Abstract
Polynucleotide phosphorylase (PNP) plays a central role in RNA degradation, generating a pool of ribonucleoside diphosphates (rNDPs) that can be converted to deoxyribonucleoside diphosphates (dNDPs) by ribonucleotide reductase. We report here that spontaneous mutations resulting from replication errors, which are normally repaired by the mismatch repair (MMR) system, are sharply reduced in a PNP-deficient Escherichia coli strain. This is true for base substitution mutations that occur in the rpoB gene leading to Rifr and the gyrB gene leading to Nalr and for base substitution and frameshift mutations that occur in the lacZ gene. These results suggest that the increase in the rNDP pools generated by polynucleotide phosphorylase (PNP) degradation of RNA is responsible for the spontaneous mutations observed in an MMR-deficient background. The PNP-derived pool also appears responsible for the observed mutations in the mutT mutator background and those that occur after treatment with 5-bromodeoxyuridine, as these mutations are also drastically reduced in a PNP-deficient strain. However, mutation frequencies are not reduced in a mutY mutator background or after treatment with 2-aminopurine. These results highlight the central role in mutagenesis played by the rNDP pools (and the subsequent dNTP pools) derived from RNA degradation.
INTRODUCTION
Elucidating the pathways that lead to mutations resulting from replication errors, arising either spontaneously or induced by chemical agents, has intrigued molecular biologists ever since the elucidation of the structure of DNA allowed one to pose this question in molecular terms (57, 58). What is the actual source of these mutations? Different tautomeric forms of the four bases in DNA or their analogs have been considered which provoke errors directly (18, 24, 28, 29, 57) or even indirectly, for instance by affecting the pools of nucleoside triphosphates (28, 29, 34). Also, the field has defined a myriad of repair systems aimed at preventing or repairing DNA damage (24) and also aimed at correcting errors of DNA replication (30, 47). In humans, defects in one of a number of repair systems leads to inherited cancer susceptibilities (e.g., see references 1 and 22). In Escherichia coli, the replicating DNA polymerase (Pol III) contains an editing subunit that corrects numerous replication errors (14, 19, 41). Directly after replication, the mismatch repair (MMR) system recognizes still-uncorrected mismatches and repairs them using the pattern of methylation to distinguish the template strand from the newly synthesized strand (30, 47). Mutants lacking any one of the components of this system (e.g., MutH, MutL, MutS, UvrD) have sharply elevated mutation rates that involve transitions (A:T→G:C or G:C→A:T) (11, 36, 54) or short indels (insertion/deletions) at repeated sequences, such as monotonous runs of G's or A's on the same strand (10, 54). The size and balance of the nucleoside triphosphate (NTP) pools are important for replication fidelity (33). Not only do unbalanced pools provoke an increase in mutagenesis (33, 38, 44), but an increase in the pools of all four dNTPs also leads to increased mutations (26, 59).
One widely used approach to studying mutational pathways is to find mutants with increased mutation rates, or “mutators” (e.g., see references 43 and 46). A more difficult approach is to detect mutants with lowered mutation rates, or “antimutators.” In a study to be reported elsewhere (E. Becket and J. H. Miller, unpublished), we screened the E. coli Keio collection of gene knockouts for mutants with a reduced rate of mutagenesis induced by the base analog 5-azacytidine (5AZ). We found that mutants deleted for the pnp gene, encoding polynucleotide phosphorylase (PNP), had lower levels of 5AZ-induced mutagenesis. Here, we report the effects of PNP deficiency on mutagenesis induced by two other base analogs, 5-bromodeoxyuridine (5BdU) and 2-aminopurine (2AP), as well as on spontaneous mutations that result from replication errors. These studies indicate that the pool of nucleoside diphosphates generated by PNP-mediated degradation of RNA drives spontaneous and certain mutagen-induced mutations.
MATERIALS AND METHODS
E. coli strains.
The Keio collection is as described by Baba et al. (3) and made from the starting strain BW25113 (13). This strain (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) is the starting strain used in the experiments reported here unless otherwise stated. Briefly, each of the 3,985 strains in the Keio collection carries a complete deletion of a different gene, with a kan insert replacing each gene. The base substitution tester strains CC102 and CC106 have been described previously (11). They carry a base substitution mutation in lacZ on the F′ plasmid, which reverts from Lac− to Lac+ only by restoring the glutamic acid codon through a reversion by a specific base substitution for each (G→A, G→C, and A→G, respectively). Additionally, these strains carry a miniTn10-cat insert (45) conferring chloramphenicol resistance for selection purposes (C. Tamae and J. H. Miller, unpublished). Also, mutL, mutT, and mutY derivatives of BW25113 were prepared by P1 transduction from strains carrying miniTn10-tet inserts (Tamae and Miller, unpublished) in either mutL, mutT, or mutY.
E. coli genetic methods.
Unless otherwise stated, all genetic methods are as described by Miller (45).
Validation controls.
A collection of close to 4,000 strains will contain some errors and some impure strains. The latter problem can be minimized by repurifying and retesting, as was done here. Yamamoto and coworkers have subjected the Keio collection to an intensive analysis aimed at uncovering errors in the collection that might arise from duplications of the target gene. They generated a list of 14 mutants that are incorrect and another 9 that might be incorrect (61). Ultimately, the most prudent use of such a large collection is to verify any mutants that are particularly important to the final results by PCR analysis, and/or sequencing, as we have done in a number of cases (4, 37). Here, using PCR and sequencing we have verified the strain with a deletion of the pnp gene. Using a primer within the kan gene and a primer outside the gene it replaced, we showed that a kan insert was at the correct position. Using internal primers for the pnp gene, we showed that there was no other copy of the pnp gene elsewhere in the chromosome. Controls with the starting wild-type (WT) strain showed that the internal primers were efficient.
We transduced the pnp deletion/kan insertion-carrying strain used in these experiments with a linked Tn10 element (from strain CAG12153 [55]). A majority of the Tetr transductants had crossed out the pnp deletion/replacement, as determined by the loss of Kanr, which was accompanied by the restoration of the normal colony size. We tested two of these transductants for the mutation frequency (Rifr) in the presence of 5BdU and found that they had restored the high mutation frequency characteristic of PNP-proficient strains (data not shown).
The major hot spot in rpoB that occurs after 5BdU treatment or in MMR-deficient strains represents two-thirds of the mutations detected (see Fig. 3 and reference 25). To verify that cells with this mutation still confer the Rifr phenotype in a PNP-deficient strain, we carried out a reconstruction experiment. We transduced this mutation from the wild type into a pnp derivative in two steps without selecting for Rifr by using a linked Tn10 in the argE gene (from strain CAG12185 [55]). First, argE∷Tn10 was transduced into the pnp deletion/kan replacement strain, scoring for retention of the Kanr phenotype. Then with a lysate prepared on the Rifr strain carrying the rpoB allele in question, the strain was transduced to Arg+. Arg+ transductants were scored for retention of the pnp/kan deletion/replacement and also for Rifr. The expected 60% of the Arg+ strains formed Rifr colonies (in the pnp background).
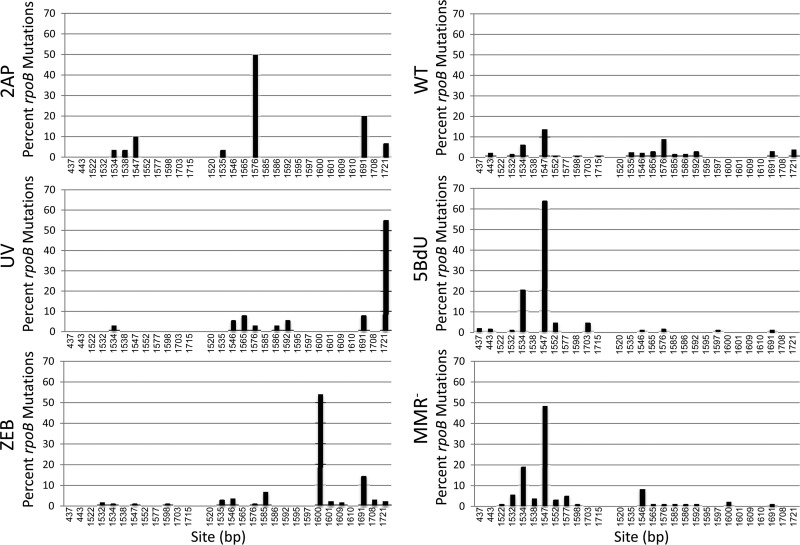
Fig 3.
Transition mutations in rpoB leading to the Rifr phenotype occurring spontaneously or after treatment with different mutagens. A:T→G:C mutations are shown in the left portion of each diagram, and G:C→A:T mutations are shown in the right portion. The height of each bar represents the percentage of all the rpoB mutations detected in that sample. For further details, see Results.
Stock solution and cell treatment.
5-Bromodeoxyuridine was prepared by dissolving in distilled water to a concentration of 2 mg/ml. Solid 2-aminopurine was added directly to LB medium for a concentration of 700 μg/ml, which was then distributed in 3-ml aliquots and followed by the addition of ∼5 × 104 cells and incubation for 24 h at 37°C with aeration.
Determination of mutation frequencies.
We inoculated 100 to 1,000 cells in a series of cultures of LB or LB plus 2-aminopurine (2AP) or 5BdU, where they were grown for 18 h at 37°C with aeration prior to plating on the appropriate medium (lactose-minimal plates, or LB plates with either 100 μg/ml rifampin or 20 μg/ml nalidixic acid). The mutation frequencies of Rifr and Lac+ revertants were determined as described previously (44). Briefly, the mutation frequency (f) was determined as the median frequency from a set of cultures (the number of cultures varied from 8 to 20), and the mutation rate (μ) was determined by the formula of Drake (17). We determined the 95% confidence limits according to the method of Dixon and Massey (15).
Chemicals.
5-Bromodeoxyuridine, 2-aminopurine, tetracycline, chloramphenicol, and kanamycin were purchased from Sigma (St. Louis, MO).
RESULTS
Base analog-induced mutagenesis.
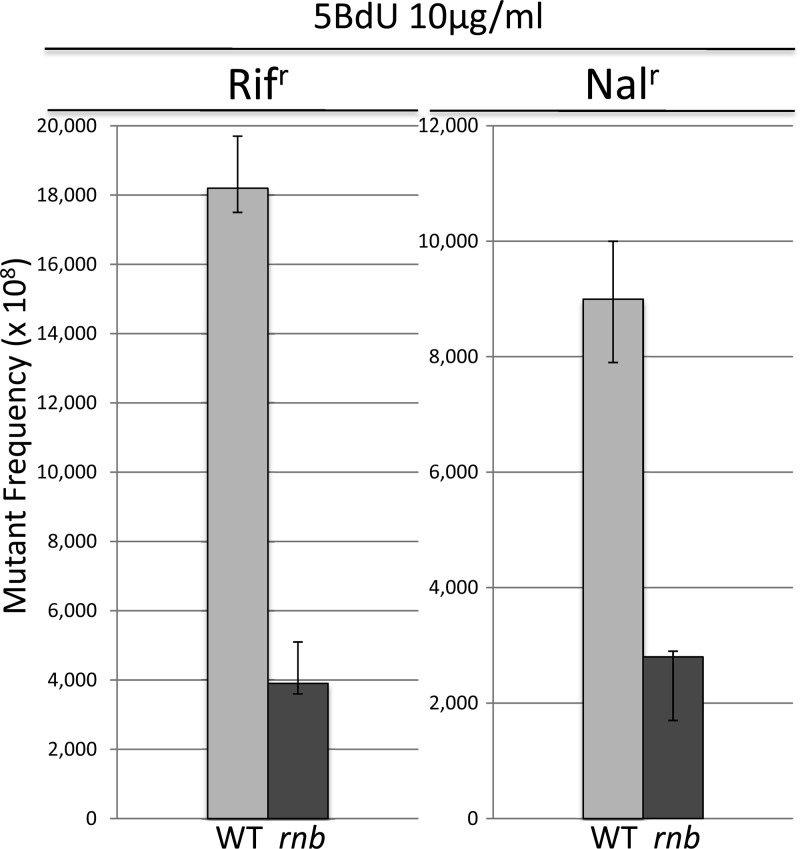
Fig. 1A shows that 5BdU mutagenesis is reduced in a pnp mutant on the order of 1,000-fold relative to the PNP+ starting strain, as measured by the frequency of Rifr mutants. However, 2AP mutagenesis is not lowered in a pnp mutant (Fig. 1B). We have verified the effect of a PNP deficiency on mutagenesis in two additional systems. Figure 1C shows that when mutagenesis was monitored by the frequency of nalidixic acid resistance (Nalr), 5BdU again displayed a reduction on the order of 1,000-fold in a pnp strain, whereas 2AP mutagenesis was not reduced (Fig. 1D). Moreover, use of the reversion of specific lacZ alleles to measure mutagenesis revealed that 5BdU-induced mutagenesis was also sharply reduced (Table 1). CC102 reverts to wild type via a specific G:C→A:T transition, and CC106 reverts via a specific A:T→G:C transition (11). Because PNP and RNase II (RNB) are the two enzymes responsible for RNA degradation and the resulting ribonucleoside diphosphate (rNDP) and rNMP pools (12, 16), respectively, derived from this degradation, we tested rnb (encoding RNB) mutants. Figure 2 shows that a deletion of rnb also resulted in a reduction of 5BdU-induced mutagenesis, although the modest 3.5- to 4.0-fold decrease was far less than that seen with pnp deletion strains.
Fig 1.
Mutagenesis in WT and PNP-deficient strains. Mutant frequencies for 5BdU- and 2AP-induced mutations in rpoB, generating Rifr mutants, and gyrA, generating Nalr strains, are shown. Error bars reflect 95% confidence limits (see Materials and Methods for further details).
Table 1.
Frequencies (f) and mutation rates (μ) in CC102 and CC106 after treatment with 5BdU at 10 μg/mla
| Strain | Frequency (f) (×108) | Mutation rate (μ) (×108) |
|---|---|---|
| CC102 WT | 470 (420–1,300)b | 60 (54–143) |
| CC102 pnp | 3 (0–7.5) | 0.86 (0–1.8) |
| CC106 WT | 760 (410–1,900) | 90 (53–206) |
| CC106 pnp | 0 (0–1.7) | 0 (0–0.57) |
The rpoB mutation frequencies (f) per cell were calculated by dividing the median number of mutants by the average number of cells in a series of cultures, and the mutation rates (μ) were determined with the method of Drake (17) (see Materials and Methods).
Values in parentheses are 95% confidence limits.
Fig 2.
5BdU mutagenesis in WT and RNB-deficient strains (see legend to Fig. 1).
Pathways of 5BdU-induced mutagenesis.
The study of mutagenic spectra has revealed that spontaneous mutations, as well as those induced by each mutagen treatment and mutator background, result in a characteristic pattern of hot spots and cold spots (e.g., see references 5, 8, 21, and 25), most probably as a result of the surrounding sequences. Analysis of the spectra of mutations in rpoB leading to Rifr is a case in point (25, 35). Figure 3 shows the pattern of transition mutations (A:T→G:C, G:C→A:T) in rpoB for the base analogs 2AP (25), zebularine (ZEB) (35), and 5BdU (this work), as well as the mutagen UV irradiation (UV) (25), the mismatch-repair-deficient mutator mutS (25), and the unmutagenized wild type (25, 60). There are 29 sites that have been detected in rpoB that lead to Rifr via a transition (60), and these are listed by base pair number in rpoB in terms of the percentage of the total number of Rifr mutants analyzed. The A:T→G:C mutations are on the left portion of each panel, and the G:C→A:T mutations are on the right portion of each panel. The total numbers of mutations analyzed in each sample were 30 (2AP), 156 (ZEB), 194 (5BdU), 40 (UV), 174 (mutS [MMR−]), and 298 (SPON). The percentage of the total mutations that resulted from transitions was close to or more than 90% for all but the SPON (WT) set. These percentages were 97% (2AP), 92% (ZEB), 99% (5BdU), 88% (UV), 98% (mutS), and 47% (SPON). What is evident from Fig. 3 is that while each mutagen or mutator shows a different pattern of transition frequencies at the available sites, 5BdU displays a pattern that is virtually identical to that of a mismatch-repair-deficient strain (mutS). Although one might argue that this pattern is coincidental, we see the same pattern among sites in the gyrB gene that lead to Nalr. Table 2 displays our analysis of gyrB mutations leading to Nalr. There are fewer sites in gyrB than in rpoB, but of the 18 sites detected, 8 involve transitions, and 4 of these represent the A:T→G:C transition that is favored by 5BdU in the rpoB gene (Fig. 3). One of these sites, at position 247, is the prominent hot spot in MMR-deficient strains (mutL) and after 5BdU treatment. Note that 2AP does not have the same hot spot. Moreover, overproducing MutL on a plasmid reduced the level of 5BdU mutagenesis up to 150-fold (data not shown). The results with 5AZ, and for mutT and mutY, are also shown here to underscore the fact that this system correctly identifies the known mutagenic specificity of these treatments or strain backgrounds. Namely, 5AZ is specific for G:C→C:G changes, mutT for A:T→C:G changes, and mutY for G:C→T:A changes (9, 11, 35, 48). Taken together, the results shown in Fig. 3 and Table 2 strongly indicate that 5BdU is not targeting mutations directly, as do other base analogs, such as 2AP, ZEB, and 5-azacytidine, but instead acts indirectly by increasing normal DNA polymerase replication errors and saturating mismatch repair.
Table 2.
Distribution of mutations in gyrA
| Site (bp) | AA change | bp change | WT | WT 5BdU | WT 2AP | mutL | mutT | mutY | WT 5AZ |
|---|---|---|---|---|---|---|---|---|---|
| 215 | D→G | AT→GC | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 245 | D→G | AT→GC | 1 | 1 | 0 | 5 | 0 | 0 | 0 |
| 247 | S→P | AT→GC | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 260 | D→G | AT→GC | 17 | 57 | 2 | 67 | 0 | 0 | 2 |
| 152 | A→V | GC→AT | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 244 | D→N | GC→AT | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| 248 | S→L | GC→AT | 17 | 0 | 8 | 10 | 0 | 1 | 0 |
| 259 | D→N | GC→AT | 5 | 0 | 8 | 1 | 0 | 0 | 0 |
| 241 | G→C | GC→TA | 2 | 0 | 0 | 0 | 0 | 1 | 0 |
| 259 | D→Y | GC→TA | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| 356 | A→E | GC→TA | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| 245 | D→A | AT→CG | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 247 | S→A | AT→CG | 1 | 0 | 0 | 0 | 7 | 0 | 0 |
| 260 | D→A | AT→CG | 1 | 0 | 0 | 0 | 19 | 0 | 0 |
| 260 | D→V | AT→TA | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 248 | S→W | CG→GC | 2 | 0 | 0 | 0 | 0 | 0 | 31 |
| 259 | D→H | CG→GC | 1 | 0 | 0 | 0 | 0 | 0 | 4 |
| 316 | Q→E | CG→GC | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 55 | 60 | 20 | 84 | 26 | 8 | 37 |
Spontaneous base substitution mutations.
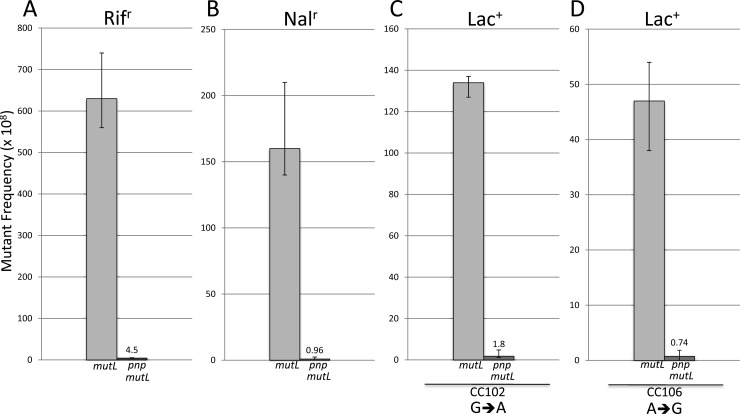
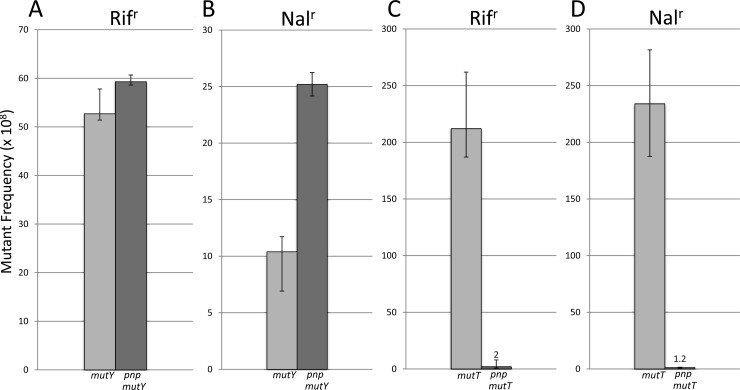
We examined the effects of deleting pnp on spontaneous mutations. While the effects on the frequency of spontaneous base substitution mutations in an MMR-proficient background are minimal (Table 3), the effects in an MMR-deficient strain are dramatic, as foreshadowed by the results with 5BdU. Figure 4 shows that in three different systems the introduction of a pnp deletion results in a decrease of Rifr mutations by 140-fold, Nalr mutations by 160-fold, and Lac+ mutations resulting from an A:T→G:C transition in lacZ by 64-fold and from a G:C→A:T transition by 78-fold. These levels are very near to those seen in an MMR-proficient strain (Table 3). To demonstrate that these reductions are not simply an artifact of the pnp background, we showed that the increase in mutation frequency that occurs in a mutY background (48) is not reduced in a mutY pnp background (Fig. 5A and B). However, the increased level of mutations in a mutT mutator background is eliminated when pnp is deleted (Fig. 5C and D), as the levels are now close to those seen in a wild-type strain.
Table 3.
Spontaneous frequencies (f) and mutation rates (μ) in wild-type and PNP-deficient strainsa
| Rifr |
Nalr |
|||
|---|---|---|---|---|
| WT | PNP-deficient strain | WT | PNP-deficient strain | |
| f (×108) | 3.7 (3.0–4.6)b | 2.9 (2.3–3.8) | 0.46 (0.23–0.69) | 0.39 (0–0.79) |
| μ (×108) | 0.81 (0.68–0.96) | 0.68 (0.57–0.85) | 0.15 (0.090–0.21) | 0.14 (0–0.24) |
The rpoB mutation frequencies (f) per cell were calculated by dividing the median number of mutants by the average number of cells in a series of cultures, and the mutation rates (μ) were determined with the method of Drake (17) (see Materials and Methods).
Values in parentheses are 95% confidence limits.
Fig 4.
Mutant frequencies of MutL-deficient strains in otherwise WT or PNP-deficient backgrounds. The frequencies of Rifr, Nalr, or Lac+ mutants are shown in each case, reflecting mutations in rpoB, gyrA, and lacZ, respectively (see legend to Fig. 1 and text).
Fig 5.
Mutant frequencies in WT and PNP strain backgrounds that are also either MutY or MutT deficient (see legend to Fig. 1 and text).
Frameshift mutations.
Frameshift mutations at repeated mono- or dinucleotides are frequent replication errors that are repaired by the MMR system (10, 30, 54). Strain CC107 reverts to Lac+ via the addition of a G in a monotonous run of 6 G's (10). Table 4 shows the frequency of this frameshift mutation in cultures of wild-type and MMR-deficient strains. 5BdU treatment also increases the level of these frameshifts to near the level of that seen in an MMR-deficient strain. Strains deleted for pnp reduce the levels of this mutation 330-fold in MMR-deficient strains and in 5BdU-treated cells down to even below the level of untreated wild-type strains, a several-thousand-fold effect (Table 4).
Table 4.
Frequencies (f) and mutation rates (μ)a of frameshift mutations with and without treatment with 5BdU at 10 μg/ml
| Strain | Frequency (f) (×108) | Mutation rate (μ) (×108) |
|---|---|---|
| Without 5BdU | ||
| CC107 WT | 56 (45–79) | 8.9 (7.4–12) |
| CC107 mutL mutant | 76,000 (69,000–78,000) | 5,900 (5,300–6,000) |
| CC107 pnp mutL mutant | 230 (180–360) | 30 (24–45) |
| With 5BdU at 10 μg/ml | ||
| CC107 WT | 27,000 (23,000–34,000) | 2,200 (1,900–2,700) |
| CC107 pnp mutant | 7 (4.5–8.8) | 1.5 (1.0–1.8) |
The rpoB mutation frequencies (f) per cell are calculated by dividing the median number of mutants by the average number of cells in a series of cultures, and the mutation rate (μ) was determined by the method of Drake (17) (see Materials and Methods). Values in parentheses are 95% confidence limits.
DISCUSSION
We show here that deleting the gene (pnp) encoding polynucleotide phosphorylase (PNP) virtually eliminates the dramatic increase of spontaneous mutations that occurs when the mismatch repair (MMR) system is inactivated (e.g., in mutS or mutL strains). This is true for base substitution mutations (Fig. 4) that occur in the rpoB gene leading to Rifr or in the gyrB gene leading to Nalr and for both base substitutions and frameshift mutations that occur in the lacZ gene (Tables 1 and 4). This is also true for base substitutions resulting from inactivation of the mutT gene (Fig. 5), which sanitizes the pool of oxidized dGTPs (39) and rGTPs (56) by converting them to monophosphates. Mutations that are elevated in a MutY-deficient background, however, are not decreased (Fig. 5), as the mutations result from oxidation of guanines in the DNA itself. Moreover, there is a similar striking reduction in mutations generated after continuous growth in 5BdU (Fig. 1; Table 4). Although 5BdU is incorporated into DNA, it has been suggested that it may cause mutations indirectly, rather than directly targeting the mutations (28, 29, 34). An inspection of the mutational spectra in rpoB argues that 5BdU induces mutations indirectly by enhancing normal replication errors and saturating mismatch repair (Fig. 3; see text). In contrast, 2AP-induced mutations are not decreased in a PNP-deficient strain (Fig. 1).
How can the absence of PNP virtually eliminate spontaneous mutagenesis under certain conditions? PNP plays a central role in RNA degradation, generating a pool of rNDPs (12, 16) that mix with the pools from de novo biosynthesis (Fig. 6) (42, 52). One should note that de novo pyrimidine biosynthesis is carried out at a low level, the rate-limiting step being the conversion of UMP/CMP to UDP/CDP, respectively (12). The primary source of pyrimidines for DNA synthesis may in fact be the pool of nucleosides derived from RNA (12). There is 10-fold more RNA in the in the cell than DNA (20), and the ribonucleoside pools have been measured to greatly exceed deoxyribonucleoside pools. For instance, in yeast, the four rNTPs are 36 to 190 times more prevalent than the four dNTPs (49). Also, because ribonucleosides are converted to deoxyribonucleosides by ribonucleoside diphosphate reductase (RNR) at the diphosphate level, the rNDPs generated by PNP-mediated degradation of RNA can be easily shunted into DNA (Fig. 6). We therefore can envision several explanations for the effect on mutagenesis of deleting pnp. One possibility is that the PNP-generated pools normally result in the incorporation of ribonucleosides into DNA and that this has mutagenic consequences. Direct measurement of ribonucleosides incorporated into DNA by yeast DNA polymerases in vitro indicates that rNMP is incorporated from once for every 625 dNMPs to once per 5,000 dNMPs, depending on the polymerase (49). These are removed in vivo by a process involving RNase H2, and mutants lacking RNase H2 in yeast have increased levels of spontaneous mutations that involve primarily short deletions or insertions of 2 to 5 bp (6, 7, 53). Models for the generation of these mutations involve processing by topoisomerase I, and misalignment (32). Interestingly, these mutations are not increased in an MMR-deficient strain (7). However, this mechanism would not account for most of the mutations that we score in the work reported here, as these are base substitution mutations that are subject to mismatch repair. Moreover, in the E. coli strain background we are using, RNase H1- or RNase H2-deficient strains (rnhA or rnhB) are not mutators for the rpoB system used here in either an otherwise wild-type background or an MMR-deficient background (data not shown).
Fig 6.
Pathways emanating from RNA biosynthesis and degradation (42, 52).
A second and related possibility is that degradation of RNA by PNP provides uracil in the form of rUDP. This is a required intermediate in the incorporation of dTTP into DNA. The rUDP is converted to dUDP and then dUTP, and then dUTPase converts dUTP to dUMP, from which it is converted into dTMP. dUTPase action prevents dUTP from being incorporated into DNA. Mutants with reduced activity of dUTPase have significant uracil incorporated into DNA, but this is normally not mutagenic (e.g., see reference 51). However, under certain circumstances the additional uracil can be mutagenic, such as when a reduced-activity dUTPase mutant also carries a mutation in the ndk gene (51). Nordman and Wright have argued that NDK is involved in removing uracil from DNA (51). It could simply be, however, that NDK-deficient cells are just beginning to saturate MMR, and the combination of NDK deficiency and reduced dUTPase activity titrate out MMR, with the result being the observed higher error rate. It is possible that, in this scenario, under normal conditions in a wild-type strain a certain amount of uracil could get into the DNA and cause replication errors that are few enough to be corrected by mismatch repair, but are revealed in an MMR-deficient strain. Then, in a PNP-deficient strain, the reduced pools of rUDP would result in greatly reduced polymerase errors, and a reduced mutation frequency. Arguing against this, however, is our demonstration that for at least the main hot spot in rpoB, the key intermediate is an A:C mispair, and not a G:T or G:U mispair (31). Also, the finding that mutT-stimulated mutations are greatly reduced in a pnp background is not explained by increases in uracil in the DNA (Fig. 5).
Perhaps the most likely model involves the concept that the levels of the pools of all four canonical bases in DNA determines the rate of DNA replication (27) and mutagenesis. Several groups have shown that plasmid overexpression of RNR results in increased levels of dNTPs and increased mutagenesis (26, 59). Moreover, a deletion of the cmk gene in E. coli results in reductions in the dCTP pools (to 30% of the wild-type level) and the dTTP pools (to 70% of the wild-type level), and a subsequent 2-fold reduction of the replication elongation rate (23). A number of studies have also shown that in higher cells the nucleotide pools affect the speed of replication. In mammalian somatic cells a reduction in nucleotide availability results in a lowering of the rate of fork progression (2). In mouse embryos, the rates of DNA synthesis are closely correlated with the intracellular deoxynucleotide pools during embryonic development (50). In synchronized populations of human HeLa cells, the addition of exogenous dNTPs accelerated the speed of replication fork movement in early S phase (40). Given the link that these studies show between nucleotide pools and both replication speed and mutagenesis, our results here suggest that in a wild-type strain, the dNDP pools emanating from rNDPs generated by PNP (Fig. 6) generate replication errors that are repaired by the MMR repair system. Elimination of the additional rNDP pool created by PNP, by deleting the pnp gene, sharply reduces these errors so that even in an MMR-deficient strain, one does not observe mutations much over the background.
Footnotes
Published ahead of print 17 August 2012
REFERENCES
- 1. Al-Tassan N, et al. 2002. Inherited variants of MYH associated with somatic G: C→ T: A mutations in colorectal tumors. Nat. Genet. 30: 227–232 [DOI] [PubMed] [Google Scholar]
- 2. Anglana M, Apiou F, Bensimon A, Debatisse M. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- 3. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becket E, Chen F, Tamae C, Miller JH. 2010. Determination of hypersensitivity to genotoxic agents among Escherichia coli single gene knockout mutants. DNA Repair (Amst.) 9: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benzer S. 1961. On the topography of the genetic fine structure. Proc. Natl. Acad. Sci. U. S. A. 47: 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen JZ, Qiu J, Shen B, Holmquist GP. 2000. Mutational spectrum analysis of RNase H (35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res. 28: 3649–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clark AB, Lujan SA, Kissling GE, Kunkel TA. 2011. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase. DNA Repair (Amst.) 10: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coulondre C, Miller JH. 1977. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J. Mol. Biol. 117: 577–606 [DOI] [PubMed] [Google Scholar]
- 9. Cox EC, Yanofsky C. 1967. Altered base ratios in the DNA of an Escherichia coli mutator strain. Proc. Natl. Acad. Sci. U. S. A. 58: 1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cupples CG, Cabrera M, Cruz C, Miller JH. 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cupples CG, Miller JH. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. U. S. A. 86: 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Danchin A. 1997. Comparison between the Escherichia coli and Bacillus subtilis genomes suggests that a major function of polynucleotide phosphorylase is to synthesize CDP. DNA Res. 4: 9–18 [DOI] [PubMed] [Google Scholar]
- 13. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Degnen GE, Cox EC. 1974. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J. Bacteriol. 117: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dixon WJ, Massey FJ., Jr 1969. Introduction to statistical analysis, 3rd ed. McGraw-Hill, New York, NY [Google Scholar]
- 16. Donovan WP, Kushner SR. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 83: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drake JW. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U. S. A. 88: 7160–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drake JW, Baltz RH. 1976. The biochemistry of mutagenesis. Annu. Rev. Biochem. 45: 11–37 [DOI] [PubMed] [Google Scholar]
- 19. Echols H, Lu C, Burgers PM. 1983. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 80: 2189–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellard GA. 1991. The use of RNA/DNA ratio measurements to assess rifampicin-induced growth inhibition of Escherichia coli. J. Antimicrob. Chemother. 28: 347–355 [DOI] [PubMed] [Google Scholar]
- 21. Farabaugh PJ, Schmeissner U, Hofer M, Miller JH. 1978. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J. Mol. Biol. 126: 847–857 [DOI] [PubMed] [Google Scholar]
- 22. Fishel R, Kolodner RD. 1995. Identification of mismatch repair genes and their role in the development of cancer. Curr. Opin. Genet. Dev. 5: 382–395 [DOI] [PubMed] [Google Scholar]
- 23. Fricke J, Neuhard J, Kelln RA, Pedersen S. 1995. The cmk gene encoding cytidine monophosphate kinase is located in the rpsA operon and is required for normal replication rate in Escherichia coli. J. Bacteriol. 177: 517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedberg EC, Walker GC, Siede W. 2006. DNA repair and mutagenesis, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 25. Garibyan L. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst.) 2: 593–608 [DOI] [PubMed] [Google Scholar]
- 26. Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. 2011. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 108: 19311–19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gon S, et al. 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goodman MF, Hopkins RL, Lasken R, Mhaskar DN. 1985. The biochemical basis of 5-bromouracil- and 2-aminopurine-induced mutagenesis. Basic Life Sci. 31: 409–423 [DOI] [PubMed] [Google Scholar]
- 29. Hopkins RL, Goodman MF. 1980. Deoxyribonucleotide pools, base pairing, and sequence configuration affecting bromodeoxyuridine- and 2-aminopurine-induced mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 77: 1801–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iyer RR, Pluciennik A, Burdett V, Modrich PL. 2006. DNA mismatch repair: functions and mechanisms. Chem. Rev. 106: 302–323 [DOI] [PubMed] [Google Scholar]
- 31. Kim M, Huang T, Miller JH. 2003. Competition between MutY and mismatch repair at A:C mispairs in vivo. J Bacteriol. 185: 4626–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim N, et al. 2011. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332: 1561–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kunz BA, et al. 1994. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 318: 1–64 [DOI] [PubMed] [Google Scholar]
- 34. Lasken RS, Goodman MF. 1984. The biochemical basis of 5-bromouracil-induced mutagenesis. Heteroduplex base mispairs involving bromouracil in G x C—-A x T and A x T—-G x C mutational pathways. J. Biol. Chem. 259: 11491–11495 [PubMed] [Google Scholar]
- 35. Lee G, Wolff E, Miller JH. 2004. Mutagenicity of the cytidine analog zebularine in Escherichia coli. DNA Repair (Amst.) 3: 155–161 [DOI] [PubMed] [Google Scholar]
- 36. Leong PM, Hsia HC, Miller JH. 1986. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J. Bacteriol. 168: 412–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu A, et al. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother. 54: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. 1995. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 254: 337–341 [DOI] [PubMed] [Google Scholar]
- 39. Maki H, Sekiguchi M. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355: 273–275 [DOI] [PubMed] [Google Scholar]
- 40. Malinsky J, et al. 2001. The supply of exogenous deoxyribonucleotides accelerates the speed of the replication fork in early S-phase. J. Cell Sci. 114: 747–750 [DOI] [PubMed] [Google Scholar]
- 41. Maruyama M, Horiuchi T, Maki H, Sekiguchi M, Gottesman M. 1983. A dominant (mutD5) and a recessive (dnaQ49) mutator of Escherichia coli. J. Mol. Biol. 167: 757–771 [DOI] [PubMed] [Google Scholar]
- 42. McMurry J, Begley TP. 2005. The organic chemistry of biological pathways. Roberts & Co, Englewood, CO [Google Scholar]
- 43. Miller JH. 1998. Mutators in Escherichia coli. Mutat. Res. 409: 99–106 [DOI] [PubMed] [Google Scholar]
- 44. Miller JH, et al. 2002. Escherichia coli strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics 162: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 46. Miller JH. 2005. Perspective on mutagenesis and repair: the standard model and alternate modes of mutagenesis. Crit. Rev. Biochem. Mol. Biol. 40: 155–179 [DOI] [PubMed] [Google Scholar]
- 47. Modrich P, Lahue R. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133 [DOI] [PubMed] [Google Scholar]
- 48. Nghiem Y, Cabrera M, Cupples CG, Miller JH. 1988. The mutY gene: a mutator locus in Escherichia coli that generates GC—-TA transversions. Proc. Natl. Acad. Sci. U. S. A. 85: 2709–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nick McElhinny SA, et al. 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U. S. A. 107: 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nordenskjold BA, Skoog L, Brown NC, Reichard P. 1970. Deoxyribonucleotide pools and deoxyribonucleic acid synthesis in cultured mouse embryo cells. J. Biol. Chem. 245: 5360–5368 [PubMed] [Google Scholar]
- 51. Nordman J, Wright A. 2008. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc. Natl. Acad. Sci. U. S. A. 105: 10197–10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Donovan GA, Neuhard J. 1970. Pyrimidine metabolism in microorganisms. Bacteriol. Rev. 34: 278–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. 1999. Saccharomyces cerevisiae RNase H (35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 19: 8361–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schaaper RM, Dunn RL. 1987. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc. Natl. Acad. Sci. U. S. A. 84: 6220–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singer M, et al. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taddei F, et al. 1997. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science 278: 128–130 [DOI] [PubMed] [Google Scholar]
- 57. Watson JD, Crick FHC. 1953. Genetical implications of the structure of deoxyribonucleic acid. JAMA 269: 1967–1969 [PubMed] [Google Scholar]
- 58. Watson JD, Crick FHC. 1953. Molecular structure of nucleic acids. Nature 171: 737–738 [DOI] [PubMed] [Google Scholar]
- 59. Wheeler LJ, Rajagopal I, Mathews CK. 2005. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst.) 4: 1450–1456 [DOI] [PubMed] [Google Scholar]
- 60. Wolff E, Kim M, Hu K, Yang H, Miller JH. 2004. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 186: 2900–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamamoto N, et al. 2009. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5: 335–335 [DOI] [PMC free article] [PubMed] [Google Scholar]