Abstract
Diaminopropionate ammonia lyase (DAPAL) is a pyridoxal-5′phosphate (PLP)-dependent enzyme that catalyzes the conversion of diaminopropionate (DAP) to pyruvate and ammonia and plays an important role in cell metabolism. We have investigated the role of the ygeX gene of Escherichia coli K-12 and its ortholog, STM1002, in Salmonella enterica serovar Typhimurium LT2, presumed to encode DAPAL, in the growth kinetics of the bacteria. While Salmonella Typhimurium LT2 could grow on dl-DAP as a sole carbon source, the wild-type E. coli K-12 strain exhibited only marginal growth on dl-DAP, suggesting that DAPAL is functional in S. Typhimurium. The expression of ygeX in E. coli was low as detected by reverse transcriptase PCR (RT-PCR), consistent with the poor growth of E. coli on dl-DAP. Strains of S. Typhimurium and E. coli with STM1002 and ygeX, respectively, deleted showed loss of growth on dl-DAP, confirming that STM1002 (ygeX) is the locus encoding DAPAL. Interestingly, the presence of dl-DAP caused a growth inhibition of the wild-type E. coli strain as well as the knockout strains of S. Typhimurium and E. coli in minimal glucose/glycerol medium. Inhibition by dl-DAP was rescued by transforming the strains with plasmids containing the STM1002 (ygeX) gene encoding DAPAL or supplementing the medium with Casamino Acids. Growth restoration studies using media lacking specific amino acid supplements suggested that growth inhibition by dl-DAP in the absence of DAPAL is associated with auxotrophy related to the inhibition of the enzymes involved in the biosynthetic pathways of pyruvate and aspartate and the amino acids derived from them.
Introduction
Diaminopropionate ammonia lyase (DAPAL) is presumed to be encoded by the ygeX gene (1,170 bp) in Escherichia coli K-12 and STM1002 (1,212 bp) in Salmonella enterica serovar Typhimurium. Nagasawa et al. (12) first reported the expression of DAPAL in various bacterial genera, such as Pseudomonas, Proteus, Salmonella, and Actinomycetes, upon growth on basal medium supplemented with 0.3% (wt/vol) diaminopropionate (DAP), a substrate for this enzyme. They successfully purified the enzyme from crude extracts of Salmonella Typhimurium and demonstrated that it is a pyridoxal 5′-phosphate (PLP)-dependent enzyme. The N-terminal sequence of this enzyme did not show any similarity with other PLP-dependent enzymes. Biochemical studies on the enzyme from Pseudomonas (17) and recombinant DAPAL from Escherichia coli (16) have shown that it catalyzes the α,β-elimination reaction with both l- and d-DAP to give pyruvate and ammonia. DAPAL can also eliminate the hydroxy group from the β carbon of d-serine, although poorly.
In an earlier study, we reported the cloning, overexpression, purification, and characterization of recombinant DAPAL enzymes from E. coli and S. Typhimurium (8). The overexpressed DAPAL from S. Typhimurium was found to be more active than that from E. coli. The kinetic parameters for the reaction with the substrates have been determined. However, the functional role of this enzyme in these organisms is not clear, as the natural substrate for DAPAL is not known.
In this study, the effect of the expression of the ygeX and STM1002 genes encoding putative DAPAL in E. coli and S. Typhimurium on the growth and bacterial physiology was investigated. It was observed that wild-type S. Typhimurium could grow on minimal medium containing dl-DAP as the sole carbon source, while wild-type E. coli showed only weak growth. Further expression analysis revealed that the poor growth of E. coli was due to the very low expression of DAPAL, insufficient for the degradation of dl-DAP.
These studies were extended further using strains of S. Typhimurium and E. coli in which STM1002 and ygeX were disrupted. The inhibition of growth of wild-type E. coli and the null strains by dl-DAP was shown to be due to the inability of these strains to degrade DAP, which remains toxic. However, the toxicity could be rescued either by complementation with plasmids containing STM1002 (ygeX) genes or by the addition of amino acids that could overcome the inhibition of specific metabolic pathways caused by dl-DAP.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Generation of the ygeX and STM1002 gene disruptions and cloning of the genes from E. coli K-12 and S. Typhimurium LT2 are described below.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Source |
|---|---|---|
| E. coli | ||
| K-12 | F− ΔlacX74 thi-1 | 10 |
| DH5α | F′ endA1 hsdR17(rK− mK+) glnV44 thi-1 recA1 gyrA (Nalr) relA1(ΔlacZYA argF) U169 deoR Φ80dlac(ΔlacZ)M15 | 18 |
| MG1655 | F− rph-1 ilvG rfb-50 rph-1 | 6 |
| HSS-EC | ygeX::cat | This work |
| HSS-EK | ygeX::kan | This work |
| BW25141 | lacIq rrnBT14 lacZWJ16 ΔphoBR580 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 galU95 end-ABT333 uidA(ΔMluI)::pir+ recA1 | 2 |
| BW25113 | lacIq rrnBT14 lacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 2 |
| S. Typhimurium | ||
| LT2 | IFO and IAM type culture collections, University of Tokyo, Tokyo | 12 |
| HSS-SC | STM1002::cat | This work |
| Plasmids | ||
| pKD3 | oriR6Kγ bla(Apr) cat rgnB(Ter) | 2 |
| pKD4 | oriR6Kγ bla(Apr) kan rgnB(Ter) | 2 |
| pKD46 | araBp-gam-bet-exo bla(Apr) repA101(Ts) ori101 | 2 |
| pACDH | Plac followed by MCS, pACYC origin of replication | 13 |
Media, chemicals, and reagents.
Media and bacteriological reagents were primarily from HiMedia, India. Other chemicals and reagents, including oligonucleotides, were purchased from Sigma-Aldrich. Diaminopropionate (dl-DAP), pyruvate, oxaloacetate, phosphoglyceric acid, phosphoenolpyruvate, lactate dehydrogenase (LDH), and reduced nicotinamide adenine dinucleotide (NADH) were purchased from Sigma. Restriction enzymes were purchased from GE (USA) and MBI Fermentas and used as per the specifications of the manufacturers. Chloramphenicol-, kanamycin-, and tetracycline-resistant transformants were selected on LB medium containing the respective antibiotic (25 μg/ml). Recovery of the E. coli ygeX and S. Typhimurium STM1002 null strains from dl-DAP toxicity was checked on M9 minimal medium containing 0.3% (wt/vol) of glucose, sucrose, mannose, fructose, arabinose, and Casamino Acids. l-Amino acids and l-ornithine were added in final concentrations ranging from 1 to 100 μg/ml. Restriction endonucleases, Taq polymerase, and murine leukemia virus (MuLV) reverse transcriptase (RT) were obtained from Fermentas.
Multiple sequence alignment of DAPAL sequences.
The amino acid sequence of DAPAL from E. coli K-12 was used for NCBI Protein BLAST to identify DAPAL sequences present in the protein database. The sequences obtained were used for CLUSTAL W multiple sequence alignment.
Construction of mutants of E. coli and S. Typhimurium with ygeX and STM1002 deleted.
The ygeX and STM1002 null strains of E. coli and S. Typhimurium, respectively, were generated by homologous recombination based on the procedure of Datsenko and Wanner (2). Hybrid primers with homology to the ygeX (STM1002) gene were constructed with sequences complementary to the chloramphenicol and kanamycin resistance genes present on the plasmids pKD3 and pKD4, respectively. The PCR products generated were used to replace the ygeX (STM1002) gene in E. coli/S. Typhimurium using the helper plasmid pKD46 expressing the phage lambda red recombinase (11).
Primers used for ygeX knockout in E. coli were as follows: forward primer, 5′ ATGTCCGTTTTCTCATTGAAGATTGATATCGCCGATGTGTAGGCTGGAGCTGCTTCG 3′; reverse primer, 5′ TTAAGGTGCTACAGCGTGTTTGCCTTCCCAGACAACCATATGAATATCCTCCTTA 3′. Primers used for STM1002 knockout in S. Typhimurium were as follows: forward primer, 5′ ATGCATGAGCTTATTAAATATCAGTTTAATACACGTGTGTAGGCTGGAGCTGCTTCG 3′; reverse primer, 5′ TTAAGCACTGCGTCCGTTCAGACTATATCTTCATACATATGAATATCCTCCTTAC 3′. Nucleotides represented in italics are the 36-nucleotide (nt) flanking sequences of ygeX and STM1002. PCR was carried out using Taq polymerase, and the conditions used were 94°C for 3 min for initial denaturation, followed by 30 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 15 min.
Cloning of the STM1002 (ygeX) gene.
The 1.2-kb STM1002 (ygeX) gene was amplified from S. Typhimurium/E. coli genomic DNA by using the forward primer and reverse primers carrying SacI and EcoRI restriction sites, respectively, as follows: STM1002 sense primer, 5′ CGAGCTCATGCATGAGCTTATTA 3′; STM1002 antisense primer, 5′ GGAATTCTTAAGCACTGCGTCCG 3′; ygeX sense primer, 5′ CGAGCTCATGTCCGTTTTCTCATTG 3′; ygeX antisense primer, 5′ GGAATTCTTAAGGTGCTACAGCGTG 3′.
The sequences in bold represent the restriction sites for SacI and EcoRI. The PCR conditions used were 94°C for 5 min for initial denaturation, the next 30 cycles at 94°C for 1 min, 54°C for 1.3 min, and 72°C for 2 min, and a final extension at 72°C for 15 min. The 1.2-kb amplified fragment was digested with SacI and EcoRI, gel purified, and ligated at SacI/EcoRI sites downstream to the inducible Plac promoter of the low-copy-number pACDH vector.
Monitoring bacterial growth.
The glycerol stock of the bacterial strains was used to streak LB agar plates with or without antibiotic, and the plates were incubated for 12 h at 37°C. A single colony was picked from the plate, inoculated into 3 ml LB broth, and incubated for 8 to 10 h (optical density at 600 nm [OD600], 0.8). An equal volume (100 μl) of the culture was taken and centrifuged to separate the broth. The pellet was dried to remove the supernatant, resuspended in M9 minimal medium, and then inoculated into 50 or 100 ml of M9 medium containing MgSO4, CaCl2, thiamine supplemented with glycerol, and DAP or in DAP alone. The absorbance at 600 nm was monitored every 1 h to determine the growth of the bacteria.
Expression of DAPAL in vivo.
S. Typhimurium and E. coli strains were cultured in M9 minimal medium containing 0.4% (vol/vol) glycerol for 2 to 3 h, and 0.3% (wt/vol) dl-DAP was added to the culture medium and further grown for 5 to 6 h at 37°C. Cells were then harvested at 4°C; the pellet was resuspended in extraction buffer containing 50 mM potassium phosphate, pH 7.4, 1 mM EDTA, 1 mM β-mercaptoethanol, and 100 μM PLP. The cells were lysed at 4°C by sonication, and the debris was pelleted by centrifugation. The soluble crude extract (3 mg of total protein) was loaded onto the SDS-PAGE gel. The fractionated proteins were transferred to a nitrocellulose membrane for Western blot analysis.
Western blot analysis.
The total soluble extract samples of E. coli and S. Typhimurium were subjected to 12% SDS-PAGE, and the separated proteins were transferred onto a nitrocellulose membrane using the transfer apparatus at 75 V for 1 h. The membrane was then blocked using 5% skimmed milk in 1× phosphate-buffered saline (PBS) for 1 h. Polyclonal antibody raised against recombinant E. coli DAPAL (1:5,000) in 1× PBS was added to the membrane and left for 1 h. The membrane was washed thrice with high-salt buffer for 10 min each, followed by washing with low-salt buffer. Secondary antibody (goat anti-rabbit IgG, 1:10,000) in PBS was added, and the membrane was incubated for 1 h. The membrane was developed using a 10-ml solution of diaminobenzidine (DAB) and H2O2 in 1× citrate buffer (pH 4.8).
Total mRNA isolation.
Overnight-grown bacterial culture (10 ml) was pelleted at 7,000 rpm for 10 min, and the pellet was resuspended in 50 to 100 μl of autoclaved water. Twice the volume of TRIzol (Sigma) was added and vortexed. This was followed by the addition of two volumes of chloroform and thorough mixing. The mix was centrifuged at 13,000 rpm for 10 min at 4°C. The upper aqueous layer was transferred into a fresh diethyl pyrocarbonate (DEPC)-treated tube. An equal volume of isopropanol was added, and the tube was centrifuged at 10,000 rpm for 10 min. The supernatant was discarded, and the pellet was dried for 5 to 10 min. The pellet was resuspended in DEPC-treated water and stored in −80°C for further use.
RT-PCR.
Total mRNA (5 μl) isolated as described above was mixed with 2 μl of a 20 pM gene-specific antisense primer and heated at 72°C for 10 min. The mix was transferred to ice for 5 min immediately. RNase inhibitor (20 units) was added to the mix, followed by 1× MuLV RT buffer and 1 mM deoxynucleoside triphosphate (dNTP) mix. MuLV RT enzyme (1 μl) was added, and the mix was incubated at 37°C for 5 min and then shifted to 42°C for 1 h to obtain the cDNA. The reaction mixture incubated in the absence of the enzyme served as a negative control.
The cDNA obtained was used for PCR using gene-specific forward and reverse primers. The conditions for PCR were as follows: initial denaturation, 98°C for 1 min; denaturation, 98°C for 30 s; annealing, 55°C for 30 s; and elongation, 72°C for 1 min for 30 cycles. The PCR products were analyzed by agarose gel electrophoresis followed by staining with ethidium bromide (EtBr).
Enzyme assays.
The α,β-elimination reaction of DAPAL with dl-DAP as the substrate was assayed by the following method. The amount of pyruvate formed during the reaction was measured spectrophotometrically by coupling the reaction to the LDH reaction. The reaction was performed at 37°C in 1 ml of a reaction mixture consisting of 50 mM potassium phosphate buffer (pH 7.4), 1 mM dl-DAP, 5 units of LDH, 0.25 μM NADH, and 50 μg or 100 μg protein from crude extracts of cells grown in the presence or absence of 0.3% (wt/vol) of dl-DAP. The assay performed without the addition of substrate dl-DAP served as a negative control to rule out residual pyruvate content in the crude extract. The reaction was started by the addition of the substrate (dl-DAP), and the decrease in absorbance at 340 nm due to the consumption of NADH was monitored (16). The molar absorption coefficient of NADH (340 nm), 6,220 M−1 cm−1, was used to calculate the activity of the enzyme. One unit of enzyme activity was defined as the amount of enzyme required to catalyze the release of 1 μmol of pyruvate min−1 at 37°C and pH 7.4.
RESULTS
Bioinformatic analysis.
The ygeX gene of E. coli and the STM1002 gene of S. Typhimurium encode DAPAL enzymes composed of 398 and 404 amino acids, respectively. Figure 1 shows the organization of the ygeX locus in E. coli K-12 and the STM1002 locus in S. Typhimurium LT2. It is evident that the context of the genes is significantly different in the two organisms, although the deduced amino acid sequences from both organisms showed 50% identity. The identity between E. coli and S. Typhimurium orthologs is generally around 95% (Table 2). There was also no similarity in the upstream or downstream sequences of the STM1002 (ygeX) genes. It was noticed that the gene upstream to ygeX in E. coli encodes a putative carbomyl transferase in the sense orientation while that in S. Typhimurium encodes asparginyl tRNA synthase in the antisense orientation. The genes downstream to ygeX and STM1002 did not show any similarity either.
Fig 1.
The genome organization corresponding to the DAPAL region was obtained from the genome databases for S. Typhimurium (A) (www.ncbi.nlm.nih.gov/nuccore/NC_003197) and E. coli (B) (www.ncbi.nlm.nih.gov/nuccore/NC_000913).
Table 2.
Percent identity of DAPAL amino acid sequences with the sequence from E. coli K-12
| Organism | % Identitya |
|
|---|---|---|
| DAPAL | GAPDH | |
| E. coli O157:H7 | 100 | 100 |
| Shigella sonnei | 99 | 100 |
| Enterobacter cloacae | 84 | 93 |
| Edwardsiella tarda | 80 | 90 |
| Vibrio shilonii | 68 | 85 |
| Acinetobacter baumannii | 53 | 88 |
| Clostridium botulinum | 50 | 78 |
| Salmonella Typhimurium | 50 | 98 |
EC number of DAPAL, 4.3.1.15; EC number of GAPDH, 1.2.1.12.
The amino acid sequence of the putative E. coli DAPAL protein was used to identify sequences similar to DAPAL from the genome database using the NCBI BLAST tool. A total of 154 sequences similar to DAPAL were identified from different prokaryotes and none from eukaryotes. It was also observed that this gene was predominantly found in pathogenic bacteria and extremophiles. Table 2 lists the top 7 organisms that showed the highest identity with the E. coli DAPAL sequence. It is evident that the DAPAL amino acid sequences show greater variability than the amino acid sequence of a housekeeping enzyme such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Table 2), which was 80 to 90% similar in all the organisms considered. It was therefore of interest to understand the functional status of DAPAL in these two organisms.
Growth of E. coli and S. Typhimurium on dl-DAP as a carbon source.
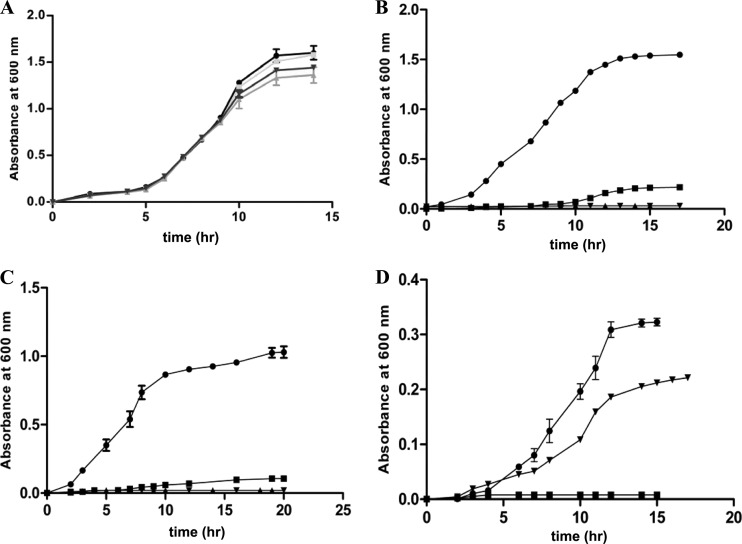
As a first step toward understanding the role of DAPAL, the growth of wild-type E. coli K-12 and S. Typhimurium LT2 in LB medium was monitored in the absence or presence of 0.3% (wt/vol) dl-DAP, and no difference in growth was observed (data not shown). When the strains were grown in M9 minimal medium containing 0.4% (vol/vol) glycerol, both strains exhibited growth, indicating that they could utilize glycerol as a carbon source (Fig. 2A). However, when the strains were grown in M9 minimal medium containing 0.3% (wt/vol) dl-DAP in addition to glycerol or 0.3% (wt/vol) dl-DAP alone, there was only marginal growth in E. coli K-12 whereas S. Typhimurium LT2 could utilize dl-DAP as a sole source of carbon and grow efficiently (Fig. 2B and C). The poor growth of E. coli K-12 on glycerol in the presence of dl-DAP suggested that the expression of DAPAL was probably low to metabolize dl-DAP and prevent DAP-mediated toxicity.
Fig 2.
Growth of strains in M9 minimal medium. (A) Growth of S. Typhimurium LT2 (●), the STM1002 null strain (▼), E. coli K-12 (■), and the ygeX null strain (▲) in M9 minimal medium with 0.4% (vol/vol) glycerol. (B) Growth of S. Typhimurium LT2 (●), the STM1002 null strain (▲), E. coli K-12 (■), and the ygeX null strain (▼) in M9 minimal medium with 0.4% (vol/vol) glycerol plus 0.3% (wt/vol) dl-DAP. (C) Growth of S. Typhimurium LT2 (●), the STM1002 null strain (▲), E. coli K-12 (■), and the ygeX null strain (▼) in M9 minimal medium with 0.3% (wt/vol) dl-DAP. (D) Growth of E. coli K-12 in M9 medium with glycerol (0.4% [vol/vol]) supplemented with 0.3% (wt/vol) l-DAP (●), 0.3% (wt/vol) d-DAP (■), and 0.3% (wt/vol) dl-DAP (▼). The graphs represent the average results of three individual experiments with standard errors. The standard error in some cases was <0.1 and hence is not visible in the graph.
It was shown earlier that the enzyme DAPAL could allow utilization of both d- and l-DAP as the substrate (7, 15). In the experiments described thus far, dl-DAP was used for induction of DAPAL. Hence, it was important to monitor the effect of the stereoisomers on the growth of the bacteria. Both wild-type E. coli and S. Typhimurium strains were grown in M9 minimal medium supplemented with either l- or d-DAP in the presence of glycerol (Fig. 2D). As observed earlier, S. Typhimurium exhibited no difference in its growth characteristics in the presence of either d- or l-DAP (data not shown). However, E. coli displayed a distinct difference in growth with l- and d-DAP. While the growth was slightly better in l-DAP than in dl-DAP, no growth was seen when only d-DAP was used.
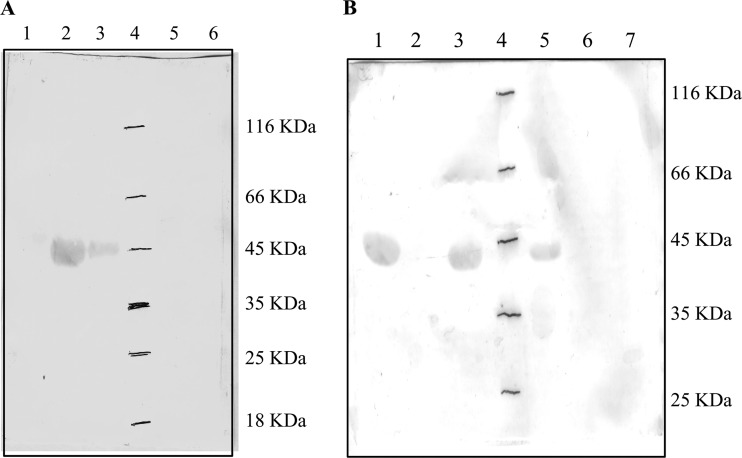
The growth patterns observed above indicated that d- and l-DAP were differentially utilized by the two organisms. In order to compare the expression of DAPAL, E. coli and S. Typhimurium were grown in minimal medium containing 0.4% (vol/vol) glycerol for 3 to 4 h. After the addition of 0.3% (wt/vol) dl-DAP to the medium, the strains were allowed to grow further for 4 to 5 h. Crude extracts of the cultures were subjected to Western blot analysis using polyclonal antibodies raised against recombinant DAPAL. A distinct band at 44 kDa, corresponding to the molecular mass of the overexpressed and purified E. coli DAPAL (eDAPAL) (Fig. 3A, lane 2), was seen in the crude extracts of S. Typhimurium LT2 (Fig. 3A, lane 3), while DAPAL was not detectable in the crude extracts of E. coli K-12 (Fig. 3A, lane 1). Since E. coli K-12 did not show the detectable presence of DAPAL, expression of the enzyme was tested in two other strains of E. coli K-12. Crude extracts from the strains DH5α and MG1655 also failed to show detectable DAPAL expression. The specific activity of DAPAL was 6.83 ± 0.4 U/mg in crude extracts of S. Typhimurium LT2 grown in the presence of 0.3% (wt/vol) dl-DAP, whereas it was not measurable in E. coli K-12.
Fig 3.
(A) Comparison of the expression of DAPAL in bacteria grown in minimal medium containing 0.4% (vol/vol) glycerol and 0.3% (wt/vol) dl-DAP by Western blot analysis using anti-DAPAL antibody. Lane 1, crude extract of E. coli K-12; lane 2, overexpressed eDAPAL; lane 3, crude extract of S. Typhimurium LT2; lane 4, unstained protein molecular mass marker; lane 5, crude extract of STM1002 null strain of S. Typhimurium; lane 6, crude extract of ygeX null strain of E. coli. (B) Western blot analysis of null strains of E. coli and S. Typhimurium transformed with pACDH-ygeX and grown in minimal medium containing 0.4% (vol/vol) glycerol and 0.3% dl-DAP (wt/vol). Lane 1, overexpressed eDAPAL; lane 2, STM1002 null strain plus pACDH-ygeX uninduced; lane 3, STM1002 null strain plus pACDH-ygeX induced with 0.5 mM IPTG; lane 4, unstained protein molecular mass marker; lane 5, ygeX null strain plus pACDH-ygeX induced with 0.5 mM IPTG; lane 6, ygeX null strain plus pACDH-ygeX uninduced; lane 7, ygeX null strain plus pACDH vector alone.
Phenotypic studies of E. coli K-12 ygeX and S. Typhimurium STM1002 knockouts.
In order to understand the role of the ygeX and STM1002 genes in the growth of E. coli and S. Typhimurium, respectively, strains carrying null alleles of ygeX and STM1002 were generated by homologous recombination using the phage λ recombination system (see Materials and Methods). The knockout strains of both organisms were grown under conditions similar to those used for the wild-type strains. As observed in the case of the wild-type strain, the E. coli knockout strain could grow on LB medium in the absence or presence of 0.3% (wt/vol) dl-DAP (data not shown) and in M9 minimal medium containing 0.4% (vol/vol) glycerol (Fig. 2A) but failed to grow in M9 minimal medium containing dl-DAP as the sole carbon source (Fig. 2C). This lack of growth could not be rescued by providing glycerol or glucose in the medium (Fig. 2B), as observed in the case of the wild-type E. coli strain, confirming the inhibitory effect of dl-DAP.
Like the E. coli strain, the STM1002 null strain of S. Typhimurium LT2 also did not show any marked difference in growth in LB medium in the absence or presence of 0.3% (wt/vol) dl-DAP (data not shown). However, there was a distinct growth difference in M9 minimal medium containing 0.3% (wt/vol) dl-DAP as a sole carbon source. While wild-type S. Typhimurium LT2 could grow utilizing dl-DAP, the STM1002 null strain could not grow in the presence of dl-DAP as the sole carbon source (Fig. 2C) and growth was not rescued by the addition of glycerol to the medium (Fig. 2B). The phenotype of the STM1002 null strain was similar to the phenotype of ygeX null mutant strains of E. coli. The crude extracts of the STM1002 null strain did not exhibit DAPAL activity, and no band corresponding to DAPAL was observed in Western blots (Fig. 3A, lanes 5 and 6). These results indicate that the wild-type S. Typhimurium LT2 strain has a functional DAPAL encoded by the STM1002 locus, deletion of which results in the loss of DAPAL activity. The wild-type E. coli strains and the null strains do not have measurable DAPAL activity and are unable to utilize glycerol due to DAP-induced toxicity.
To test whether the ygeX (STM1002) loci are transcriptionally active, the total mRNA from all the strains grown in minimal medium containing glycerol in the presence and absence of dl-DAP was isolated. RT-PCR was carried out using the reverse primer specific to either eDAPAL or sDAPAL. The PCR carried out with cDNA using gene-specific primers suggested that the DAPAL gene did not get transcribed when the strains were grown in medium with glycerol, as no band of corresponding size was observed (data not shown). The 16S rRNA PCR product was used as a loading control. On the other hand, when the strains were grown on medium with dl-DAP, a band corresponding to the expected size (∼1 kb) was observed in S. Typhimurium and a very faint band was noticed in E. coli (Fig. 4B). As expected, RT-PCR products were not observed in ygeX (STM1002) null strains. These results suggest that the RNA for DAPAL is induced by the presence of DAP. The levels of DAPAL transcript in E. coli are much too low, and therefore it is unable to overcome the DAP-mediated inhibition of growth.
Fig 4.
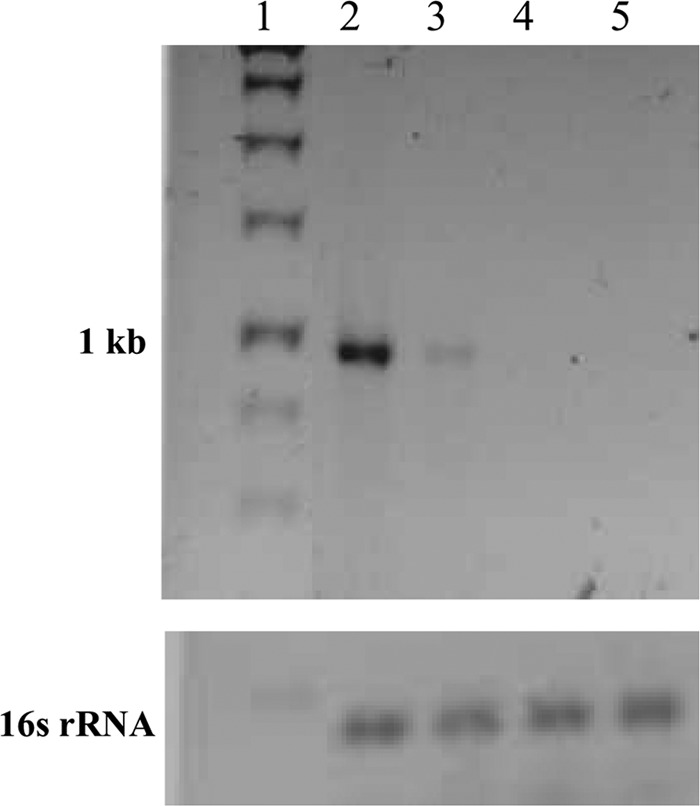
RT-PCR using total mRNA isolated from E. coli, S. Typhimurium, ygeX null strain of E. coli, and STM1002 null strain of S. Typhimurium grown in minimal medium plus glycerol (0.4% [vol/vol]) plus dl-DAP (0.3% [wt/vol]) Top, PCR using primers for gene encoding DAPAL; bottom, PCR using primers for 16S rRNA. Lane 1, 1-kb ladder; lane 2, S. Typhimurium; lane 3, E. coli; lane 4, STM1002 knockout of S. Typhimurium; lane 5, ygeX knockout of E. coli.
Complementation studies using STM1002 and ygeX on low-copy-number plasmids.
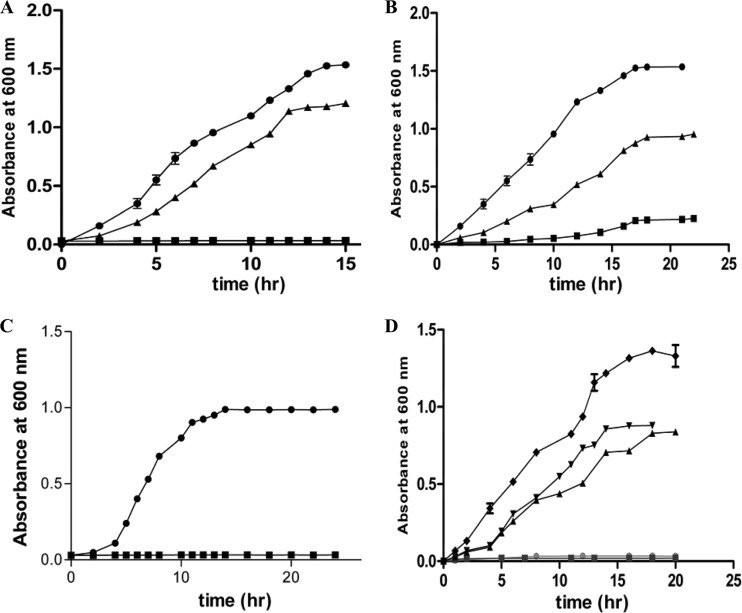
From the results obtained thus far, it is evident that the wild type and the ygeX null mutant of E. coli as well as the STM1002 null strain of S. Typhimurium show identical growth properties in the presence of dl-DAP in minimal medium. Since the E. coli ygeX gene was overexpressed earlier, it was of interest to monitor the growth of these strains when the gene encoding DAPAL was present in trans. The wild-type S. Typhimurium STM1002 gene was cloned in a low-copy vector, pACDH, under the lac promoter and was used to transform the STM1002 null strain of S. Typhimurium, wild-type E. coli K-12, and the ygeX null strain of E. coli K-12. The transformants were screened on LB agar plates containing tetracycline. The strains were grown in minimal medium with 0.4% (vol/vol) glycerol for 3 to 6 h (until the OD600 reached 0.4). DAPAL expression was induced by the addition of 0.5 mM IPTG, and the cultures were allowed to grow for 1 h. The medium was then supplemented with 0.3% (wt/vol) dl-DAP, and growth was monitored after the addition of dl-DAP (Fig. 5). The STM1002 null strain of S. Typhimurium harboring the plasmid carrying the STM1002 gene exhibited growth kinetics similar to those of the wild-type strain in the presence of 0.3% (wt/vol) dl-DAP and 0.4% (vol/vol) glycerol (Fig. 5A). Similarly, the inhibition of E. coli K-12 growth by dl-DAP was rescued by transformation with pACDH-STM1002 (Fig. 5B). The growth inhibition in the presence of dl-DAP of the ygeX null mutant of E. coli was also rescued posttransformation with pACDH-STM1002 (Fig. 5C). In the presence of 0.3% (wt/vol) dl-DAP and 0.4% (vol/vol) glycerol, the time taken for attaining stationery phase was similar to that of E. coli K-12 (Fig. 5 B,C). The ability of E. coli transformants expressing STM1002 to grow on dl-DAP confirms that E. coli can transport DAP efficiently.
Fig 5.
Complementation studies using the STM1002 (ygeX) gene in the pACDH vector. (A) Growth of S. Typhimurium LT2 (●) and the STM1002 null strain transformed with the pACDH vector alone (■) and the STM1002 null strain transformed with the pACDH-STM1002 plasmid (▲) in M9 minimal medium with 0.4% (vol/vol) glycerol plus 0.3% (wt/vol) dl-DAP and 0.5 mM IPTG. (B) Growth of E. coli K-12 transformed with the pACDH vector alone (■), E. coli K-12 transformed with the pACDH-STM1002 plasmid (▲), and S. Typhimurium in M9 minimal medium with 0.4% (vol/vol) glycerol plus 0.3% dl-DAP and 0.5 mM IPTG (●). (C) Growth of the ygeX null mutant of E. coli transformed with the pACDH vector alone (■) and the pACDH-STM1002 plasmid in M9 minimal medium with 0.4% (vol/vol) glycerol and 0.3% (wt/vol) dl-DAP and 0.5 mM IPTG (●). (D) Growth of the ygeX null mutant of E. coli with the pACDH vector alone (●), the STM1002 null strain of S. Typhimurium with the pACDH alone (■), the STM1002 null strain of S. Typhimurium transformed with the pACDH-ygeX plasmid postinduction with 0.5 mM IPTG (▲), the ygeX null mutant of E. coli with the pACDH-ygeX plasmid (▼) postinduction with 0.5 mM IPTG, and wild-type S. Typhimurium LT2 in M9 minimal medium containing 0.4% (vol/vol) glycerol and 0.3% (wt/vol) dl-DAP (◆). The graphs represent the average results of three individual experiments with standard errors. The standard error in some cases was <0.1 and hence is not visible in the graph.
Complementation was also carried out by transforming the plasmid pACDH-ygeX harboring the ygeX gene from E. coli into ygeX (STM1002) null strains. The transformants were grown on minimal medium with 0.3% (wt/vol) dl-DAP in the presence of glycerol (0.4% vol/vol), and the expression of ygeX was induced by the addition of 0.5 mM IPTG. The growth of STM1002 (ygeX) null strains of S. Typhimurium/E. coli was restored upon induction with IPTG (Fig. 5D). We could also observe activities of 0.9 ± 0.06 μmol/min/mg and 0.37 ± 0.13 μmol/min/mg of the total protein in the null strains of S. Typhimurium and E. coli, respectively, postinduction with IPTG. Expression of ygeX was also confirmed using Western blot analysis (Fig. 3B), which suggests that the expression levels are similar in the two strains. These results indicate that ygeX from E. coli can be expressed under an inducible promoter, resulting in the production of functional DAPAL.
These results confirm that the genes encoding DAPAL from E. coli and S. Typhimurium are functional and can act in trans in E. coli K-12 and Salmonella strains and protect the cells from dl-DAP toxicity in the absence of the chromosomally encoded DAPAL.
DAP-mediated inhibition is not due to inhibition of sugar uptake.
Since the growth inhibition of wild-type E. coli K-12, as well as that of the ygeX (STM1002) null mutants of E. coli/S. Typhimurium, by dl-DAP was not rescued upon supplementing the minimal medium with glycerol, the dl-DAP-induced growth inhibition might be acting at the level of blocking glycerol uptake. To address this possibility, we used a series of carbon sources such as glucose, mannose, maltose, fructose, galactose, and arabinose. Different sugars were chosen to check whether sugar uptake was blocked in the presence of dl-DAP, as the sugars chosen have specific transporters in the cell. The DAP-mediated inhibition was not rescued by any of the sugars tested (data not shown), indicating that the dl-DAP-mediated inhibition of growth is not likely to be at the level of blocking carbohydrate uptake.
Rescue of dl-DAP-mediated inhibition of growth in the presence of amino acids.
As the next step, we tested the possibility of dl-DAP inhibiting amino acid pathways by monitoring growth after supplementing the minimal medium containing dl-DAP and glycerol with a mixture of all amino acids (Casamino Acids). The strains were cultured in M9 minimal medium containing 0.3% (wt/vol) dl-DAP and 0.4% (wt/vol) Casamino Acids. Growth of all strains could be rescued in the presence of 0.4% (wt/vol) Casamino Acids (Fig. 6A and B), suggesting that inhibition by dl-DAP is exerted at the level of amino acid metabolism when DAP is not degraded by DAPAL.
Fig 6.
Growth of bacteria in minimal medium containing dl-DAP (0.3% [wt/vol]). (A) E. coli K-12 (●) and the ygeX null strain of E. coli (■) in the presence of Casamino Acids. (B) S. Typhimurium LT2 (●) and the STM1002 null strain in the absence (■) and presence (▲) of Casamino Acids.
Identification of specific amino acids involved in the rescue of dl-DAP-mediated inhibition.
Since the growth inhibition was rescued in the presence of Casamino Acids, we attempted to identify the specific amino acid(s) involved in the rescue of growth in the presence of dl-DAP. The growth properties of all the strains were tested in M9 medium devoid of defined sets of amino acids (Table 3). By sequential elimination of amino acids from the mixture, a set of eight amino acids—isoleucine, leucine, threonine, asparagine, methionine, arginine, glutamic acid, and cysteine–-were found to be essential for maximal recovery of growth in the presence of dl-DAP (Table 3). In the absence of the set of eight amino acids, the bacterial strains failed to grow in the presence of dl-DAP (Table 3), suggesting that dl-DAP might have an inhibitory effect on the biosynthetic pathways of the identified amino acids.
Table 3.
OD600 values of S. Typhimurium and E. coli strains in the absence of various sets of amino acids
| Omitted amino acidsa | Final OD600b |
|||
|---|---|---|---|---|
| S. Typhimurium | E. coli | ygeX knockout | STM1002 knockout | |
| C, S, M, T, N, Q | 1.5 ± 0.22 | 0.73 ± 0. 11 | 0.68 ± 0. 07 | 0.52 ± 0. 05 |
| C, M, T, N, I | 1.5 ± 0.16 | 0.75 ± 0.05 | 0.70 ± 0.03 | 0.53 ± 0.03 |
| C, D, R, V, A | 1.5 ± 0.21 | 0.99 ± 0.04 | 0.97 ± 0.06 | 0.87 ± 0.06 |
| C, D, R, L, E | 1.5 ± 0.21 | 0.44 ± 0.05 | 0.31 ± 0.042 | 0.29 ± 0.02 |
| C, R, M, T, N, L, I | 1.5 ± 0.14 | 0.08 ± 0.03 | 0.058 ± 0.003 | 0.052 ± 0.004 |
| L, E, C, M, R, N, T, I | 1.5 ± 0.2 | 0.01 ± 0.006 | 0.01 ± 0.002 | 0.01 ± 0.004 |
| All amino acids added to medium | 1.5 ± 0.18 | 1.32 ± 0.18 | 1.28 ± 0.30 | 0.95 ± 0.25 |
| Eight amino acids added to mediumc | 1.42 ± 0.1 | 1.3 ± 0.16 | 1.2 ± 0.25 | 0.9 ± 0.2 |
Medium contained 0.4% (vol/vol) glycerol and 0.3% (wt/vol) DL-DAP and was subjected to alterations of amino acids as shown.
Final OD600 (after 12 h) ± standard error.
Isoleucine, leucine, threonine, aspargine, methionine, arginine, glutamic acid, and cysteine.
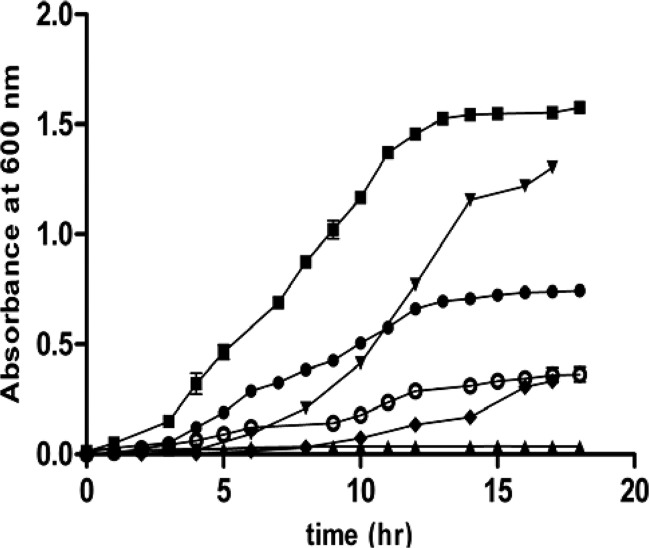
The identified eight amino acids are derived from intermediates associated with glycolysis. Since the growth restoration was observed in the presence of these amino acids, it was relevant to check for the effect of the intermediates in the glycolysis pathway. The wild-type E. coli alone was used to study the growth pattern, as it had no antibiotic selection constraints. Growth was monitored in M9 minimal medium containing 0.4% (vol/vol) glycerol and dl-DAP (0.3% [wt/vol]) and supplemented with various glycolytic intermediates. To check if pyruvate synthesis influences the growth pattern, we monitored the growth in the presence of 0.3% (wt/vol) pyruvate. The presence of pyruvate could rescue the growth marginally, suggesting that the synthesis of pyruvate might be the target of DAP-mediated inhibition. To further identify the steps in glycolysis which are arrested due to DAP, 3-phosphoglyceric acid (PGA) and phosphoenolpyruvate (PEP) were used as supplements. There was no rescue with PGA in the medium, whereas marginal rescue similar to that observed with pyruvate was seen in the presence of PEP. This observation suggests that indeed the upstream steps involved in pyruvate synthesis, especially after the formation of 3-PGA, might be the targets of DAP-induced toxicity. Since providing pyruvate resulted in minimal growth, we checked the growth in the presence of downstream intermediates like oxaloacetic acid (OAA) and aspartate (Fig. 7). Individual addition of these intermediates did not show rescue of growth. However, addition of pyruvate along with these intermediates resulted in convincing rescue of bacterial growth, suggesting that apart from inhibition of pyruvate synthesis, dl-DAP also inhibits synthesis of amino acids, especially those derived from pyruvate and aspartate. Since cysteine and glutamic acid are derived from other intermediates of glycolysis, using them as supplements in addition to pyruvate and aspartate resulted in maximum recovery of growth (Fig. 7).
Fig 7.
Recovery of growth in the presence of metabolic intermediates. Growth of wild-type E. coli in the minimal medium containing glycerol (0.4% vol/vol) and dl-DAP (0.3% wt/vol) supplemented with 0.3% (wt/vol) phosphoglyceric acid (▲), 0.3% (wt/vol) phosphoenolpyruvate (◆), 0.3% (wt/vol) pyruvate (○), 0.3% (wt/vol) pyruvate plus 0.3% (wt/vol) aspartate (●), 0.4% (wt/vol) pyruvate plus 0.3% (wt/vol) aspartate plus cysteine (100 μg/ml) plus glutamic acid (100 μg/ml) (▼), and 0.4% (wt/vol) Casamino Acids (■). The graphs represent the average results of three individual experiments with standard errors. The standard error in some cases was <0.1 and hence is not visible in the graph.
DISCUSSION
The results presented in this report show that although E. coli K-12 possesses an intact ygeX gene that encodes DAPAL and that shares 50% amino acid identity with the enzyme from S. Typhimurium (Table 2 and Fig. 1), different strains of E. coli tested could not efficiently utilize dl-DAP when it was provided as the sole source of carbon (Fig. 2 and 3A). On the other hand, the growth of S. Typhimurium under the same condition indicates that it can efficiently utilize dl-DAP as a sole source of carbon and the utilization of dl-DAP was dependent on the expression of DAPAL, which was not detectable in E. coli (Fig. 3A). Further, the toxicity induced due to the presence of dl-DAP in E. coli could be rescued by the introduction of functional DAPAL expressed from the S. Typhimurium STM1002 gene (Fig. 5A, B, and C). Interestingly, the ygeX gene of E. coli when expressed in trans using a heterologous promoter in ygeX (STM1002) null strains could rescue them from dl-DAP toxicity (Fig. 5D), suggesting that the ygeX gene in E. coli is intact but is expressed at very low levels and hence does not support the growth of the bacteria in dl-DAP. This was confirmed by RT-PCR analysis which showed the presence of a low level of product of approximate size even from the total RNA extracts of E. coli grown in the presence of DAP and glycerol (Fig. 4). It is possible that the ygeX gene of E. coli is expressed under a different set of conditions that are unknown.
Earlier reports have shown that E. coli and S. Typhimurium carry genes that require different conditions for their induction (9). A review (4) suggests that the regulation of certain genes involved in oxidative-stress response is different in E. coli and S. Typhimurium. It was observed that the MnSOD gene is induced in response to H2O2 levels in S. Typhimurium whereas there was no significant induction of the gene in E. coli under the same condition (1, 15). It was also shown that the ahpF gene is induced by heat shock in S. Typhimurium and the same gene cannot be induced by heat shock in E. coli or when introduced into S. Typhimurium (14). The position of the gene in S. Typhimurium is nt 1093858, while that in E. coli is at nt 3005532. E. coli encodes for preprotein translocase at the position of the DAPAL locus in S. Typhimurium, while S. Typhimurium encodes membrane-bound lytic murein transcarboxylase at the position of the DAPAL locus in E. coli. Differential expression of DAPAL in E. coli and S. Typhimurium may be due to differences in the mechanism of regulation of the genes in the two organisms, consistent with the differences observed in terms of their organization within the genome.
It was shown earlier that DAP has an inhibitory effect on the growth of Corynebacterium diphtheriae (6), which does not contain the gene for DAPAL. The inability of STM1002 (ygeX) null strains and wild-type E. coli to grow in minimal medium in the presence of dl-DAP is correlated with the absence of functional DAPAL (Fig. 2 and 3A). This inhibition could not be rescued by providing a variety of sugars in the medium. Interestingly, rescue of growth in the presence of Casamino Acids (Fig. 6) suggests that dl-DAP might be interfering with the metabolism of specific amino acids. By a process of elimination, a set of eight amino acids consisting of isoleucine, leucine, threonine, asparagine, methionine, arginine, glutamic acid, and cysteine was identified to be essential for the rescue of dl-DAP-mediated growth inhibition (Table 3). Growth of wild-type E. coli was rescued moderately in the presence of both pyruvate and aspartate, consistent with the fact that some amino acids from the set belong to the pyruvate family and the others to the aspartate family (Fig. 7). The complete inhibition exhibited by d-DAP suggests that the d isomer of DAP is a more potent inhibitor than l-DAP, which also holds true for other amino acids like d-serine (3).
The possible mechanism of growth inhibition could be that dl-DAP inhibits the enzymes involved in the synthesis of pyruvate and aspartate. Metabolism of sugars in bacteria primarily involves glycolysis that yields pyruvate. Since the growth was inhibited in the presence of all the sugars and glycerol, dl-DAP might be inhibiting the reactions taking place after the merging of the respective pathways with glycolysis and thereby arrest pyruvate synthesis. It is possible that the steps after the formation of 3-phosphoglyceric acid are inhibited in glycolysis by dl-DAP, as growth was not rescued by either PGA or PEP. The enzymes that might get inhibited by dl-DAP are phosphoglycerate mutase, enolase, and pyruvate kinase. Pyruvate and aspartate are substrates for most of the enzymes involved in the biosynthesis of the amino acids. Aspartate is not an essential amino acid for bacteria. It is synthesized in prokaryotes from pyruvate. Pyruvate carboxylase adds a carboxyl group to pyruvate using CO2 to form oxaloacetate. The oxaloacetate then undergoes a transamination reaction by aspartate aminotransferase to give aspartate. We have shown that OAA alone did not rescue growth while together with pyruvate it did, suggesting that the enzymes pyruvate carboxylase and aspartate aminotransferase are also on the list of enzymes inhibited by DAP. Hence, external addition of pyruvate or aspartate provides the substrate for the downstream pathways and restores growth.
Figure 7 shows the complete restoration of growth in the presence of Casamino Acids, which was also observed when the medium was supplemented with pyruvate, aspartate, cysteine, and glutamic acid. Since cysteine and glutamic acid are derived from intermediates other than pyruvate and aspartate, we suggest that dl-DAP might also inhibit the enzymes which are involved in biosynthesis of cysteine and glutamic acid. DAP, being a three-carbon molecule, was shown to inhibit the activity of a few PLP-dependent enzymes which are involved in amino acid metabolism, such as cystathionine β-lyase, O-acetylserine sulfhydrylase, and aminotransferases. It has also been shown earlier that analogues of dl-DAP were used to study regulation of threonine biosynthesis (19), and dl-DAP has also been shown to inhibit enzymes involved in the biosynthesis of amino acids belonging to the aspartate family (7).
In conclusion, the results presented in this communication indicate that the STM1002 gene of S. Typhimurium is functional and can degrade dl-DAP and thereby prevent its toxicity. In contrast, the ygeX gene of E. coli is expressed at very low levels and therefore cannot rescue the growth inhibition by dl-DAP. However, expression of ygeX in trans using a heterologous promoter can rescue the inhibition of growth. The observed negative effect on growth could be due to the inhibition of enzymes involved in the synthesis of pyruvate and aspartate and the amino acids derived from them as well as the enzymes involved in the synthesis of glutamate and cysteine.
ACKNOWLEDGMENTS
We thank the Department of Biotechnology, Department of Science and Technology, New Delhi, India, and the Indian Institute of Science for financial support.
Footnotes
Published ahead of print 17 August 2012
REFERENCES
- 1.Christman MF, Morgan RW, Jacobson FS, Ames BN. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753–762 [DOI] [PubMed] [Google Scholar]
- 2.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durham NN. 1963. Inhibition of microbial growth and separation by d-serine, vancomycin, and mitomycin C. J. Bacteriol. 86:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reference deleted .
- 6.Jensen KK, Schmidt V. 1945. The antigrowth effects of diaminopropionic acid on Corynebacterium diphtheriae. Acta Pharmacol. Toxicol. 1:346–350 [Google Scholar]
- 7.Jin JH, et al. 2004. Regulatory analysis of amino acid synthesis pathway in Escherichia coli: aspartate family. Enzyme Microb. Technol. 35:694–706 [Google Scholar]
- 8.Khan F, Jala VR, Rao NA, Savithri HS. 2003. Characterization of recombinant diaminopropionate ammonia-lyase from Escherichia coli and Salmonella typhimurium. Biochem. Biophys. Res. Commun. 306:1083–1088 [DOI] [PubMed] [Google Scholar]
- 9.Miticka H, et al. 2003. Transcriptional analysis of the rpoE gene encoding extracytoplasmic stress response sigma factor sigmaE in Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 226:307–314 [DOI] [PubMed] [Google Scholar]
- 10.Moorthy S, Mahadevan S. 2002. Differential spectrum of mutations that activate the Escherichia coli bgl operon in an rpoS genetic background. J. Bacteriol. 184:4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy KC. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagasawa T, Tanizawa K, Satoda T, Yamada H. 1988. Diaminopropionate ammonia-lyase from Salmonella typhimurium. Purification and characterization of the crystalline enzyme, and sequence determination of the pyridoxal 5′-phosphate binding peptide. J. Biol. Chem. 263:958–964 [PubMed] [Google Scholar]
- 13.Singh NS, Das G, Seshadri A, Sangeetha R, Varshney U. 2005. Evidence for a role of initiation factor 3 in recycling of ribosomal complexes stalled on mRNAs in Escherichia coli. Nucleic Acids Res. 33:5591–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storz G, et al. 1989. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J. Bacteriol. 171:2049–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touati D. 1988. Transcriptional and posttranscriptional regulation of manganese superoxide dismutase biosynthesis in Escherichia coli, studied with operon and protein fusions. J. Bacteriol. 170:2511–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uo T, Yoshimura T, Nishiyama T, Esaki N. 2002. Gene cloning, purification, and characterization of 2,3-diaminopropionate ammonia-lyase from Escherichia coli. Biosci. Biotechnol. Biochem. 66:2639–2644 [DOI] [PubMed] [Google Scholar]
- 17.Vijayalakshmi KR, Rao DR, Rao MR. 1975. Studies on a 2,3-diaminopropionate:ammonia-lyase from a pseudomonad. Hoppe Seylers Z. Physiol. Chem. 356:193–201 [DOI] [PubMed] [Google Scholar]
- 18.Woodcock DM, et al. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wormser EH, Pardee AB. 1958. Regulation of threonine biosynthesis in Escherichia coli. Arch. Biochem. Biophys. 78:416–432 [DOI] [PubMed] [Google Scholar]