects of sensory experience. Trends Neurosci. 2005;28:248–254. doi: 10.1016/j.tins.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- 58.Carleton A, Petreanu LT, Lansford R, et al. Becoming a new neuron in the adult olfactory bulb. Nat Neurosci. 2003;6:507–518. doi: 10.1038/nn1048. [DOI] [PubMed] [Google Scholar]
- 59.Alvarez-Buylla A, Garcia-Verdugo JM. Neurogenesis in adult subventricular zone. J Neurosci. 2002;22:629–634. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alvarez-Buylla A, Temple S. Stem cells in the developing and adult nervous system. J Neurobiol. 1998;36:105–110. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Wt and ROSA-tomRITVA ES cells show normal chromosomal consistency. (A) and (B): Karyotype of wt 129SvJ parental and ROSA-tomRITVA ES cells.
Supplementary Figure S2. ROSA-tomRITVA ES cell-derived neurons show expression of pre- and postsynaptic protein markers. (A): Neurons express Synapsin, (B): Gephyrin, and (C): Bassoon. Scale bars: 60 μm.
Supplementary Figure S3. ROSA-tomRITVA ES cell-derived neurons form synapses with primary cortical neurons. (A) DIC image of synaptic pairs targeted for paired whole cell recordings (scale bar: 10 μm). Presynaptic cell is ROSA-tomRITVA ES cell-derived SADΔG-EGFP infected neuron. Postsynaptic is primary cortical neuron (B): Representative traces of synaptic coupling. Top, presynaptic changes in membrane potential with current injection; bottom, postsynaptic current responses. Responses are blocked via bath application of CNQX, APV, and Bicuculline. Washout rescues synaptic responses (traces are averaged over 15 sweeps of the same neuron).