Abstract
Understanding the aggregation mechanism of amyloid fibrils and characterizing their structures are important steps in the investigation of several neurodegenerative disorders associated with the misfolding of proteins. We report a simulation study of coherent two-dimensional chial signals of three NMR structures of Aβ protein fibrils associated with Alzheimer’s Disease: two models for Aβ (8–40) peptide wild-type (WT) and one for the Iowa (D23N) Aβ (15–40) mutant. Both far ultraviolet (FUV) signals (λ = 190–250 nm) which originate from the backbone nπ* and ππ* transitions, and near ultraviolet (NUV) signals (λ ≥ 250 nm) associated with aromatic side-chains (Phe and Tyr) show distinct cross-peak patterns that can serve as novel signatures for the secondary structure.
Introduction
A large number of neurodegenerative diseases is linked to the formation and deposition of mis-folded proteins to form amyloid fibrils. Probing the aggregation kinetics and fibril formation pathways had drawn a considerable experimental and computational effort (1–14). Alzheimer’s Disease (AD) is a progressive neurodegenerative disorder whose pathology is characterized by the accumulation of senile plaques and neurofibrillar tangles in the brain. These plaques are made of fibrillar aggregates of the amyloid β-protein (Aβ). This protein comes from the cleavage of the amyloid precursor protein (APP) and is present in the body predominantly in two alloforms Aβ 40 and Aβ 42 which has two additional amino-acids in the C-terminal (15, 16).
Determination of the structure of amyloid fibrils is a challenging task. X–ray diffraction is a common tool used for the determination of global atomistic structures of proteins in crystals whereas nuclear magnetic resonance (NMR) techniques can yield structures in solution. The information obtained from these conventional structural biology techniques is limited since amyloid fibrils are insoluble and do not crystallize. High-resolution molecular structure of amyloid fibrils is not available but geometrical models of amyloid fibrils based on solid NMR and others experimental data (17–20). Recent CD and NMR studies of Aβ monomers and dimers in solutions were used to characterize global properties such as secondary structures at different solutions concentrations, pH and temperatures (11, 21, 22). Ultraviolet (UV) spectroscopy provides an alternative structural tool. Both the peptide backbone and some side chains are active in this regime. The nπ* and ππ* transitions of the peptide backbone in the far ultraviolet (FUV) region (λ = 190–250 nm) and used for estimating secondary structure content. Circular dichroism (CD) spectroscopy in this region is a useful probe for protein secondary structures such as α-helices and β-strands. In the near ultraviolet (NUV) region (λ ≥ 250 nm), aromatic side chains (Phe, Tyr, and Trp) usually provide more details about local interactions. Absorption and CD spectra of proteins are usually calculated using the Frenkel exciton matrix model which provides an efficient parametrization scheme for their electronic excitations (23–27).
Two-dimensional (2D) optical spectroscopy is a novel tool that can be used for structure refinement. Sequences of femtosecond laser pulses are employed to excite vibrational or electronic states and determine correlations between events which occur within two controlled time intervals. These correlations are displayed in two dimensional maps that reveal the molecular motions and fluctuating structures of a protein. These help to characterize local dynamics and determine intermediate states present in Aβ fibril formation process (28–31). Two-dimensional (2DIR) photon echo experiments combined with molecular dynamics simulations (MD) have been employed to test the quality of the MD methods and probe protein structures and fast folding processes. Previous simulation studies have demonstrated the potential of two-dimensional ultraviolet spectroscopy (2DUV) in the far and near UV regions for characterizing secondary structure in Aβ amyloid. A simulation protocol based on the Exciton Hamiltonian with Electrostatic Fluctuations (EHEF) was developed (32–34). The present computational study employs that protocol and is aimed at demonstrating how the cross-peaks in chiral 2DUV signals may be used to distinguish between three Aβ amyloid fibrils geometrical models suggested recently by the NMR studies of Tycko and co-workers (17–20).
In Materials and Methods section, we explain the EHEF protocol, provide some technical details and present the amyloid fibril models used. In Results and Discussion section, we present the one and two-dimensional results for the near and far UV spectra. Finally, we provide the conclusions.
Materials and Methods
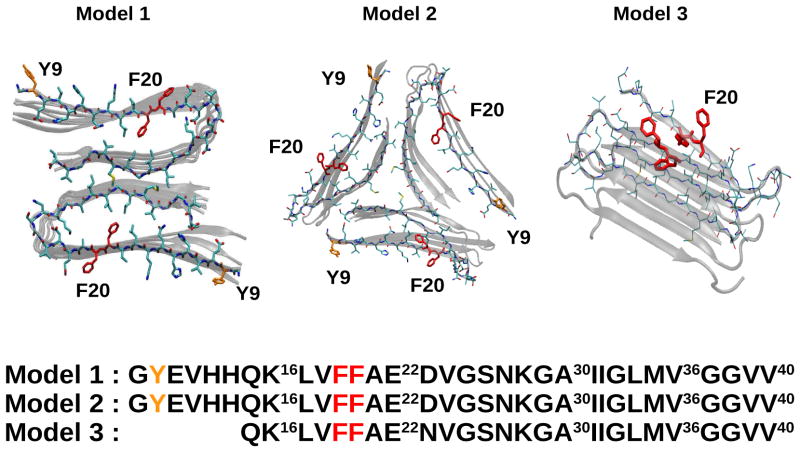
We consider the three Aβ peptides models shown in Figure 1. Models 1 and 2 correspond to the Aβ (8–40) peptide with the sequence GYEVHHQK16LVFFAE22DVGSNKGA30IIGLMV36GGVV40. Model 1, first suggested by Petkova et al. (18), consists of parallel β-sheets growing along the fibril growth axis. Model 2 which has a three-fold symmetry about that axis is based on solid state NMR and electron microscopy performed by Paravastu et al. (19). We used three peptides in each block making a three-fold symmetry complex composed by 9 peptides. Model 3 is the Aβ (15–40) segment of the above protein with the point D23N mutation, known as the Iowa mutant. Its geometry consists of an anti-parallel β-sheet growing along the fibril axis (20).
Figure 1.
Structure and sequence of the three Aβ amyloid fibril models reported by Tycko and coworkers (18–20). The aromatic residues are marked: Phenylalanine (red); and Tyrosine (orange).
Molecular dynamics (MD) trajectories of the above models were obtained by using the NAMD Package 2.7 (35) with the CHARMM27 force field (36). Simulations were carried out at in aqueous solutions (TIP3P water model) with cubic periodic boundary conditions. The particle-mesh Ewald (PME) protocol was used to calculate the long-range electrostatic interactions and a 12 cutoff distance was used for non-bonded interactions. 2.0 ns runs were used to minimize the proteins at 300 K and 1250 MD snapshots were harvested for the 2DUV calculations. The 2DUV spectra were computed using the EHEF protocol (32–34, 37) and the SPECTRON homemade program (38).
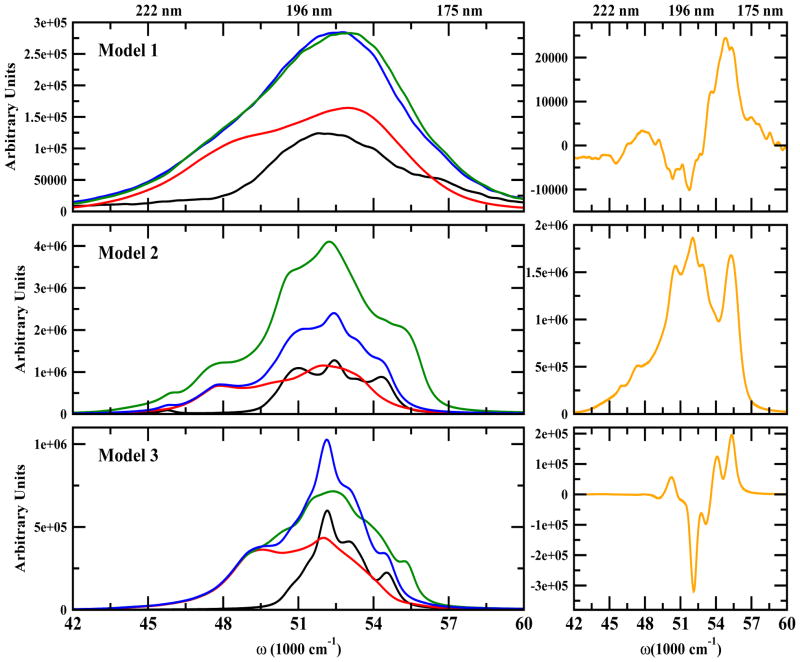
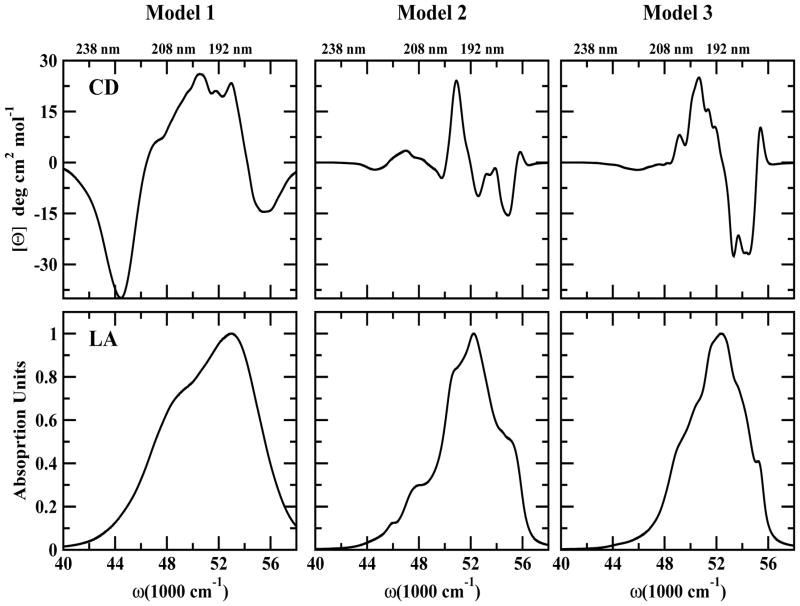
The simulated Linear Absorption (LA) and Circular Dichroism (CD) spectra in the entire UV regime (λ = 180–250 nm) for the three models are displayed in Figure 2 and Figure 3 respectively. We show the spectra of the side chain alone (black), the backbone (red), the additive spectrum (their sum in blue) and the total spectrum (green). The difference of the additive and the total spectrum shown in the right column in Figure 2 and Figure 3 indicates interactions between the backbone and side chains. The strongest differences are around 52000 cm−1. The NUV region (above 250 nm) covers transitions of aromatic side chains (Phe, Tyr and Trp). Each has four valence electronic excitations known in Platt’s notation as: 1La, 1Lb, 1Ba, and 1Bb. 1La and 1Lb which give the strong NUV peaks. The interactions of the aromatic residues may be used to characterize the structure. NUV signals of Model 1 have been studied earlier (34, 37). The FUV spectra (190–250 nm) originate from the peptide backbone nπ* and ππ* transitions and are sensitive to secondary structure as well. Even though aromatic residues are rare, their contributions to the FUV spectra are not much weaker than those of the backbone since they have larger transition dipole moments. The difference spectrum is much stronger for LA than CD. The CD backbone spectra in all models (Figure 3, red plots) show a clear FUV signatures of a typical β-strand conformation. In the coming sections, we shall analyze the NUV and FUV spectra in detail.
Figure 2.
Left column: Simulated LA spectra. Total signal (green), backbone alone (red), side chain alone (black), additive spectrum (backbone + side chain) (blue). Right column: Difference of the Total and the Additive LA spectra (orange)
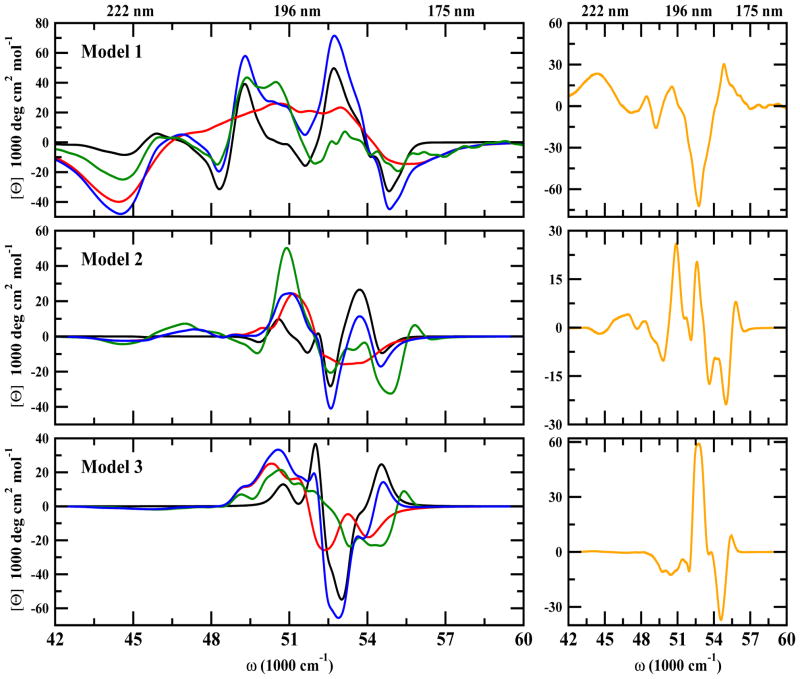
Figure 3.
Same as in Figure 2 but for CD spectra
Results and Discussion
Near Ultraviolet (NUV) Signals
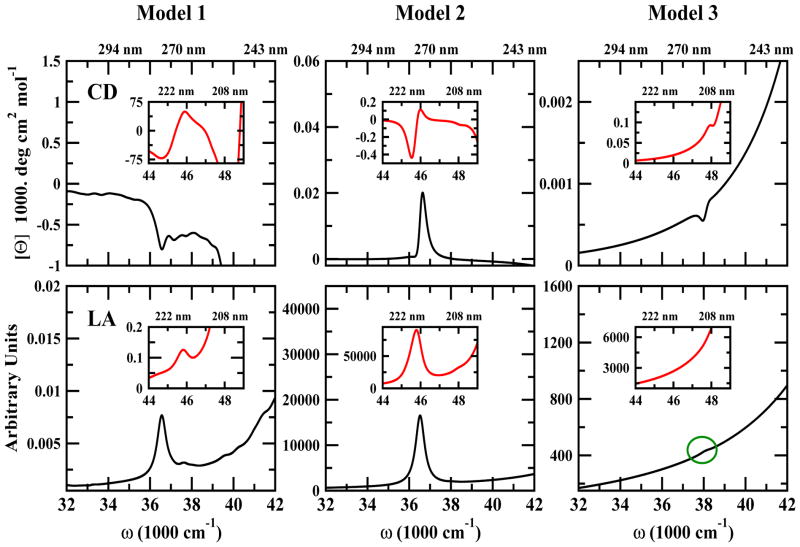
The simulated LA signals in the NUV regime for all three models are shown in Figure 4 (bottom). for Models 1 and 2. They exhibit sharp peaks around 37000 cm−1 while Model 3 only has a modest bump (indicated by green circle) at 38000 cm−1. Another peak is observed in the 46000 cm−1 for Model 1 and 2. This peak is missing in Model 3 due to its shorter sequence that does not have the Tyrosine Y9. We expect that a full sequence for Model 3 should show the aromatic side chain transitions at 46000 cm−1.
Figure 4.
Simulated NUV CD (top row) and linear absorption (LA) (bottom row) of the three Aβ amyloid models. Inserts show the region [44000, 48000] cm−1.
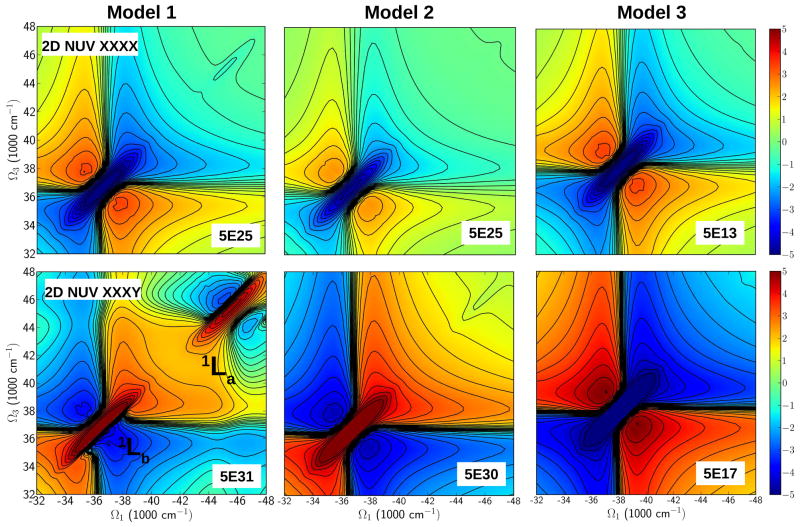
The 2DNUV spectra computed with laser pulses centered at 37000 cm−1 and 3754 cm−1 bandwidth are displayed in Figure 5. All 2D spectra in this article are plotted on a nonlinear scale that interpolates between logarithmic for small values and linear for large values. It thus reveals both the strong and weak signals (34). The non-chiral xxxx signals (All pulses have parallel polarization) are shown in the top row. For Model 1, the signal is symmetric with respect to the diagonal. The 1Lb transitions dominate and show a negative diagonal peak (blue) centered at 37000 cm−1. This is accompanied by two positive (red) side bands. The 1La transitions which originate from the coupling between the aromatic residues Phe and Tyr appear as a peak in Model 1 around 46000 cm−1 (blue), a weaker peak appear in than Model 2 and no peak is present in Model 3 where 1La transitions are invisible due to the lack of Tyrosine Y9. In Models 2 and 3, the 1Lb transitions shifted to 36000 cm−1 dominate with their respective side bands but Model 2 seems to have a weaker 1Lb transition than Models 1 and 3.
Figure 5.
Simulated 2DNUV photon echo signals of the three Aβ amyloid models. Top row: The non-chiral (xxxx) polarization configuration. The signals show a similar pattern but different intensities. Peaks centered at 37000 cm−1 is similar in intensity for Models 1 and 2 while in Model 3 is shifted away from the center and less intense. Bottom row: chirality induced (xxxy) polarization configuration. The spectra show some differences in the 1Lb region (37000 cm−1). Model 1 has a strong 1La transition at 46000 cm−1 while in Model 2 is weaker. Spectra are displayed on the non-linear scale as defined in ref. (34). The s parameter is shown at the bottom right of each panel.
We next turn to chirality-induced signals. The CD spectra of all models shown in Figure 4 have aromatic side chain transition peaks at 37000 cm−1 and 46000 cm−1. Model 3 also shows tiny peaks which could be attributed to aromatic side chain transition between Phe of a peptide with the other Phe in another peptide on the same layer.
The chirality-induced xxxy 2DUV signal of Model 1, where the last pulse is polarized perpendicular to the other three (bottom row in Figure 5) is non-symmetric with respect to the diagonal. The 1Lb and 1La transitions appear as two positive (red) peaks with their negative side bands centered around at 37000 cm−1 and 46000 cm−1, respectively. The 1Lb peak has a strong side band in the lower triangle, shifted from the diagonal while a similar pattern is seen in the upper side band of the 1La transition peak. This indicates that the 1Lb and 1La transitions are coupled. The signals of Model 2 and 3 are very different from Model 1. Both are symmetric but with different intensities. The 1Lb transition of Model 2 is stronger than in Model 3. This may be due to the nearest-neighbor interactions between Phe in the Aβ peptides in Model 2 that exist also in Model 1. 1La is weak in models 2 and 3, suggesting weak coupling between 1Lb and 1La transitions. A possible explanation could be the fact that intermolecular interactions of aromatic residues Phe-Tyr, ie Phe and Tyr interact each other in one Aβ peptide in a given layer, and generates coupling between 1Lb and 1La transitions. The geometry of Model 2 allows Tyr to interact with the Phe of another Aβ peptide in another layer. Our previous 2DNUV studies of the amyloid fibril growth in Model 1 (39) showed how the intensities in the signals correlate with the complex size. Intramolecular interaction of aromatic residues Phe-Tyr in one Aβ peptide is seen only in Model 1 while aromatic residue couplings are missed in Model 3 which lacks the Tyr residue (Y9) in its sequence.
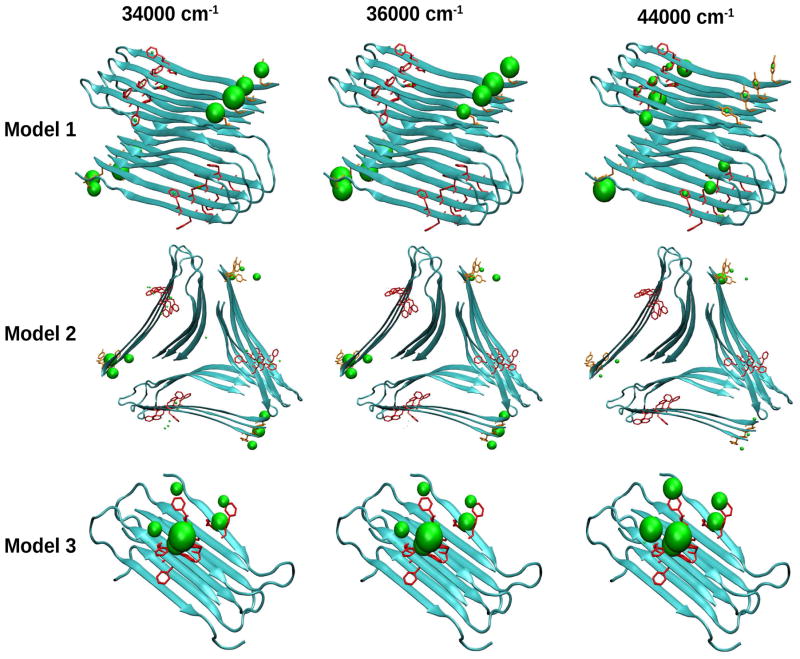
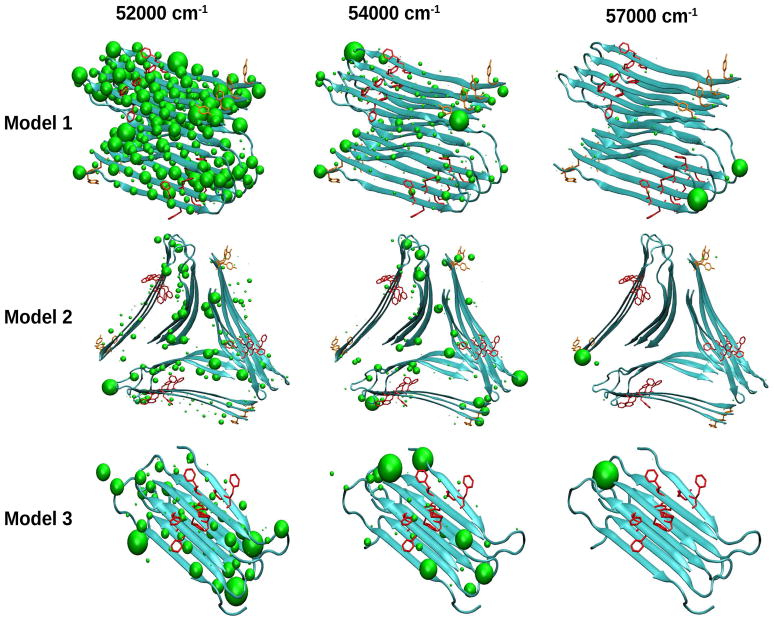
In order to trace the origin of these spectral features, we have computed the transition populations. These are defined as the squares of the exciton wave function expansion coefficients, in the basis of the local amide excitations and are displayed in Figure 6. We see that the NUV xxxy signals (bottom row in Figure 5) of all models are induced by the aromatic residues (Phe and Tyr) and are well localized in the sequence. In Model 2, 1Lb transitions is observed as a red peak in the 34000–36000 cm−1 region but no coupling (perhaps very weak) between 1Lb and 1La transitions are observed in the 44000–46000 cm−1 region. Model 3 shows a blue peak at 34000–36000 cm−1 region while models 1 and 2 have red peaks that can be ascribed to the complex geometry since it has only Phe–Phe intermolecular interactions between two peptides. Similarly, the 44000–46000 cm−1 region shows signals for Models 1 and 2 and not in Model 3.
Figure 6.
Transition populations for three NUV spectral regimes of the Aβ amyloid fibril models.
Far Ultraviolet (FUV) Signals
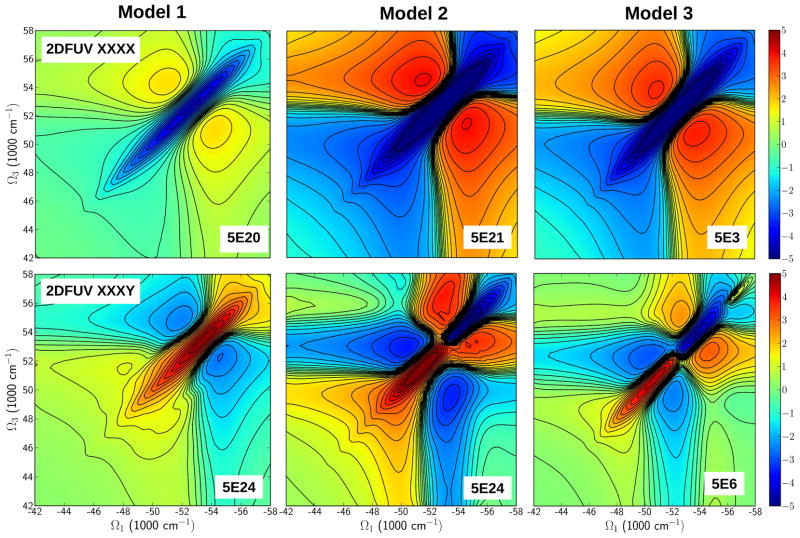
The simulated LA and CD spectra of the three Aβ peptide amyloid fibril models in the FUV region are shown in Figure 7. All spectra are similar with peaks centered around at 52000 cm−1. The 2DFUV spectra generated by laser pulses centered at 52000 cm−1 and bandwidth at 3754 cm−1 are displayed in Figure 8. The non-chiral 2DUV xxxx spectra (top row) show the characteristic features of a β-sheet: a weak peak centered around at 52000 cm−1 on the diagonal maps (Ω3 = −Ω1) for all three models.
Figure 7.
Simulated FUV CD (top row) and linear absorption (LA) spectra (bottom row) of the three Aβ amyloid models.
Figure 8.
Same as in Figure 5 but for the 2DFUV regime. Top row: The non-chiral (xxxx) polarization configuration. All models display similar pattern but different intensities. Bottom row: chirality induced (xxxy) polarization configuration. Model 1 displays a strong peak at 52000 cm−1 while in Models 2 and 3 display butterfly-like shapes with two peaks in the 50000 cm−1 to 54000 cm−1 regime. Model 3 shows a small additional peak at 57000 cm−1.
We next turn to the chiral signals. The CD signals are distinguishable and have the characteristic of a β-strand rich-structure profile, in agreement with experiments (21, 22, 40). Model 1 has stronger CD signals in the region 50000–54000 cm−1 whereas signal for models 2 and 3 show peaks around 50000 cm−1 and decay beyond 52000 cm−1. The (xxxy) chiral 2DUV signals are much richer than the xxxx spectra (Figure 8, bottom row). Model 1 has a symmetric strong peak at 52000 cm−1. Models 2 and 3 show a symmetric butterfly-like spectra with a positive peak near 52000 cm−1 and a negative peak at 54000 cm−1. Model 3 has an additional positive peak near 57000 cm−1. The transition populations in the FUV regime are displayed in Figure 9. These reveal that at 52000 cm−1, the signals for all three models come from the electronic transition of all peptide groups and reflect the strongest peak of the CD spectra (See Figure 9 top row and Figure 9 left column). At 54000 cm−1, weak contributions from the backbone are seen only in Model 1 CD and LA spectra. In contrast, that region shows a broad feature that overlaps with the 52000 cm−1 peak (Figure 9 top row left panel). The middle column in Figure 9 shows that this is mainly due to interactions between the turn loop of a Aβ peptide located in a β-sheet layer with the terminal of a Aβ peptide located in the other β-sheet layer. The 2DUV signals of models 2 and 3 are different. They show an additional weak peak at 54000 cm−1 on the 2DFUV spectra originating from the interactions between turns-ends between different fibrils layers. The CD spectra of both models in the 52000 cm−1 and 54000 cm−1 regime are different. We expect a similar 2DUV spectrum to Models 1 and 3 since their global geometry consists in a β-sheet layer with alternate loops between Aβ peptides. However, the two show significant differences. The 2DFUV spectra (Figure 8 lower-right plot) shows also a butterfly-like spectrum as in Model 2, indicating that in addition to the interactions between turns of a Aβ peptide from a β-sheet layer with the ends of a Aβ peptide from another β-sheet layer as in Model 2, the complex geometry is important as well. Both models show interactions between turn and termini among peptides. In Model 2, these are between stacks while in Model 3 they are among peptides in the same stack. This could be the origin of the butterfly shape of 2DFUV signal.
Figure 9.
Transition populations for three FUV spectral regimes of the Aβ amyloid fibril models.
Conclusions
We have employed the EHEF protocol to predict the 2DUV spectra of three amyloid fibril structure models proposed by Tycko and collaborators. The 1D (CD and LA) spectra of the three models suggest that the side chains and backbone transitions are coupled. 2DUV spectra of the aromatic side chains show clear signatures of 1Lb and 1La transitions and their respective couplings in all models. The 2DFUV chirality-induced xxxy signals associated with aromatic residue couplings between transitions can describe the variations of interactions between residues which strongly depend on the complex geometry. The 1Lb and 1La couplings can be hidden in the 2DFUV maps by either intermolecular interactions of aromatic residues Phe-Tyr as seen in Model 2 or there is a correlation between number of peptides in a Aβ sheet with the 2DUV intensity. Since the sequence of the Iowa mutant Aβ peptide shown in Model 3 was shorter, the 1Lb and 1La transitions were undetectable. However, its geometry (peptides forming an anti-parallel β-strand sheets) had resulted in a butterfly-like chiral-induced signals in the FUV similar to Model 2.
Acknowledgments
We wish to thank Dr. Robert Tycko for providing the three Aβ amyloid models.
Funding Information: National Institutes of Health (Grant GM059230 and GM091364) and National Science Foundation (Grant CHE-1058791)
References
- 1.Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow D. Amyloid β-protein fibrillogenesis: detection of a protofibrillar intermediate. J Biol Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 2.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer’s Disease and other degenerative disorders: the emerging role of the oligomers assemblies. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 3.Nostrand WEV, Melchor JP, Cho HS, Greenberg SM, Rebeck GW. Pathogenic Effects of D23N Iowa Mutant Amyloid β-Protein. J Biol Chem. 2001;276:32860–32866. doi: 10.1074/jbc.M104135200. [DOI] [PubMed] [Google Scholar]
- 4.Crescenzi O, Tomaselli S, Guerrini R, Salvatori S, D’Ursi AM, Temussi PA, Picone D. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 5.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc Natl Acad Sci USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. In silico study of amyloid β-protein (Aβ) folding and oligomerization. Proc Natl Acad Sci USA. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glabe CG. Amyloid accumulation and pathogensis of Alzheimer’s disease: significance of monomeric, oligomeric and fibrillar Aβ. Subcell Biochem. 2005;38:167–177. doi: 10.1007/0-387-23226-5_8. [DOI] [PubMed] [Google Scholar]
- 8.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. On the nucleation of amyloid β-protein monomer folding. Prot Sci. 2005;14(6):1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Shen J, Luo X, Zhu W, Chen K, Ma J, Jiang H. Conformational Transition of Amyloid β-peptide. Proc Natl Acad Sci USA. 2005;102:5403–5407. doi: 10.1073/pnas.0501218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumketner A, Bernstein SL, Wyttenbach T, Bitan G, Teplow DB, Bowers MT, Shea JE. Amyloid β-protein monomer structure: a computational and experimental study. Prot Sci. 2006;15:420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim KH, Collver HH, Le YTH, Nagchowdhuri P, Kenney JM. Characterization of distinct amyloidogenic conformations of the Aβ (1–40) and Aβ (1–42) peptides. Biochem and Biophys Res Com. 2006;353:443–449. doi: 10.1016/j.bbrc.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 12.Teplow DB. Preparation of amyloid β-protein for structural and functional studies. Methods Enzymol. 2006;413:20–33. doi: 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- 13.Grant MA, Lazo ND, Lomakin A, Condron MM, Arai H, Yamin G, Rigby AC, Teplow DB. Familial Alzheimer’s disease mutations alter the stability of the amyloid β-protein monomer folding nucleus. Proc Natl Acad Sci USA. 2007;104(42):16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam AR, Teplow DB, Stanley HE, Urbanc B. Effects of the Arctic (E(22)-> G) Mutation on Amyloid β-Protein Folding: Discrete Molecular Dynamics Study. J Am Chem Soc. 2008;130(51):17413–17422. doi: 10.1021/ja804984h. [DOI] [PubMed] [Google Scholar]
- 15.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 16.Hardy J. Testing times for the “amyloid cascade hypothesis”. Neurobiol Aging. 2002;23:1073–1074. doi: 10.1016/s0197-4580(02)00042-8. [DOI] [PubMed] [Google Scholar]
- 17.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A Structural Model for Alzheimer’s β-Amyloid Fibrils Based on Experimental Constraints from Solid State NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petkova AT, Yau WM, Tycko R. Experimental Constraint on Quaternary Structure in Alzheimer’s β-Amyloid Fibrils. Biochemistry. 2006;45:598–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paravastu A, Leapman RD, Yau WM, Tycko R. Molecular Structural Basis for Polymorphism in Alzheimer’s β-Amyloid Fibrils. PNAS. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tycko R, Sciarretta KL, Orgel JPRO, Meredith SC. Evidence for Novel β-Sheet Structures in Iowa Mutant β-Amyloid Fibrils. Biochemistry. 2009;48(26):6072–6084. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrow CJAY, Kenny PTM, Zagorski MG. Solution Conformations and Aggregational Properties of Synthetic Amyloid β-Peptides of Alzheimer’s Disease: Analysis of Circular Dichroism Spectra. J Mol Biol. 1992;225(4):1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 22.Gursky O, Aleshkov S. Temperature-dependent β-sheet formation in beta-amyloid Aβ1–40 peptide in water: Uncoupling β-structure folding from aggregation. Biochim Biophys Acta. 2000;1476:93–102. doi: 10.1016/s0167-4838(99)00228-9. [DOI] [PubMed] [Google Scholar]
- 23.Frenkel J. On the Transformation of Light into Heat in Solids. I Physical Review. 1931;37:17–44. [Google Scholar]
- 24.Besley NA, Hirst JD. Theoretical Studies toward Quantitative Protein Circular Dichroism Calculations. J Am Chem Soc. 1999;121(41):9636–9644. [Google Scholar]
- 25.Woody RW. The Exciton Model and the Circular Dichroism of Polypeptides. Monatsh Chem. 2005;136(3):347–366. [Google Scholar]
- 26.Bulheller BM, Rodger A, Hirst JD. Circular and Linear Dichroism of Proteins. Phys Chem Chem Phys. 2007;9:2020–2035. doi: 10.1039/b615870f. [DOI] [PubMed] [Google Scholar]
- 27.Abramavicius D, Palmieri B, Voronine DV, Sanda F, Mukamel S. Coherent Multidimensional Optical Spectroscopy of Excitons in Molecular Aggregates; Quasiparticle versus Supermolecule Perspectives. Chemical Review. 2009;109(6):2350–2408. doi: 10.1021/cr800268n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith AW, Tokmakoff A. Amide I two-dimensional infrared spectroscopy of β-hairpin peptides. J Chem Phys. 2007;125:045109–1–10. doi: 10.1063/1.2428300. [DOI] [PubMed] [Google Scholar]
- 29.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Two-dimensional Infrared Spectra of Isotopically Diluted Amyloid Fibrils from Aβ40. Proc Natl Acad Sci USA. 2008;105(22):7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strasfeld DB, Ling YL, Shim SH, Zanni MT. Tracking Fiber Formation in Human Islet Amyloid Polypeptide with Automated 2D-IR Spectroscopy. J Am Chem Soc. 2008;130:6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim S-H, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc Natl Acad Sci USA. 2009;106(16):6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, Abramavicius D, Bulheller BM, Hirst JD, Mukamel S. Ultraviolet Spectroscopy of Protein Backbone Transitions in Aqueous Solution: Combined QM and MM Simulations. J Phys Chem B. 2010;114:8270–8277. doi: 10.1021/jp101980a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J, Mukamel S. Two-Dimensional Ultraviolet (2DUV) Spectroscopic Tools for Identifying Fibrillation Propensity of Protein Residue Sequences. Ang Chem. 2010;49(50):9666–9669. doi: 10.1002/anie.201005093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang J, Mukamel S. Two-Dimensional Near-Ultraviolet Spectroscopy of Aromatic Residues in Amyloid Fibrils: A First Principles Study. Phys Chem Chem Phys. 2011;13:2394–2400. doi: 10.1039/c0cp02047h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips JCR, Braun WW, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comp Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKerell AD, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Comp Chem. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Abramavicius D, Falvo C, Bulheller BM, Hirst JD, Mukamel S. Simulation of Two Dimensional Ultraviolet (2DUV) Spectroscopy of Amyloid Fibrils. J Phys Chem B. 2010;114:12150–12156. doi: 10.1021/jp1046968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuang W, Abramavicius D, Mukamel S. Dissecting Coherent Vibrational Spectra of Small Proteins into Secondary Structural Elements by Sensitivity Analysis. Proc Natl Acad Sci USA. 2005;102(7):7443–7448. doi: 10.1073/pnas.0408781102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang J, Mukamel S. Probing Amyloid Fibril Growth by Two-Dimensional Near-Ultraviolet Spectroscopy. J Phys Chem B. 2011;115:6321–6328. doi: 10.1021/jp201164u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Nostrand WE, Melchor JP, Ruffini L. Pathologic amyloid β-protein cell surface fibril assembly on cultured human cerebrovascular smooth muscle cells. Journal of Neurochemistry. 1998;70(1):216–223. doi: 10.1046/j.1471-4159.1998.70010216.x. [DOI] [PubMed] [Google Scholar]