Abstract
Phage lysis is a ubiquitous biological process, the most frequent cytocidal event in the biosphere. Lysis of Gram-negative hosts has been shown to require holins and endolysins, which attack the cytoplasmic membrane and peptidoglycan, respectively. Recently, a third class of lysis proteins, the spanins, was identified. The first spanins to be characterized were λ Rz and Rz1, an integral cytoplasmic membrane protein and an outer membrane lipoprotein, respectively. Previous work has shown that Rz and Rz1 form complexes that span the entire periplasm. Phase-contrast video microscopy was used to record the morphological changes involved in the lysis of induced λ lysogens carrying prophages with either the λ canonical holin-endolysin system or the phage 21 pinholin-signal anchor release (SAR) endolysin system. In the former, rod morphology persisted until the instant of an explosive polar rupture, immediately emptying the cell of its contents. In contrast, in pinholin-SAR endolysin lysis, the cell began to shorten and thicken uniformly, with the resultant rounded cell finally bursting. In both cases, lysis failed to occur in inductions of isogenic prophages carrying null mutations in the spanin genes. In both systems, instead of an envelope rupture, the induced cells were converted from a rod shape to a spherical form. A functional GFPΦRz chimera was shown to exhibit a punctate distribution when coexpressed with Rz1, despite the absence of endolysin function. A model is proposed in which the spanins carry out the essential step of disrupting the outer membrane, in a manner regulated by the state of the peptidoglycan layer.
INTRODUCTION
For nearly all double-stranded DNA phages, it has long been established that lysis involves two essential components: the holin and the endolysin (Fig. 1). Holins are grouped into two general types, canonical holins and pinholins, both of which cause temporally regulated, lethal membrane permeabilization that terminates the infection cycle. In both cases, the timing of lysis is defined by a sudden transition in the subcellular localization of the holin, based on green fluorescent protein (GFP) fusion studies (30; T. Pang, T. Fleming, K. Pogliano, and R. Young, submitted for publication). During the latent period, the holins accumulate in the cytoplasmic membrane in a dispersed, highly mobile form until suddenly aggregating into large, immobile foci. This sudden aggregation, designated “holin triggering,” occurs at an allele-specific time and is associated with the formation of lethal membrane lesions, or holes. Cryo-electron microscopy (cryo-EM) and tomography revealed that λ S105, the prototype canonical holin, forms holes of unprecedented size, near-micrometer scale (12). In contrast, pinholins, like the holin S2168 of lambdoid phage 21, form small heptameric channels of ∼2 nm (20). Endolysins are enzymes that degrade the peptidoglycan (PG). Two functional types of endolysin have been identified: canonical endolysins and signal anchor release (SAR) endolysins (Fig. 1). The former, exemplified by λ R, are soluble enzymes that accumulate fully folded and active in the cytoplasm until released through the membrane lesion formed by a canonical holin. In contrast, the SAR endolysins use the host sec system and are exported as inactive, membrane-tethered enzymes; e.g., R21 of phage 21 or Lyz of phage P1 (16, 26, 31, 32). In these cases, when the holin, either a canonical holin or a pinholin, depolarizes the membrane, the SAR endolysin escapes from the bilayer, refolds into an enzymatically active form, and attacks the PG.
Fig 1.
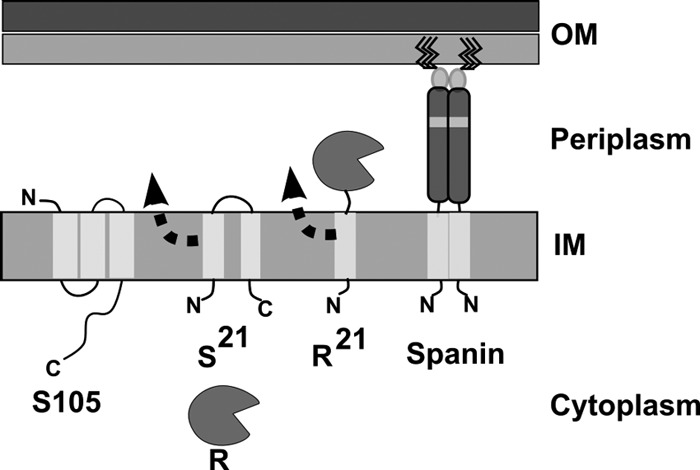
Topology and localization of λ and phage 21 lysis genes. The λ holin (S105) and endolysin (R) are localized to the IM and cytoplasm, respectively. The holin protein oligomerizes in the IM to form large lesions that enable the R protein to diffuse into the periplasmic space, where its substrate, the PG, is located. The lipopolysaccharide leaflet of the OM is indicated by a darker shade of gray. For simplicity, the PG layer and IM holes are not illustrated here. The phage 21 pinholin (S21) and SAR endolysin (R21) are both initially localized to the IM. Both S21 and R21 exhibit dynamic membrane topology, as indicated by an arrowhead and dotted line. The first transmembrane domain of S21 is a SAR domain, and it exits the bilayer as a result of S21 hole formation. Irreversible hole formation by S21 collapses the IM energy gradient, thus triggering global release of the single N-terminal SAR domain of R21. Release of the R21 SAR domain from the IM results in a periplasmic localization of the R21 enzyme. For simplicity, the spanin subunits of both λ and phage 21 are represented by the same illustration, as the respective IM and OM subunit proteins sequences are 97% and 95% identical. The spanin is illustrated here as a dimer based on unpublished results and reference 3. The IM spanin (λRz or Rz21) has a single N-terminal transmembrane domain with an Nin/Cout topology and an extended and primarily alpha-helical periplasmic domain. The approximate position of a predicted hinge region for the periplasmic domain of the IM spanin is indicated by a gray shaded box. The OM spanin (λRz1 or Rz121) is a small (40-aa) OM lipoprotein that interacts directly with the C-terminal end of the IM spanin via its own periplasmic domain. The OM spanin subunit is tethered to the inner leaflet of the OM following the posttranslational addition of 3 N-terminal fatty acid moieties (solid black lines). The function of the spanin is OM disruption.
Recently, a third functional class of lysis proteins, the spanins, was identified (25). The best-characterized spanins are the Rz and Rz1 proteins of λ. Rz is an inner membrane (IM) protein with an N-terminal transmembrane domain and a periplasmic domain of 130 residues, dominated by two predicted alpha-helical, coiled-coil subdomains separated by a hinge region. Rz1 is a small outer membrane (OM) lipoprotein, with a periplasmic domain of 40 amino acids (aa), of which 10 residues are prolines. During the morphogenesis period, Rz and Rz1 accumulate in their respective membranes and form complexes by C-terminal interactions (4). The term “spanin” derives from the fact that these complexes span the distance between the IM and OM; Rz and Rz1 are thus spanin subunits. Biochemical studies using the soluble periplasmic domains, sRz and sRz1, of the spanin subunits revealed that the formation of the complex is associated with a large increase in alpha-helical content and the formation of multimeric rod-shaped bundles (3).
The Rz and Rz1 genes first attracted attention because the latter is entirely embedded within the former in the +1 reading frame, thus constituting the only known instance where the same DNA sequence encodes two different proteins required for the same biological function (25). Rz-Rz1-like gene pairs have been identified in nearly all phages of Gram-negative hosts. These genes are highly diverse and are also found in two other gene architectures different from the embedded arrangement found in λ. For example, in the paradigm coliphage P2, the Rz1 equivalent lysC extends beyond the end of the Rz equivalent, lysB; this is designated an overlapped architecture. In T4, the coding sequence of the Rz and Rz1 equivalents are separated. In addition to these remarkably distinct architectures for the genes encoding spanin subunits, there is also a completely different type of spanin, represented by gp11 of coliphage T1. gp11 has an N-terminal OM lipoprotein signal and also a C-terminal transmembrane domain and is therefore predicted to span the periplasm as a single, covalent chain. The product of gene 11 has been shown to complement λ Rz and Rz1 mutations and has thus been designated a unimolecular spanin.
Despite the ubiquity, diversity, and intriguing molecular behavior of the spanins, the lysis phenotype associated with Rz and Rz1 defects is rather subtle. Lysis of induced λ lysogens in liquid culture does not require Rz and Rz1 unless the medium is supplemented with millimolar concentrations of divalent cations (35). In the presence of millimolar concentrations of divalent cations, lysis is blocked at a spherical-cell stage. This has been taken to mean that the spanins are auxiliary lysis proteins that serve to destabilize the OM where that barrier has enhanced stability even in the absence of the murein layer. Here we re-examined the process of phage lysis and the phenotypes of spanin mutants using time lapse phase-contrast microscopy. The results are discussed in terms of a model in which the spanins effect a third step in lysis, disrupting the OM after holins and endolysins have attacked the IM and PG, respectively.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The following strains were used in the phase microscopy portion of this study and are described elsewhere: MC4100 tonA::Tn10 λSRRzRz1 bor::kan, MC4100 tonA::Tn10 λSRRzamRz1 bor::kan, MC4100 tonA::Tn10 λSRRzRz1am bor::kan, and MC4100 tonA::Tn10 λSRRzamRz1am bor::kan (4). Construction of the λ21 hybrid phages is described elsewhere (20). Production of the MC4100 λ21 single lysogens MC4100 tonA::Tn10 λ(SRRzRz1)21 bor::kan and MC4100 tonA::Tn10 λ(SRRzamRz1am)21 bor::kan was achieved by infection at a low multiplicity of infection followed by selection for resistant colonies by plating on LB-Kan (40 μg/ml kanamycin). For the GFP-based localization of Rz, two strains, MC4100 tonA::Tn10 λSam7RRzamRz1 bor::kan and MC4100 tonA::Tn10 λSam7RRzamRz1am bor::kan (4), were transformed with plasmid pJB_GFPΦRz.
The gfpΦRz fusion allele was generated through splicing by overlapping extension (SOE) (13) using the following primers: FOR1, 5′-TCCCGAATTCATGAGTAAAGGAGAAGAACTTTT-3′; FOR2, GAACTATACAAACCCGGGAGCAGAGTCACCGCGATT; REV1, GGGGATCCCTATCTGCACTGCTCATTAAT; and REV2, CCCGGGTTTGTATAGTTCATCCATGCCATGTGTAAT. The PCR product was treated with EcoRI and BamHI (NEB Biolabs) and ligated into the pMLB1113 backbone, generating pJB_GFPΦRz. The GFPΦRz fusion consists of the entire GFPA206K protein sequence plus a two-amino-acid ProGly linker fused to the His residue at position 2 of the Rz protein sequence. The pMLB1113 vector, which is described elsewhere (9), is a pBR322 derivative and carries a single copy of the lacIq gene under the control of a constitutive promoter and a single polylinker site downstream of the lac promoter. The pJB_GFPΦRz plasmid was transformed into the lysogenic strains indicated above. Complementation of the pJB_GFPΦRz construct was confirmed under nonpermissive conditions, i.e., LB plus 10 mM MgCl2 with strain MC4100 tonA::Tn10 λSRRzamRz1 bor::kan.
Bacterial cultures were grown in standard LB medium supplemented with MgCl2 (10 mM), ampicillin (100 μg/ml), and kanamycin (40 μg/ml) when appropriate. Culture growth and lysis profiles were monitored as previously described (24). Briefly, 100 μl of a fresh overnight culture was diluted into 25 ml of LB and grown, unless indicated otherwise, with aeration by shaking at a 250 rpm. The temperature for growth was 30°C for lysogenic cultures and 37°C for nonlysogens. Lysogens were thermally induced at an A550 of ∼0.3 by aeration at 42°C for 15 min, followed by continued growth at 37°C. When indicated, isopropyl β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM.
Time lapse phase-contrast microscopy.
Each trial was initiated by a 1/300 dilution of an 18-h starter culture. Each starter culture was inoculated from a single colony. In a 250-ml Erlenmeyer flask with constant vigorous aeration, the time between the onset and end of lytic events, as determined by A550 dropping below 0.05, is ∼10 min at 37°C. At 50 min after thermal induction, the time marking the onset of lysis, a 10-microliter sample of the appropriate culture was placed on a glass slide and then covered with a coverslip, both of which were pre-equilibrated to 37°C. Cells were immediately imaged using an EC Plan-Neofluor 40×/0.75 Ph2 objective installed on a Zeiss Axio Observer A1 microscope equipped with a heated stage, a PECON temperature control unit, and an Axiocam HSM camera. In order to achieve a stable temperature of 37°C for the mounted slide for the duration of the experiment, the heated stage was equilibrated to 42°C and a stream of air heated to 42°C was directed under the slide. Time lapse video was captured at 2 to 5 frames per second for 10 min. The A550 at the time of lysis and working magnification allowed the capture of ∼4 lytic events per lysogenic induction. Video was edited and scaled using the Axio vision 4 software package (Zeiss). All images were stored as .zvi files and converted to a TIFF or JPEG format as necessary. Each lysis event was scored based on the observed location of lysis and morphology prior to the lysis event. Points of lysis that were determined to occur within a total distance from one pole that accounted for no more than 20% of the total cell length based on scaled measurements were scored as polar lysis. The rate of spherical conversion was determined by scaled measurements of transforming cells, with the sphere-rod boundary serving as one reference point and the untransformed pole the other. The conversion of 20 cells of various lengths was determined, and the values were averaged to arrive at a conversion rate estimate.
Fluorescence microscopy.
Fluorescence microscopy was performed as described previously (18). Briefly, the appropriate strains were grown in LB medium supplemented with appropriate antibiotics to an A550 of ∼0.3 and supplemented with 1 mM IPTG to coincide with the time of thermal induction of resident λ prophages. At 60 min postinduction, 20 μl of each culture was combined with 0.5 μl of the membrane dye FM 4-64 (1 mg/ml) (Invitrogen). Ten μl of this mixture was then placed on an agarose pad (1.2% agarose, tryptone broth, 1 μg/ml FM 4-64). Images were captured for 10 z sections, 0.2 μm apart, for each sample with an Applied Precision optical sectioning microscope system. The system was equipped with an Olympus IX70 microscope, an Olympus Plan Apo 100× oil immersion objective (numerical aperture [NA], 1.4), a Photometrics Cool Snap HQ digital camera, and Delta Vision standard fluorescence filters: fluorescein isothiocyanate (FITC) for GFP visualization (excitation, blue 490/20 nm; emission, green 528/38 nm) and rhodamine-Texas Red phycoerythrin (PE) for FM 4-64 visualization (excitation, green 550/28 nm; emission, orange 617/73 nm). SoftWoRx software (Applied Precision, Inc.) was used to deconvolve images using the constrained iterative deconvolution algorithm. ImageJ was used to adjust the brightness and contrast of each fluorochrome (2).
SDS-PAGE and Western blotting.
At 60 min after induction of plasmid, temperature-sensitive prophage, or both, total cellular protein was precipitated by addition of 10% (final concentration) ice-cold trichloroacetic acid (TCA) to an aliquot of culture. Protein was collected in a microcentrifuge at 18,000 × g for 5 min and washed twice by resuspension in cold acetone. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as described elsewhere with a few exceptions (3, 4) and described here briefly. Protein pellets were air dried and resuspended in 1× SDS-PAGE buffer (50 mM Tris-HCl, 2% SDS, 5% glycerol, 100 mM β-mercaptoethanol [pH 6.8]). Resuspension volume was normalized to 10 μl per 0.1 OD unit of culture. Thirty μl of sample was then loaded and resolved on a 16% polyacrylamide SDS-Tricine gel. Protein was transferred to a polyvinylidene difluoride (PVDF) membrane using a Hoefer TE unit at 0.5 mA for 2 h at room temperature. The transferred protein was probed with a polyclonal antibody against Rz, described previously (4), at a 1:1,000 dilution or an anti-GFP antibody, rabbit polyclonal serum (A6455; Invitrogen), at a 1:1,000 dilution. The secondary antibody, goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (Thermo-Scientific), was used at a 1:5,000 dilution. Imaging of chemiluminescence was achieved with the Bio-Rad XR Gel Doc system. Densitometry of bands was performed with ImageJ (2).
RESULTS
Different morphological transitions in canonical and pinholin-SAR endolysin lysis.
The gross morphological changes of the λ lysis process have not been subjected to systematic study, despite the long-term paradigm status of the system. Our first goal was to obtain a clear perspective on the morphology associated with the canonical λ holin-endolysin lysis event and the differences, if any, from those characteristic of the pinholin-SAR endolysin system. We applied phase-contrast videomicroscopy to capture images of cells undergoing lysis following thermal induction of λ prophages. The observation of eight independent cultures of λSRRzRz1 resulted in the acquisition of 40 individual lysis events, three of which are shown in Fig. 2A and B as time lapse series. The typical result (34/40 events) was that no gross changes in cellular morphology are observed until the periphery of the cell abruptly ruptures in a restricted zone near or at a pole (Fig. 2A; also, see Movie S1 in the supplemental material). The immediate consequence of this localized rupture is the expulsion of the cytoplasmic contents, as detected by loss of refractivity, leading to a transient nonrefractile ghost that retains substantial rod shape. At a relatively low frequency the same type of rupture was observed (Fig. 2B; also, see Movie S2 in the supplemental material) at locations other than the cell pole.
Fig 2.
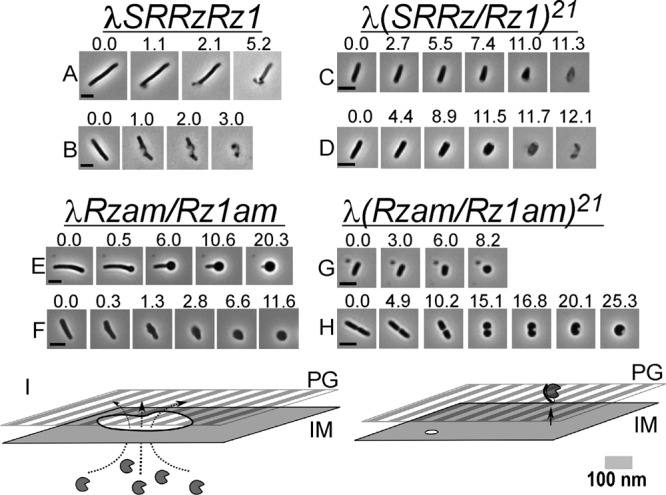
The spanin complex is required for rapid lysis. (A to H) MC4100 lysogens were thermally induced, and 1 min prior to the onset of lysis, a field of cells was observed with phase-contrast microscopy in order to document the morphology of phage lysis. Time lapse series of individual cells beginning immediately prior to and continuing through completion of morphological conversion are shown. Prophages and relevant genotypes are indicated above each group of time series. Time (in seconds) is displayed above each individual frame, with time zero representing the first available frame prior to an observable morphological change or lysis. Bars, 5 μm. (I) Proposed models for holin-endolysin (left) and pinholin-SAR endolysin (right) action prior to spanin function and lysis. The holin accumulates and forms near-micrometer-scale lesions in the IM (irregular white circle with black border), resulting in a localized release of R (notched circles). The pinholin forms a heptameric pore (small white circle) approximately 2 nm in diameter. Pore formation collapses the membrane potential and causes release of the IM-tethered SAR endolysin (notched circle with black bar) to the periplasm. See the text for further details. Note that the PG peptide cross-links and OM are omitted for simplicity.
To address the morphology of pinholin-SAR endolysin lysis in an isogenic system, we repeated these experiments with a λ hybrid, λ(SRRzRz1)21, in which the native lysis cassette has been replaced with that of the lambdoid phage 21. This chimera, which encodes the prototypical pinholin, S21, and SAR endolysin, R21 (Fig. 1), has been shown to be normal in terms of plaque formation and lysis kinetics (21). Induction of eight independent cultures resulted in the capture of 45 lytic events. All events were distinctly different from that of λ in two respects: first, there was a progressive reduction in cell length and thickening of the cell diameter during the lysis process, and second, the location of lysis at the cell envelope was primarily between the pole and midcell (23/45 events) (Fig. 2C and D; also, see Movies S3 and S4 in the supplemental material), rather than a blowout localized to a restricted polar zone. The common morphology at the time of lysis was more typically of a cone or prolate spheroid shape. Furthermore, λ(SRRzRz1)21 lysis also ultimately resulted in the production of phase-light cell ghosts, although unlike the ghosts generated in λ lysis, the rod morphology was less apparent after lysis.
The different morphological changes during lysis of induced λ and λ(SRRzRz1)21 lysogens can be interpreted according to the current understanding of the molecular mechanisms in each system. We postulate that the abrupt, localized rupture of the envelope seen in the λ induction reflects the sudden S105-dependent formation of the large membrane lesions, averaging ∼340 nm in diameter, which we previously reported (12). This would presumably expose >100,000 nm2 of the peptidoglycan to the pool of fully active endolysin in the cytoplasm (Fig. 2I). The predominance of polar events is consistent with the real-time observations using the GFP-tagged version of the λ holin S105; after triggering, the majority of S105-GFP foci were polar (see Figures 2, 4, and 5 of reference 30). Moreover, although the cryo-EM tomographic studies that identified, quantified, and characterized the S105-dependent lesions found them to be present in one to three instances per cell, randomly distributed along the long axis, it must be noted that the cell poles were excluded from the analysis (12).
Fig 4.
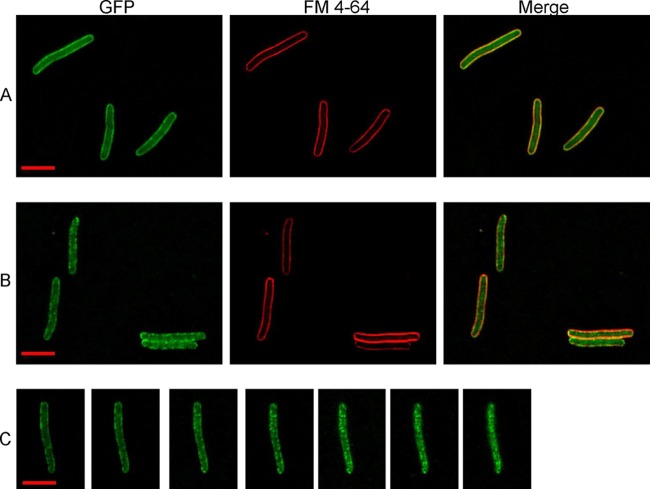
Deconvolution fluorescence imaging of GFPΦRz reveals the oligomeric state prior to PG degradation. Rows A and B represent samples collected 60 min after induction of GFPΦRz and λSam7Rzam/Rz1am or λSam7Rzam/Rz1, respectively. Row C is a deconvoluted z-stack series (step size, 0.2 μm) of GFPΦRz fluorescence following λRzam/Rz1 induction 60 min earlier. Bar (inset), 5 μm.
Similarly, the progressive loss of rod-shaped morphology following induction of λ(SRRzRz1)21 lysis can also be understood as reflecting the completely different mode of lysis control conferred by the pinholin-SAR endolysin system. Although studies with a GFP-tagged pinholin show a redistribution from a uniform, high-mobility state to a immobile punctate distribution, associated with the lethal triggering of the pinholin, the foci formed are more numerous, smaller, and not disproportionately associated with the poles (Pang et al., submitted). Other than that it is tethered to the membrane, nothing is known about the distribution of the SAR endolysins in the envelope prior to holin triggering. For S21, it has been estimated that there are ∼7,000 pinholins at the time when the wild type is triggered (20), presumably uniformly distributed and laterally mobile in the membrane. Thus, when the pinholin pool is triggered to form ∼103 heptameric pinholes and massively depolarize the membrane, it would be expected that the SAR endolysins are globally released from the bilayer and become activated throughout the periplasm (Fig. 2I). According to this view, the progressive loss of the rod shape reflects the generalized degradation of the peptidoglycan, which by virtue of the glycosidase activity of the endolysin occurs by the progressive shortening of the average glycosidic chain length in the murein. As for what causes the lytic burst in both cases, it is been widely considered to occur solely as a consequence of both endolysin-mediated cell wall degradation and turgor pressure. This question, however, has never been addressed directly.
Spherical cells: the terminal morphology of spanin-defective mutants.
With the morphological transitions leading to lysis of the canonical holin-endolysin and pinholin-SAR endolysin systems in hand, we next addressed the role of the spanins by repeating these analyses with spanin mutants. Forty-eight individual induced cells carrying the λRzam/Rz1am prophage were monitored, two of which are displayed in Fig. 2E and F. In both cases, instead of the sudden, localized rupture near or at a pole, the cells are converted to a spherical form, without loss of refractivity. The conversion begins as a single localized deformation on the cell periphery, which occurs usually at the pole (Fig. 2E) and more rarely at the midcell (Fig. 2F; 4 of 48) (see Movies S5 and S6 in the supplemental material). A similar result regarding conversion initiating at or near the pole was also observed in the case of λRzam and λRz1am inductions (39/44 and 40/46 events, respectively). Polar deformations always progressed toward the opposite pole in a diffusive fashion, in the form of a narrow boundary distinguishing the native rod morphology from the developing spherical morphology. For most of the conversion, the conversion boundary moves at ∼0.5 μm/s. During the observation period, only ∼5% of induced cells actually progressed to lysis (data not shown).
Under the same experimental conditions, induction of lysogens carrying λ(SRRzamRz1am)21 also led to development of spherical cells (Fig. 2G and H; also, see Movies S7 and S8 in the supplemental material). In contrast to the λRzam/Rz1am inductions, there was no sharp boundary marking the progress of the morphological conversion. Instead, the induced cells followed the same progression of gradual loss of rod shape noted above for pinholin, SAR endolysin, and spanin induction, except that in each case the spherical conversion was fully realized, with no lytic event observed.
We interpret these results as indicating that in the absence of spanin function, neither the holin-endolysin nor the pinholin-SAR endolysin lysis program can effect lysis of the host cell and thus release of the progeny virions on a time scale useful to the phage. The morphological changes that lead to the formation of spherical cells following λRzam/Rz1am or λ(RzamRz1am)21 induction reflect the degradative activity of the R endolysin or the R21 SAR endolysin, respectively, on the PG. Specifically, in the case of λRzam/Rz1am, the progression of the spherical cell development down the long axis of the cell (Fig. 2E) would reflect the diffusion of the fully active R endolysins from a site of localized release near the pole through the periplasmic space until the opposing pole is reached. In contrast, global R21 release effected by the triggering of the pinholin would lead to a much larger surface area of the PG being initially subject to degradation. As a result, a larger surface area of the PG layer is weakened uniformly, leading to oblate spheroid shape prior to culminating in a spherical form. In turn, the activity of the endolysins mandates that the S105 holin or the S21 pinholin have been triggered. Since in at least the case of the induced λRzamRz1am lysogen, the deformation that initiates the rod-to-sphere conversion occurs at approximately the same time and location as the localized rupture does for the wild-type lysogen, the temporal program of lysis appears to be normal. Thus, the spanin lysis defect is in a step functionally downstream from holin triggering and murein degradation, i.e., OM disruption.
Shearing forces attendant on aeration complement the RzRz1 lysis defect in vivo.
Previously, spanin function was shown to be dispensable for lysis in batch aerobic culture unless the medium was supplemented with a 10 mM concentration of divalent cations, under which conditions the terminal phenotype was spherical cells (7, 35). In light of the above results, we revisited λ lysis in the context of shaker flasks, with the hypothesis that shearing forces attendant on aerobic growth might complement the RzRz1 lysis defect. To test this idea, liquid cultures of λ and λRzamRz1am lysogens were induced normally but were removed from agitation 10 min prior to the normal onset of lysis. The results clearly showed that lysis was defective in the RzamRz1am culture but not the wild type (Fig. 3A), with, as expected, spherical cells accumulating (data not shown). Several physiological parameters were shown to affect the degree of the lysis defect, which presumably reflects the stability of the spherical cells: lowering the temperature of the culture to 30°C 10 min prior to lysis (Fig. 3B) and delaying inductions until later in exponential phase (Fig. 3C) both exacerbated the spanin lysis defect, whereas increasing the shearing forces by the use of a baffled flask suppressed the lysis defect even in the presence of stabilizing Mg2+ ions (Fig. 3B). Delaying induction to mid-log phase also produced a partial Mg2+-independent phenotype for the RzamRz1am culture (Fig. 3C).
Fig 3.
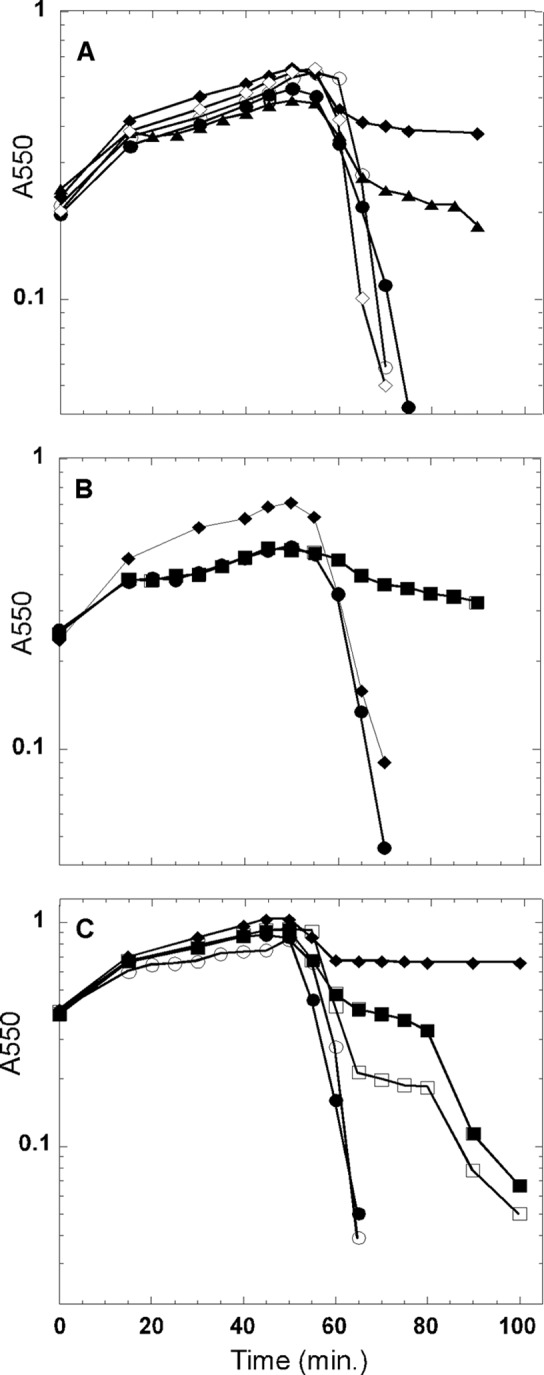
Saltatory λRzam/Rz1am lysis is an artifact. The effect of various physiological parameters on cell lysis was assessed following induction of λ prophages. (A) Shearing forces attendant on flask shaking complement the λRzam/Rz1am lysis defect. The following prophage inductions were conducted with constant shaking: λ (open circles), λRzam/Rz1am in LB plus 10 mM Mg2+ (closed diamonds), and λRzam/Rz1am (open diamonds). For the following prophage inductions, culture shaking was ceased at 45 min after induction: λ (closed circles) and λRzam/Rz1am (closed triangles). (B) The Mg2+-dependent lysis defect is complemented by growth in a baffled flask. For the λ (circles) and λRzam/Rz1am (squares) inductions, shaking was ceased and the temperature decreased to 30°C 45 min postinduction. The λRzam/Rz1am (diamonds) prophage was induced in a baffled flask in the presence of 10 mM Mg2+. (C) Late-log-phase induction leads to a Mg2+-independent lysis defect. For the following prophage inductions, culture shaking was ceased at 45 min after induction: λ (closed circles) and λRzam/Rz1am (closed squares). The following prophage inductions were subjected to constant shaking: λ (open circles), λRzam/Rz1am in LB plus 10 mM Mg2+ (closed diamonds) and λRzam/Rz1am (open squares).
Spanin complexes accumulate in foci before holin or endolysin action.
Information regarding the subcellular localization of the spanin during the lysis pathway is needed to better understand how its role in OM disruption contributes to phage lysis. To address this issue, a chimera was constructed in which GFP was fused to the cytoplasmic N terminus of Rz. The GFPΦRz chimera was fully functional as an IM spanin subunit, as judged by its ability to complement the lysis defect of λRzam and its accumulation as a single species at a level approximately 2-fold lower than that of the wild-type protein (see Fig. S1 in the supplemental material). To avoid cell lysis associated with expressing the full complement of lysis genes, we chose to monitor GFPΦRz localization in the absence of a functional holin protein by using a null Sam7 allele. Expression of the plasmid-borne copy of GFPΦRz was induced to coincide with thermal induction of λSam7RRzamRz1am or λSam7RRzamRz1. At 60 min postinduction, in the λSam7RRzamRz1am background, GFPΦRz exhibited generally diffuse fluorescence with some distinct foci also localized to the IM (Fig. 4A). At the same point in the isogenic Rz1+ induction, GFPΦRz fluorescence was much more punctate (Fig. 4B and C). Thus, in the absence of holin function and, by extension, endolysin-mediated degradation of the PG, the presence of Rz1 in the OM causes Rz to assemble predominantly in foci. Previously, Rz-Rz1 complex formation had been demonstrated to form efficiently in the undisturbed envelope (3). Moreover, when mixed together, the purified periplasmic domains of Rz and Rz1 bind, undergo major conformational changes generating increased helical structure, and form rod-shaped oligomeric bundles (3). Taken together, these findings suggest that the punctate distribution assumed by GFPΦRz in the presence of Rz1 reflects multimeric Rz-Rz1 complexes that have formed by C-terminal interactions following lateral oligomerization of the IM and OM spanin subunits.
DISCUSSION
Rz and Rz1 are essential for λ lysis of E. coli.
Many genetic, physiological, and environmental perturbations of the cell envelope can lead to lysis of the bacterial cell. For example, when cell wall antibiotics block the incorporation of cell wall precursors into the PG in a growing E. coli cell, the cell ultimately lyses when it attempts to progress through septation, despite no direct effects of the antibiotic on the other components of the envelope, the cytoplasmic membrane and outer membrane. Phage lysis is qualitatively different from the lysis derived from such derangements, principally in that it must be effected in a temporally regulated manner. Compelling arguments that the capacity to adjust the timing of lysis, and thus the length of the infection cycle, is an important fitness factor, especially in terms of optimizing phage fecundity in the face of changing availability of the host bacterium, have been presented (1, 5, 27, 28). Moreover, it is obvious that the lysis system should have a minimal effect on the intracellular rate of accumulation of progeny virions until the scheduled time of lysis, after which the most rapid and efficient dispersion of the progeny virions should be effected (29). These considerations have led to a widespread acceptance of the key role of holins and endolysins in phage lysis (33), with the former controlling the timing of lysis and the permeabilization of the cytoplasmic membrane, which immediately leads to attack on the PG by the latter. However, it was only recently that it began to be appreciated that disruption of the OM would also be important for rapid and efficient host lysis. Initially, the RzRz1 genes of λ were designated auxiliary lysis genes based on the fact that the RzRz1-null phenotype was a cation-dependent lysis defect, suggesting that the spanin complex, the components of which are encoded by RzRz1, was required only when the OM was artificially stabilized (35). Here we have shown that in fact λ lysis, on a time scale useful to the phage, actually requires functioning RzRz1 genes without artificial stabilization of the OM. Moreover, we provide evidence that the terminal phenotype of RzRz1-null mutants is the formation of a spherical cell sufficiently fragile that the shearing forces derived from aerobic growth in a shaker flask artifactually complement the lysis defect (Fig. 3A to C), which led to the inaccurate designation of RzRz1 as auxiliary lysis genes (15, 17, 23, 34).
The cation-dependent spherical cell phenotype has also been demonstrated for mutants in the two-component spanin genes of diverse phages, including the coliphage P2 (19) and the cystovirus PRD1 (15). Although videomicroscopy of infections in the absence of stabilizing cations needs to be done for a few of these systems, the nearly universal presence of either two-component or single-component spanin genes in the genomes of phages that grow on Gram-negative hosts (25) makes it likely that the lessons derived from λ lysis will be generally applicable. This conclusion leads to a model that lysis of the Gram-negative host is a three-step process, mediated by holins, endolysins, and spanins, subverting, in turn, the IM, the cell wall, and the OM, respectively. In this three-step process, each step depends on the previous step, although there is no evidence for any direct physical interaction between the holins, endolysins, or spanins. In fact, the diversity and interchangeability of holins, endolysins, and spanins indicate that direct interaction is unlikely. For example, the λ holin and endolysin functions equally well with the phage P2 LysBC two-component spanins, which are encoded by an overlapped spanin gene architecture and have no sequence similarity with the lambda proteins (19). Likewise, the P22 endolysin, a glycosidase of the T4 lysozyme family, can substitute for the lambda endolysin, a transglycosylase, although the lambda and P22 holins are nearly identical (7).
At this point, two interesting questions about the third step and the spanins remain. (i) Why is spanin function necessary for host lysis? (ii) How does the spanin complex effect disruption of the OM?
The spherical-cell phenotype.
It has generally been thought that destruction of the PG was sufficient to lyse the host, with estimations of ∼3 atm turgor pressure resulting from the differences in osmolyte concentrations in the cytoplasm and most growth media (8, 14). However, here we found that the spherical cells produced by holin-endolysin action in the absence of spanin function are stable, at least in terms of the time scales of the phage infection cycle, without osmotic protection, as long as they are not subjected to the shearing forces extant in a shaker flask culture. The simplest interpretation is that the OM has sufficient tensile strength to withstand the intracellular turgor pressure. We are not aware of any measurements of the tensile strength of the OM in vivo or in vitro. Moreover, recent direct measurements obtained by using atomic force microscopy on intact cells have indicated that the actual turgor pressure is ∼0.3 atm, an order of magnitude lower than previously accepted (11). It is also possible that there is some residual PG in the spherical cells. This seems unlikely, considering that the same spherical-cell phenotype has been demonstrated for spanin deficiency in diverse phage systems with diverse endolysins with different levels of expression and enzymatic activities. There is no known mechanism to inactivate the endolysins once they are active in the periplasm, either by passage through the lesion generated by a canonical holin or by release from the bilayer after membrane depolarization by a pinholin. Bacteria produce periplasmic proteins like Ivy that complex with and inhibit mammalian lysozymes and have been shown to have some activity on some phage endolysins (6), but there would be no opportunity to produce more of these proteins once the holin or pinholin has been triggered to terminate macromolecular metabolism in the host cell.
The gross morphology of the spherical cells produced by the lethal action of the λ holin-endolysin lysis system in the absence of spanin function has been examined by cryo-electron microscopy (12). In this case, the spherical cells were stabilized by 10 mM MgCl2. Assuming that the cation supplement has no effect on the gross morphology, it is striking that the OM and IM appear to be completely independent spherical surfaces separated in much of the cell by hundreds of nanometers. It should be noted that the spherical cells can also be produced without any damage to the IM by simply expressing a SAR endolysin (4). Each SAR endolysin has a rate of spontaneous release from the membrane and thus activation, even in the absence of holin function to depolarize the bilayer. Thus, inducing moderate overexpression of a SAR endolysin, in the absence of spanins, leads to conversion of an entire culture of cells to spherical forms. In these situations, macromolecular synthesis is not compromised, so SAR endolysin production and release and activation in the periplasm are constitutive, making the persistence of any murein structures unlikely. The properties of these “wall-less cells” may make them useful for exploring the ability of IM and OM components of biosynthetic and regulatory proteins to communicate.
Mechanism of OM disruption by the spanin complex.
The ultrastructure of the spherical cells produced in spanin-defective lysis, the ability of low concentrations of divalent cations to stabilize these forms, and the instantaneous sensitivity of these cells to chelators all indicate that the OM is the main, and likely the only, barrier to lysis and release of the progeny virions. This leads to the question of how the spanin proteins disrupt the OM. We assume that the basic mechanism is the same for the two-component spanins encoded by the embedded, overlapped, and separated gene architectures as well as for the single-component spanin, again relying on the interchangeability of holins, endolysins, and spanins cited above. It must be noted that spanins act independently of holin function; i.e., disruption of the OM does not depend on hole formation in the IM (4). The only functional constraint useful for contemplating the mechanism of spanin function is the absolute requirement for endolysin action. Before degradation of the murein by the endolysin, each complex formed by the IM and OM spanin components would have to be threaded through the intact meshwork of the PG. Indeed, evidence was previously provided that the Rz-Rz1 complexes accumulate from the beginning of the morphogenesis phase, which occupies most of the phage infection cycle, and that, indeed, the periplasmic domain of Rz is subject to proteolytic cleavage in the absence of Rz1 (3). There are as yet no comparable studies of the unimolecular spanin, T1 gp11, but presumably the lipobox signal and the presence of Ser and Thr at the +2 and +3 positions direct the generation of a fully lipoylated N-terminal Cys residue and its transfer to the inner leaflet of the OM. Again, the periplasmic domain of gp11 would have to thread through the PG meshwork.
Based on these considerations, we propose that the PG barrier acts as a negative regulator of spanin function. Considering the disparity of the molecular topology of the single- and two-component spanin systems, the simplest notion is that the inhibition stems from the entrapment of the spanin proteins within the PG, preventing their lateral mobility. Accordingly, the removal of the PG by endolysin action would allow lateral oligomerization of the spanins. In support of this notion, we have shown that, when mixed in free solution, the purified periplasmic domains of Rz and Rz1, designated sRz and sRz1, form equimolar complexes and undergo a significant conformational change leading to increased alpha-helical content (3). This is associated with the formation of rod-like, 24-nm-by-5.7-nm bundles which exhibit coiled-coil morphology and are estimated to contain 8 to 10 copies of both Rz and Rz1 per complex. These observations fit well with the predicted extensive alpha-helix and coiled-coil proclivity of the Rz periplasmic domain. Interestingly, the long axis of the rod complex is in close agreement with the distance between the periplasmic leaflets of both membranes (25.5 nm) as reported previously (3), while the width is consistent with the experimentally determined value for the diameter of the PG lacunae (4.12 nm) (10). Although gp11 has little predicted helical structure, the purified periplasmic domain of gp11 seems to form a highly oligomeric structure, as judged by a gross change in viscosity of the protein upon elution from a chromatographic matrix (J. Berry, unpublished data).
Here we show that a GFPΦRz chimera is fully functional as an IM spanin subunit and that it accumulates in foci in an Rz1-dependent fashion, even in the absence of PG degradation (Fig. 4B and C). The simplest interpretation of these images is that immobile Rz-Rz1 complexes form from pools of mobile Rz and Rz1 proteins in the IM and OM, respectively, and that there is a pronounced tendency of the complexes to nucleate higher-order, lateral oligomers within the lacunae of the meshwork of the PG, possibly reflecting the formation of rod-like structures observed in vitro and discussed above. In the case of the canonical holin-endolysin system, the sudden, localized access of the soluble endolysin opposite the holin-mediated IM holes would cause localized degradation of the PG and thus liberate the sequestered spanin complexes. Once freed from the constraints of the PG, the spanins could assemble into further higher-order oligomers that would exert the disruptive effect on the OM. However, it is not clear whether the localized oligomers would be the diffusible unit or whether the oligomers would dissociate into monomer complexes and redistribute accordingly in the murein-free neighborhood. In any case, no matter where considerable damage to the PG occurs within the envelope, presumably spanin complexes or free spanin subunits are always nearby.
As for the question of how the spanin effects OM disruption, there are multiple possibilities. The coiled-coil propensity of the IM spanin subunits like Rz is reminiscent of the coiled-coil transitions of the SNARE proteins that underlie the fusion events of the vesicular protein trafficking system in mammalian cells (22). According to this perspective, the higher level oligomerization of the spanin complexes, or the single-molecule gp11-like spanins, leads to a collapsing conformational change and ultimately localized fusion of the IM and OM, thus removing the OM barrier to the release of the virions. Other possibilities include membrane tearing at the point of spanin complex foci, where the two membranes are massively linked and are thus subject to thermal fluctuations in physical forces without the stable support structure of the PG. Experiments with purified IM and OM spanin subunits reconstituted into lipid vesicles may provide insight into the mechanistic details.
Collectively these results define the three-step process by which temporally regulated lysis is achieved by bacteriophages of Gram-negative hosts. The process requires the sequential but autonomous action of the each lysis protein, the holin, the endolysin, and the spanin, in the disruption of the inner membrane, peptidoglycan, and outer membrane, in rapid succession. Consequently, the spanins can no longer be considered auxiliary lysis proteins.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by grant GM27099from NIH to R Y. and by the Center for Phage Technology, a Texas A&M University Initial University Multidisciplinary Research Initiative program.
We also thank Kit Pogliano for her assistance in executing the fluorescence microscopy portion of this work and acknowledge the clerical assistance of Daisy Wilbert.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abedon ST. 1989. Selection for bacteriophage latent period length by bacterial density: a theoretical examination. Microbiol. Ecol. 18:79–88 [DOI] [PubMed] [Google Scholar]
- 2.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 3.Berry J, Savva C, Holzenburg A, Young R. 2010. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 19:1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry J, Summer EJ, Struck DK, Young R. 2008. The final step in the phage infection cycle: the Rz and Rz1 lysis proteins link the inner and outer membranes. Mol. Microbiol. 70:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull JJ, Pfennig DW, Wang IN. 2004. Genetic details, optimization and phage life histories. Trends Ecol. Evol. 19:76–82 [DOI] [PubMed] [Google Scholar]
- 6.Callewaert L, et al. 2005. Purification of Ivy, a lysozyme inhibitor from Escherichia coli, and characterisation of its specificity for various lysozymes. Enzyme Microb. Technol. 37:205–211 [Google Scholar]
- 7.Casjens S, Eppler K, Parr R, Poteete AR. 1989. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology 171:588–598 [DOI] [PubMed] [Google Scholar]
- 8.Cayley DS, Guttman HJ, Record MT., Jr 2000. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys. J. 78:1748–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649 [DOI] [PubMed] [Google Scholar]
- 10.Demchick P, Koch AL. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178:768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Y, M Sun, JW Shaevitz. 2011. Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys. Rev. Lett. 107:158101. [DOI] [PubMed] [Google Scholar]
- 12.Dewey JS, et al. 2010. Micron-scale holes terminate the phage infection cycle. Proc. Natl. Acad. Sci. U. S. A. 107:2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932 [DOI] [PubMed] [Google Scholar]
- 14.Koch AL. 1998. The biophysics of the gram-negative periplasmic space. Crit. Rev. Microbiol. 24:23–59 [DOI] [PubMed] [Google Scholar]
- 15.Krupovic M, Cvirkaite-Krupovic V, Bamford DH. 2008. Identification and functional analysis of the Rz/Rz1-like accessory lysis genes in the membrane-containing bacteriophage PRD1. Mol. Microbiol. 68:492–503 [DOI] [PubMed] [Google Scholar]
- 16.Kuty GF, Xu M, Struck DK, Summer EJ, Young R. 2010. Regulation of a phage endolysin by disulfide caging. J. Bacteriol. 192:5682–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim JA, Shin H, Kang DH, Ryu S. 2012. Characterization of endolysin from a Salmonella Typhimurium-infecting bacteriophage SPN1S. Res. Microbiol. 163:233–241 [DOI] [PubMed] [Google Scholar]
- 18.Liu NJ, Dutton RJ, Pogliano K. 2006. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol. Microbiol. 59:1097–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markov D, et al. 2004. P2 growth restriction on an rpoC mutant is suppressed by alleles of the Rz1 homolog lysC. J. Bacteriol. 186:4628–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang T, Savva CG, Fleming KG, Struck DK, Young R. 2009. Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. U. S. A. 106:18966–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park T, Struck DK, Dankenbring CA, Young R. 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J. Bacteriol. 189:9135–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risselada HJ, Grubmuller H. 2012. How SNARE molecules mediate membrane fusion: recent insights from molecular simulations. Curr. Opin. Struct. Biol. 22:187–196 [DOI] [PubMed] [Google Scholar]
- 23.Schmidt C, Velleman M, Arber W. 1996. Three functions of bacteriophage P1 involved in cell lysis. J. Bacteriol. 178:1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DL, Chang CY, Young R. 1998. The λ holin accumulates beyond the lethal triggering concentration under hyper-expression conditions. Gene Expr. 7:39–52 [PMC free article] [PubMed] [Google Scholar]
- 25.Summer EJ, et al. 2007. Rz / Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373:1098–1112 [DOI] [PubMed] [Google Scholar]
- 26.Sun QK, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC. 2009. Regulation of the phage 21 endolysin by topological dynamics and membrane proximity. Nat. Struct. Mol. Biol. 16:1192–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang IN. 2006. Lysis timing and bacteriophage fitness. Genetics 172:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang IN, Dykhuizen DE, Slobodkin LB. 1996. The evolution of phage lysis timing. Evol. Ecol. 10:545–558 [Google Scholar]
- 29.Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 30.White R, et al. 2011. Holin triggering in real time. Proc. Natl. Acad. Sci. U. S. A. 108:798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, et al. 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307:113–117 [DOI] [PubMed] [Google Scholar]
- 32.Xu M, Struck DK, Deaton J, Wang IN, Young R. 2004. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U. S. A. 101:6415–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young R, Wang IN. 2006. Phage lysis, p 104–126. In Calendar R. (ed), The bacteriophages, 2nd ed. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 34.Young R, Way J, Way S, Yin J, Syvanen M. 1979. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J. Mol. Biol. 132:307–322 [DOI] [PubMed] [Google Scholar]
- 35.Zhang N, Young R. 1999. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol. Gen. Genet. 262:659–667 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.