Abstract
Sugar phosphorylation is an indispensable committed step in a large variety of sugar catabolic pathways, which are major suppliers of carbon and energy in heterotrophic species. Specialized sugar kinases that are indispensable for most of these pathways can be utilized as signature enzymes for the reconstruction of carbohydrate utilization machinery from microbial genomic and metagenomic data. Sugar kinases occur in several structurally distinct families with various partially overlapping as well as yet unknown substrate specificities that often cannot be accurately assigned by homology-based techniques. A subsystems-based metabolic reconstruction combined with the analysis of genome context and followed by experimental testing of predicted gene functions is a powerful approach of functional gene annotation. Here we applied this integrated approach for functional mapping of all sugar kinases constituting an extensive and diverse sugar kinome in the thermophilic bacterium Thermotoga maritima. Substrate preferences of 14 kinases mainly from the FGGY and PfkB families were inferred by bioinformatics analysis and biochemically characterized by screening with a panel of 45 different carbohydrates. Most of the analyzed enzymes displayed narrow substrate preferences corresponding to their predicted physiological roles in their respective catabolic pathways. The observed consistency supports the choice of kinases as signature enzymes for genomics-based identification and reconstruction of sugar utilization pathways. Use of the integrated genomic and experimental approach greatly speeds up the identification of the biochemical function of unknown proteins and improves the quality of reconstructed pathways.
INTRODUCTION
Various carbohydrates play a key role in the diet of many diverse microbial species as a primary source of carbon and energy. The extensive sugar utilization machinery that has evolved to keep up with this nutritional diversity is characterized by a matching level of species-to-species variations. Among them are alternative biochemical routes, nonorthologous gene replacements, and variations of substrate specificity within large and divergent protein families (28). Therefore, although many components of the microbial sugar utilization machinery can be seamlessly recognized at the level of general class function (e.g., a putative sugar kinase of the FGGY family and a sugar transporter of the MFS family), an accurate assignment of their substrate specificity remains a challenging task. Incorrect and imprecise gene annotations in genomic databases hamper our ability to accurately reconstruct and model metabolic networks beyond a few well-studied species. To address this important challenge, we use a subsystems-based approach (31) which combines comparative genomic reconstruction of respective pathways with the analysis of genome context (operons, regulons). The efficiency of this approach was illustrated by our recent studies, in which the sugar utilization machinery in the group of Shewanella species was reconstructed and a number of novel gene functions were predicted (33, 41). An ability to recognize signature components (e.g., specific enzymes) of metabolic pathways is one of the key factors of successfully applying this approach. Since phosphorylation is an indispensable enzymatic reaction in nearly all sugar catabolic pathways, we would like to address the question whether sugar kinases are a good choice for signature enzymes. It is expected that accurate assignment of substrates for all sugar kinases in any genome (termed here a “sugar kinome”) would help us to confidently infer the entire carbohydrate catabolic machinery in previously unexplored microbial species and communities.
Substrate preferences of sugar kinases can easily be assessed by in vitro biochemical assays, and many representatives of this popular class of enzymes have been extensively characterized. The remarkable diversity of carbohydrate-specific kinases includes several structurally distinct protein families with various partially overlapping as well as yet unknown substrate specificities (4–6). The most versatile family, FGGY, characterized by the RNase H-like fold (5), includes sugar kinases with experimentally demonstrated preferences toward various C3 to C6 substrates, such as glycerol, l-fuculose, d-gluconate, d-xylulose, l-xylulose, l-rhamnulose, l-ribulose, and erythritol (45). Among the substrates of sugar kinases from the PfkB family, featuring the Rossmann-like fold, are d-ribose, d-glucose, d-fructose, d-fructose-6-phosphate, d-fructose-1-phosphate, 2-dehydro-3-deoxy-d-gluconate, and d-tagatose-6-phosphate (5, 40). The GHMP kinase family displays a broad range of substrate preferences, including carbohydrates (d-galactose, l-arabinose, l-fucose) and other intermediary metabolites, such as homoserine, shikimate, and mevalonate (1, 5). The ROK family includes two functionally distinct groups of proteins: (i) sugar kinases that are catalytically active on either aldohexoses (d-glucose, d-allose, d-mannose) or amino sugars (N-acetyl-d-glucosamine, N-acetyl-d-mannosamine) and (ii) sugar-responsive transcriptional regulators that are characterized by an additional N-terminal DNA-binding domain (37).
Although the families to which diverse kinases belong are easily recognized by homology, the assignment of their substrates often requires additional analysis of genomic/functional context and/or experimental characterization of representatives of previously unassigned subfamilies (or cohesion groups on the family tree [3]). Indeed, biochemical in vitro assays as well as the analysis of close sequence similarity may be useful to identify substrate specificities of enzymes. On the other hand, the available genetic data along with the genomic context and pathway reconstruction point to actual roles of these enzymes in metabolic pathways. In that regard, an important and frequently debated question is, to what extent do in vitro substrate preferences of enzymes from functionally heterogeneous families reflect their actual physiological substrates in the respective pathways (21, 22, 45)? A systematic mapping of substrate preferences across the entire microbial sugar kinome is expected to yield a reference data set that will enable automated accurate reconstruction of associated pathways from genomic and metagenomic data.
Here we addressed this problem in the context of the genome-scale reconstruction of the carbohydrate utilization machinery of a marine hyperthermophilic bacterium, Thermotoga maritima (24). The choice of this model organism was dictated by several considerations. First, the analysis of this deep-branched extremophile with a peculiar evolutionary history, which apparently included massive lateral gene transfer from the Archaea and Firmicutes (24, 46), would extend the reference set of sugar kinases characterized by novel phylogenetically diverse enzymes. Second, genes for carbohydrate metabolism account for an unusually large fraction of the T. maritima genome, reflecting access to a large variety of carbohydrates in its ecological niche (7). Third, a published genome-scale reconstruction and structural survey of the T. maritima metabolic network (44) along with recently sequenced additional genomes from the group of Thermotogales (10, 36) provided a solid structure-function and genomic context for this analysis.
To explore the relationship between the physiological roles deduced from the genomic context with their substrate specificities determined by in vitro assays, we profiled the enzymatic activity of 14 tentatively assigned purified recombinant sugar kinases from T. maritima with a panel of 45 different mono- and disaccharides. Remarkably, nearly all of the tested enzymes from the FGGY and PfkB families displayed a strong preference toward a single physiological substrate consistent with their tentative pathway assignments. A single enzyme from the ROK family, previously known as the glucose-specific kinase Glk (13), displayed wide substrate specificity on a panel of tested hexoses and hexosamines, pointing to its actual role in the carbohydrate utilization network of T. maritima. Overall, the obtained results strongly supported the general utility of sugar kinome analysis for genomics-based reconstruction of carbohydrate utilization machinery and extended the reference set of confidently assigned subfamilies of sugar kinases. These findings are expected to impact the analysis of rapidly amassing genomic data on new microbial species and communities.
MATERIALS AND METHODS
Genome resources and bioinformatics tools.
Genome sequences of the Thermotogales species were obtained from GenBank (2). Candidate sugar kinase genes in the T. maritima genome were identified by similarity searches using representatives of protein families of known sugar kinases (FGGY, PfkA, PfkB, GHMP, ROK) using NCBI BLAST (43). Identification of orthologs in closely related genomes and gene neighborhood analysis were performed using the MicrobesOnline (8) and SEED (31) web resources. Phylogenetic analyses of proteins from the FGGY and PfkB families were performed by the maximum likelihood method implemented in PhyML (11). The constructed phylogenetic trees were visualized using Dendroscope (16). Comparative genomic analysis of sugar utilization pathways in 11 Thermotogales species with completely sequenced genomes was performed and captured in the SEED subsystem Sugar Utilization in Thermotogales, available online (http://pubseed.theseed.org/seedviewer.cgi?page=SubsystemSelect). Identification of candidate regulatory sites in genomic sequences and regulon reconstruction were performed using RegPredict software (27). The details of the reconstructed regulons for sugar catabolic genes are displayed in the RegPrecise database in the collection of regulons in the Thermotogales taxonomic group (http://regprecise.lbl.gov/) (26).
Bacterial strains, plasmids, and reagents.
Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) was used for gene cloning, and E. coli BL21(DE3) (Gibco-BRL, Rockville, MD) was used for protein expression. The E. coli DL41 strains that carry a pMH2T7-derived plasmid harboring the T. maritima gene TM1469, TM0067, TM0443, TM1073, TM0828, TM0209, or TM0296 and E. coli HK100 strains that harbor the gene TM0116, TM0284, TM0415, or TM1280 in the same vector under the control of the arabinose-inducible T7 promoter were a kind gift from S. Lesley at the Joint Center for Structural Genomics (20). In addition, the TM0960, TM0952, TM1430, and TM1190 genes were amplified using specific primer pairs (see Table S1 in the supplemental material) from T. maritima MSB8 genomic DNA and cloned into the pET28a vector (Novagen, Madison, WI) or the pET-derived vector pODC29 (29). The resulting plasmids were transformed into E. coli BL21(DE3) and confirmed by DNA sequencing. All recombinant proteins were expressed as fusions with the N-terminal His6 tag. Strains overexpressing E. coli genes araA and rhaA, encoding l-arabinose and l-rhamnose isomerases, respectively, for the coupled assays (see below) were from the ASKA collection (19). Enzymes for PCR and DNA manipulations were from New England BioLabs Inc. (Beverly, MA). Oligonucleotides were synthesized by Sigma-Genosys (Woodlands, TX). Plasmid purification kits were from Promega (Madison, WI). Other chemicals, including the enzyme assay components NADH, ATP, phosphoenolpyruvate (PEP), lactate dehydrogenase (LDH), pyruvate kinase (PYK), and all tested sugars, were from Sigma-Aldrich (St. Louis, MO).
Protein purification.
Recombinant proteins containing an N-terminal His6 tag were overexpressed in E. coli and purified using Ni2+-chelating chromatography. The E. coli strains DL41and HK100 carrying the respective expression plasmids were grown in Terrific broth (TB) medium (50 ml containing 24 g/liter yeast extract and 12 g/liter tryptone) supplemented with 1% glycerol and 50 mM MOPS (morpholinepropanesulfonic acid; pH 7.6), induced by 0.15% l-arabinose at 37°C, and harvested after 4 h of shaking. The E. coli BL21(DE3) strains carrying other expression plasmids were grown in LB medium (50 ml), induced by 0.2 mM isopropyl-β-d-thiogalactopyranoside, and harvested after 12 h of shaking at 20°C. A rapid purification of recombinant proteins on Ni-nitrilotriacetic acid (NTA) agarose minicolumns was performed as described previously (30). Briefly, cells were harvested and resuspended in 20 mM HEPES buffer, pH 7, containing 100 mM NaCl, 2 mM β-mercaptoethanol, and 0.03% Brij 35 with 2 mM phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Sigma-Aldrich). Cells were lysed by incubation with lysozyme (1 mg/ml) for 30 min, followed by a freeze-thaw cycle and sonication. After centrifugation, Tris-HCl buffer (pH 8) was added to the supernatant to a final concentration of 50 mM. The supernatant was then loaded onto an Ni-NTA agarose minicolumn (0.2 ml) from Qiagen Inc. (Valencia, CA). After bound proteins were washed with 50 mM Tris-HCl buffer (pH 8) containing 1 M NaCl and 0.3% Brij 35, they were eluted with 0.3 ml of the same buffer supplemented with 250 mM imidazole. Protein size, expression level, distribution between soluble and insoluble forms, and extent of purification were monitored by SDS-PAGE (see Fig. S1 in the supplemental material). All proteins were obtained with high yield (>1 mg) and purity (80 to 90%).
Enzyme assays.
Activities of the purified recombinant T. maritima enzymes were routinely assayed in a 96-well plate format at 37°C using the standard enzymatic coupling assay as described previously (42). Briefly, an ATP-dependent sugar kinase activity was assayed by coupling the formation of ADP to the oxidation of NADH to NAD+ via PYK and LDH with continuous monitoring at 340 nm in a Beckman DTX-880 plate reader. Typically, 0.2 to 0.4 μg of the tested kinase was added to 200 μl of a reaction mixture containing 50 mM Tris-HCl buffer (pH 7.5), 10 mM MgSO4, 1.2 mM ATP, 1.2 mM PEP, 0.3 mM NADH, 1.2 U of PYK, 1.2 U of LDH, and 5 mM individual sugar substrate. The observed rates (calculated using an NADH extinction coefficient of 6.22 mM−1 cm−1) were compared to those for the two sets of control samples: one control without the tested enzyme and one without the sugar substrate. The initial screening for potential substrates was performed using the following series of cocktails: (i) pentoses, (ii) hexoses, (iii) amino sugars; (iv) phosphosugars, (v) sugar acids, (vi) sugar polyols, and (vii) disaccharides (see Table 2). The final concentration of each sugar in the reaction mixture was 5 mM. Individual substrates from the cocktails revealing enzymatic activity were retested one by one using the same protocol. l-Ribulose and l-rhamnulose, which were not commercially available, were obtained in situ from l-arabinose and l-rhamnose, respectively, by adding excessive amounts of recombinant purified arabinose isomerase (AraA) or rhamnose isomerise (RhaA) from E. coli.
Table 2.
Distribution of genes encoding sugar kinases in 12 Thermotogales genomes
| Family | Protein name | T. maritima locus taga | Presence or absence of ortholog inb: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Thermotoga sp. strain RQ-2 | Thermotoga petrophila RKU-1 | Thermotoga naphthophila RKU-10 | Thermotoga neapolitana DSM 4359 | Thermotoga lettingae TMO | Thermotoga thermarum DSM 5069 | Thermosipho africanus TCF52B | Thermosipho melanesiensis BI429 | Fervidobacterium nodosum Rt17-B1 | Petrotoga mobilis SJ95 | Kosmotoga olearia TBF 19.5.1 | |||
| FGGY | GntK | TM0443 | + | + | + | + | − | − | − | − | − | − | − |
| AraB | TM0284 | + | + | + | + | + | + | − | − | − | + | − | |
| XylB | TM0116 | + | + | + | + | + | + | − | − | − | + | − | |
| GlpK | TM1430 | + | + | + | + | + | + | + | + | + | + | + | |
| DrlK | TM0952 | − | − | − | + | + | − | − | − | − | − | − | |
| RhaB | TM1073 | + | + | + | + | − | − | − | − | − | − | − | |
| PfkB | RbsK | TM0960 | − | + | + | + | + | + | + | − | − | + | + |
| ScrK | TM0296 | + | + | + | + | − | − | − | − | − | + | + | |
| KdgK | TM0067 | + | + | + | + | + | − | − | − | − | − | − | |
| IolK | TM0415 | − | + | + | + | − | + | − | − | − | − | + | |
| LacC | TM0828 | + | + | + | + | + | + | + | + | + | + | + | |
| TM0795 | TM0795 | − | + | + | + | + | + | − | − | − | + | + | |
| COG2971 | BglK | TM1280 | + | + | + | + | + | + | − | − | − | − | − |
| GHMP | GalK | TM1190 | + | + | + | + | + | + | + | + | + | + | + |
| ROK | Glk | TM1469 | + | + | + | + | + | + | + | + | + | + | + |
| COG0469 | Pyk | TM0208 | + | + | + | + | + | + | + | + | + | + | + |
| COG0126 | Pgk | TM0689 | + | + | + | + | + | + | + | + | + | + | + |
| COG2379 | GckA | TM1585 | + | + | + | + | + | + | + | + | + | + | + |
| PfkA | PfkA1 | TM0209 | + | + | + | + | + | + | + | + | + | + | + |
| PfkA2 | TM0289 | + | + | + | + | + | + | − | − | − | − | − | |
Genomic locus tags of sugar kinases in Thermotoga maritima MSB8 are listed. Detailed information on the locus tags of all sugar kinase orthologs is given in Table S2 in the supplemental material.
The presence or absence of orthologs of the T. maritima kinases in other Thermotogales genomes is shown by + and −, respectively.
For selected sugar kinases (e.g., Glk/TM1469), the activity was measured at 80°C using a discontinuous assay. After incubation of the assay mixture containing 50 mM Tris-HCl, 20 mM MgCl2, 2 mM ATP, and 5 mM substrate (at a 200-μl volume) for 5 min at 80°C, the reaction was stopped by the addition of 20 μl of ice-cold 50% (vol/vol) perchloric acid. After vortexing, the mixture was kept on ice for 10 min, neutralized with 30% KOH, and centrifuged. One hundred microliters of the supernatant was mixed with 400 μl of 50 mM Tris-HCl buffer (pH 7.5) containing 2 mM PEP and 0.25 mM NADH. The amount of ADP released by the sugar kinase reaction was quantified by the endpoint measurement of NADH formed after incubation with a mixture of the PYK and LDH coupling enzymes at 340 nm.
RESULTS AND DISCUSSION
Combined bioinformatics and experimental analysis of T. maritima sugar kinome.
Homology-based scanning of the T. maritima genome revealed the presence of at least 20 genes encoding possible sugar kinases (Table 1). Among them, the most abundant were members of the FGGY and PfkB families, including six previously uncharacterized subfamilies within each family. The remaining eight enzymes from other protein families included representatives of six well-known subfamilies mainly implicated in the central carbon metabolism (Glk, PfkA1, PfkA2, Pgk, Pyk, and GckA) and two enzymes of unknown specificity, TM1280 and TM1190. The scope of our bioinformatics analysis of these enzymes was in tentative assignment of physiological roles (e.g., their likely physiological substrates) by combining homology with functional and genomic context analyses in 12 Thermotogales species (Table 2). We used the subsystems-based approach (illustrated in reference 33) to reconstruct the pathway context for 14 putative sugar kinases in T. maritima (see Fig. 1 for an example of pentose utilization pathways). These proteins, as well as the Glk and PfkA1 enzymes, were individually purified and tested for enzymatic activity using in vitro assays.
Table 1.
Sugar kinases encoded in the T. maritima genome
| Family | Gene | Name | Functional role | EC no. | Sugar utilization pathway | Homologs with known functiona | In vitro specificity | Source or reference(s) |
|---|---|---|---|---|---|---|---|---|
| FGGY/COG1070 | TM0116 | xylB | Xylulokinase | 2.7.1.17 | Xylose | Xylulokinase, XYLB_BACSU (38) | d-Xylulose | This work |
| FGGY/COG1070 | TM0284 | araB | l-Ribulokinase | 2.7.1.16 | Arabinose | Gluconokinase, GNTK_BACSU (32) | l-Ribulose | This work |
| FGGY/COG1070 | TM0443 | gntK | Gluconokinase | 2.7.1.12 | 5-Ketogluconate | Gluconokinase, GNTK_BACSU (37) | d-Gluconate | This work |
| FGGY/COG1070 | TM1073 | rhaB | Rhamnulokinase | 2.7.1.5 | Rhamnose | Rhamnulokinase, RHAB_ECOLI (36) | l-Rhamnulose | This work |
| FGGY/COG0554 | TM1430 | glpK | Glycerol kinase | 2.7.1.30 | Glycerol | Glycerol kinase, GLPK_BACSU (67) | d-Glycerol | This work |
| FGGY/COG0554 | TM0952 | drlK | Putative d-ribulokinase | 2.7.1.47? | Putative d-arabinose | Glycerol kinase, GLPK_BACSU (45) | Not determined (weak activity on d-glycerol) | This work |
| PfkB/COG0524 | TM0067 | kdgK | KDG kinase | 2.7.1.45 | Glucuronate | 5-Dehydro-2-deoxyglucono-kinase, IOLC_BACSU (24) | KDG | This work |
| PfkB/COG0524 | TM0960 | rbsK | Ribokinase | 2.7.1.15 | Ribose | Ribokinase, RBSK_ECOLI (39) | d-Ribose | This work |
| PfkB/COG0524 | TM0296 | scrK | Fructokinase | 2.7.1.4 | Mannitol | Fructokinase (SCRK1_ARATH (40) | d-Fructose | This work |
| PfkB/COG0524 | TM0415 | iolK | Putative sugar kinase | Inositol | Not determined | This work | ||
| PfkB/COG0524 | TM0795 | Putative sugar kinase | Unknown | Ribokinase, RBSK_BACSU (24) | Not tested | |||
| PfkB/COG1105 | TM0828 | lacC | Putative tagatose-6-phosphate kinase | 2.7.1.144 | Unknown | Tagatose-6-phosphate kinase, LACC_STRPN (32) | Not determined (weak activity on ManNac) | This work |
| GHMP/COG0153 | TM1190 | galK | Galactokinase | 2.7.1.6 | Galactose | Galactokinase, GAL1_PYRFU (42) | d-Galactose | This work |
| ROK/COG1940 | TM1469 | glk | Glucokinase, hexokinase | 2.7.1.2 | Glycolysis | Glucokinase, GLK_BACSU (34) | d-Glucose, d-mannose, GlcNAc, GlcN | 13 and this work |
| PfkA/COG0205 | TM0209 | pfkA1 | 6-Phosphofructokinase | 2.7.1.11 | Glycolysis | 6-Phosphofructokinase, K6PF_BACSU (55) | d-Fructose-6-phosphate | 9, 12, and this work |
| PfkA/COG0205 | TM0289 | pfkA2 | 6-Phosphofructokinase, polyphosphate dependent | 2.7.1.90 | Glycolysis, gluconeogenesis | 6-Phosphofructokinase, K6PF_BACSU (35) | d-Fructose-6-phosphate | 9 |
| COG2971 | TM1280 | bglK | Glucosamine kinase | 2.7.1.8 | β-Glucoside? | N-Acetylglucosamine kinase, NAGK_RAT (22) | GlcN, d-gluconate | This work |
| COG0469 | TM0208 | pyk | Pyruvate kinase | 2.7.1.40 | Glycolysis | Pyruvate kinase, KPYK1_ECOLI (42) | Pyruvate | 17 |
| COG0126 | TM0689 | pgk | Phosphoglycerate kinase | 2.7.2.3 | Glycolysis | Phosphoglycerate kinase, PGK_BACSU (62) | 3-Phosphoglycerate | 34 |
| COG2379 | TM1585 | gckA | Glycerate-2-kinase | 2.7.1.165 | Serine utilization pathway | Glycerate-2-kinase, GLCK_PYRHO (45) | d-Glycerate | 42 |
The best homologs with known function were determined by BLAST analysis versus the UniProt database. The functional role, UniProt identifier (percent identity with the T. maritima kinase, in parentheses) are shown. Organism abbreviations: BACSU, Bacillus subtilis; ECOLI, Escherichia coli; ARATH, Arabidopsis thaliana; STRPN, Streptococcus pneumoniae; PYRFU, Pyrococcus furiosus; RAT, Rattus norvegicus; PYRHO, Pyrococcus horikoshii.
Fig 1.
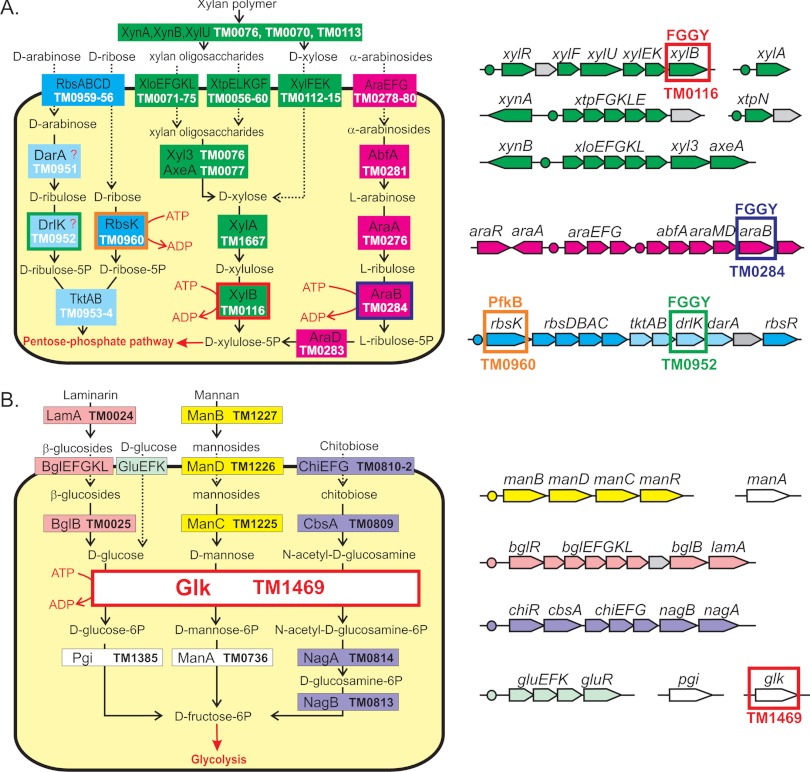
Genomic reconstruction of carbohydrate catabolic pathways in T. maritima. (A) Catabolism of pentoses and pentose-containing sugar polymers; (B) catabolism of selected hexoses and hexose-containing sugar polymers. Sugar catabolic genes and transcription factor binding sites (operators) for sugar-responsive regulons are shown by arrows and circles of matching colors, respectively. Sugar kinases are highlighted by colored rectangles. 5P, 5-phosphate; 6P, 6-phosphate.
The substrate specificity of the 14 proteins was characterized by screening with a panel of 48 commercially available pentoses, hexoses, amino sugars, phosphosugars, sugar acids, and sugar polyols, as well as three disaccharides. We used a general colorimetric assay coupling ADP production to enzymatic conversion of NAD to NADH, which was carried out in a 200-μl volume in a 96-well plate format. The screening results for 14 enzymes are summarized in Table 3.
Table 3.
In vitro specificities of T. maritima sugar kinasesa
| Tested substrate | CM | Sp act (U/mg)b |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KdgK (TM0067) | XylB (TM0116) | PfkA1 (TM0209) | AraB (TM0284) | ScrK (TM0296) | GntK (TM0443) | LacC (TM0828) | DrlK (TM0952) | RbsK (TM0960) | RhaB (TM1073) | GalK (TM1190) | BglK (TM1280) | GlpK (TM1430*c) | Glk (TM1469) | ||
| d-Ribose | (i) | 8.5 | |||||||||||||
| d-Xylulose | (i) | 68 | |||||||||||||
| d-Lyxose | (i) | 3.3 | |||||||||||||
| l-Ribulose | (i) | 4.3 | |||||||||||||
| d-Glucose | (ii) | 15 | |||||||||||||
| 2-Deoxy-d-glucose | (ii) | 7 | |||||||||||||
| d-Mannose | (ii) | 3.4 | |||||||||||||
| d-Galactose | (ii) | 3.4 | |||||||||||||
| d-Fructose | (ii) | 0.27 | |||||||||||||
| l-Rhamnulose | (ii) | 1.3 | |||||||||||||
| N-Acetylmannosamine | (iii) | 0.28 | 0.6 | ||||||||||||
| N-Acetylglucosamine | (iii) | 4.5 | |||||||||||||
| d-Galactosamine | (iii) | 0.25 | |||||||||||||
| d -Glucosamine | (iii) | 6.54 | 4.4 | ||||||||||||
| d-Mannosamine | (iii) | 0.7 | |||||||||||||
| d-Fructose 6-phosphate | (iv) | 2.4 | |||||||||||||
| d-Gluconate | (v) | 31 | 0.12 | 5.45 | |||||||||||
| 5-keto-d-gluconate | (v) | 0.12 | |||||||||||||
| KDG | (v) | 0.2 | |||||||||||||
| d-Glycerate | (v) | 0.2 | |||||||||||||
| Arabinitol | (vi) | 0.55 | |||||||||||||
| Xylitol | (vi) | 0.37 | |||||||||||||
| Ribitol | (vi) | 0.1 | |||||||||||||
| Glycerol | (vi) | 0.53 | 14 | ||||||||||||
Other potential substrates from six cocktail mixtures (CM) that were tested and demonstrated no specific activity include (i) pentoses (2-deoxy-d-ribose, l-xylose, l-arabinose, d-arabinose); (ii) hexoses (d-allose, l-sorbose, l-fucose, d-tagatose, l-rhamnose); (iii) amino sugars (N-acetylgalactosamine); (iv) phosphosugars (d-fructose 1-phosphate, d-glucose-6-phosphate); (v) sugar acids (2-keto-d-gluconate, d-glucuronate); (vi) sugar polyols (erythritol, sorbitol, mannitol, inositol); and (vii) β-glucosides (chitobiose, cellobiose, gentiobiose).
Activities of the purified recombinant enzymes were assayed using the standard enzymatic coupling assay at 37°C (see Materials and Methods) and were recalculated in the enzyme values (U/mg) shown in the table.
Repaired recombinant protein TM1430* was obtained by site-directed mutagenesis (see Results and Discussion).
For the scope of this study, the comparison of specific activities (in lieu of genuine steady-state kinetic parameters) provided a reasonable approximation of the relative substrate preferences of each tested T. maritima kinase. The initial activity screen was performed using mixtures (cocktails) of several similar sugars (such as hexoses and pentoses; Table 3). Proteins with identified catalytic activity against cocktails then passed on to secondary screens with individual substrates. Although T. maritima enzymes are usually thermophilic, most of the analyzed sugar kinases were sufficiently active to collect the necessary data at 37°C using the continuous coupled assay format. For selected enzymes (including those showing only low or no activity at 37°C), the measurements were repeated at 80°C using a discontinuous endpoint assay. As illustrated for the example of TM1469, the substrate selectivity profile at 80°C generally remained the same as that at 37°C, despite the ∼10-fold higher observed activity (Table 4).
Table 4.
Specific activity of TM1469 on various hexoses at 37°C and 80°C
| Substratea | Sp act (U mg−1) |
|
|---|---|---|
| 37°C | 80°C | |
| d-Glucose | 10.69 ± 4.03 (100)b | 104.9 ± 3.5 (100) |
| 2-Deoxy-d-glucose | 3.74 ± 0.52 (35) | 33 (32) |
| N-Acetyl-d-glucosamine | 0.95 ± 0.57 (8.8) | 18.68 (18) |
| d-Glucosamine | 2.29 ± 1.52 (21) | 12.13 ± 1.59 (11.5) |
| d-Mannose | 0.99 ± 0.13 (9.2) | 4.99 ± 1.41 (1.9) |
| d-Mannosamine | 0.25 (2.3) | 0.57 ± 0.36 (0.5) |
| N-Acetyl-d-mannosamine | 0.58 (5.4) | 0.082 (0.07) |
Substrates were used at 5 mM.
Data in parentheses represent percentage of total activity.
Overall, for nine analyzed kinases (TM0116, TM0284, TM0443, TM1073, TM1430, TM0067, TM0960, TM0296, and TM1190), the experimentally observed narrow substrate specificity matched precisely the physiological role inferred by the bioinformatics analysis, whereas for a single kinase (TM1280), the experimental screening determined a novel specificity that was not predicted by bioinformatics (Table 1). Among them, TM0284 and TM1280 are novel kinases that are only distantly related to characterized enzymes according to the phylogenetic analysis (see below). For Glk (TM1469), the observed broader substrate specificity allowed us to suggest a previously unknown dual physiological role in the d-mannose and N-acetyl-d-glucosamine utilization pathways. For PfkA1 (TM0209), we confirmed the specific activity on d-fructose-6-phosphate that was reported previously (9, 12). For two enzymes, TM0952 and TM0828, the predicted activity could not be tested because of substrate availability. Below, we describe the results of both bioinformatics and experimental analyses for all enzymes grouped by sugar kinase families.
FGGY family.
We revealed orthologs of each of six FGGY-family kinases from T. maritima in the genomes of other Thermotogales (Table 2) and also determined their best similarity hits in the UniProt database of experimentally characterized proteins (Table 1). Phylogenetic analysis of the identified proteins from the FGGY family performed here, as well as in our previous study (45), provided first clues on their possible substrate preferences (Fig. 2). These results, supplemented by genome context analysis, allowed us to reconstruct the cognate sugar utilization pathways and predict the specificities of the associated sugar kinases (Table 1) (45). In vitro enzymatic assays over a panel of diagnostic sugar substrates confirmed stringent preferences toward the respective physiological substrates for five FGGY kinases in T. maritima, namely, AraB, XylB, GlpK, GntK, and RhaB (Table 3).
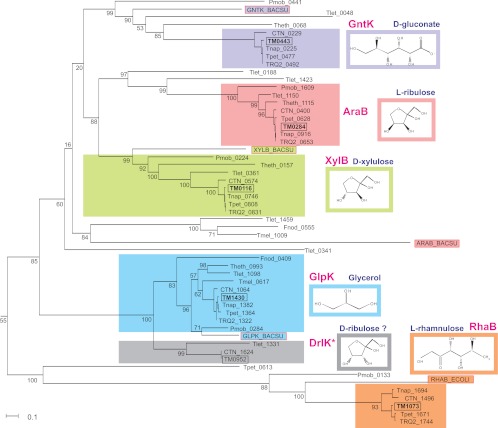
Fig 2.
Maximum likelihood phylogenetic tree of FGGY-family kinases in the Thermotogales. The numbers represent bootstrap values (in percent) obtained from 1,000 replicates. TRQ2, Thermotoga sp. strain RQ-2; Tpet, Thermotoga petrophila RKU-1; Tnap, Thermotoga naphthophila RKU-10; CTN, Thermotoga neapolitana DSM 4359; Tlet, Thermotoga lettingae TMO; Theth, Thermotoga thermarum DSM 5069; Tmel, Thermosipho melanesiensis BI429; Fnod, Fervidobacterium nodosum Rt17-B1; Pmob, Petrotoga mobilis SJ95.
AraB/TM0284 and XylB/TM0116 kinases are involved in the catabolism of two pentose sugars, l-arabinose and d-xylose, respectively. The proposed metabolic pathways for utilization of these sugars and their oligosaccharide/polymer precursors are illustrated in Fig. 1A. Genomic loci encoding the arabinose and xylose utilization genes are present in all analyzed Thermotoga spp. (Table 2), where they are controlled by the cognate transcriptional regulators AraR and XylR, respectively (Fig. 1A). T. maritima XylE is a component of the ABC-type xylose transporter that has substrate specificity to d-xylose (23). The ortholog of araA in Thermotoga neapolitana encodes functional l-arabinose isomerase (18). Our experimental results confirmed the proposed activity of the recombinant T. maritima AraB and XylB enzymes on l-ribulose and d-xylulose, respectively (Table 3). Interestingly, the Thermotoga-specific branch of AraB proteins is distant from other known AraB proteins on the phylogenetic tree, suggesting their early divergence in the evolution of FGGY kinases (45).
GlpK/TM1430 kinase is encoded in the glycerol utilization gene cluster containing glycerol-3-phosphate dehydrogenase and glycerol operon antiterminator. GlpK is the only kinase from the FGGY family that is conserved in all Thermotogales genomes (Table 2). On the phylogenetic tree, GlpK proteins from the Thermotogales cluster with known glycerol kinases from other bacterial lineages such as Bacillus subtilis (Fig. 2) (45). However, the initial screening of TM1430 enzymatic activity on all tested substrates revealed very low activity on d-glycerol (0.32 U/mg). Alignment of orthologous GlpK proteins from Thermotoga spp. showed that the T. maritima TM1430 gene is truncated due to a single nucleotide nonsense mutation resulting in a protein product that lacks the last 14 amino acids (see Fig. S2 in the supplemental material). We confirmed this mutation by resequencing from genomic DNA, showing that it was not due to gene cloning or sequencing error. The premature stop codon TGA in the TM1430 gene (a likely result of isolation or cultivation under nonselective conditions) was replaced by TGG (which occurs in the orthologs from other Thermotogales) using site-directed mutagenesis. The repaired recombinant protein, TM1430*, was purified, and its activity and substrate specificity were assessed in vitro. The TM1430* protein has 43-fold higher activity on d-glycerol than the original truncated TM1430 protein (14 U/mg) and is not active on other tested substrates (Table 3).
GntK/TM0443 and RhaB/TM1073 kinases are encoded by gene clusters that are implicated in the catabolism of 5-keto-d-gluconate and l-rhamnose, respectively. The d-gluconate kinase GntK is functionally coupled with 5-keto-d-gluconate reductase TM0441 (I. A. Rodionova, unpublished data). The l-rhamnulose kinase RhaB is linked to the characterized l-rhamnose isomerase RhaA/TM1071 (32). Orthologs of both T. maritima kinases were identified in four closely related Thermotoga genomes, where they have the same genomic and functional context (Table 2). Our experimental results confirmed the proposed substrate specificities of the recombinant T. maritima GntK and RhaB enzymes on d-gluconate and l-rhamnulose, respectively (Table 3).
Predicted d-ribulose kinase DrlK/TM0952 is encoded by a putative d-arabinose catabolic gene cluster, which is colocalized with the d-ribose utilization genes (Fig. 1). The putative two-subunit transketolase TktAB encoded in this gene cluster is solely dedicated to the predicted d-arabinose utilization pathway, as the primary transketolase from the pentose phosphate pathway is also present in all Thermotogales genomes (TM1762 in T. maritima). Interestingly, the predicted d-arabinose catabolic genes are present in 3 out of 9 Thermotogales genomes possessing the d-ribose catabolic operons (see Fig. S3 in the supplemental material). Experimental testing of TM0952 did not reveal activity on any tested substrates except for a relatively low activity on d-glycerol and d-glycerate (Table 3). However, the predicted d-ribulose kinase activity could not be tested, as this compound was not commercially available. The observed low activity of DrlK on d-glycerol is consistent with the observation that DrlK is the closest paralog of the glycerol-specific GlpK kinases (Fig. 2).
PfkB family.
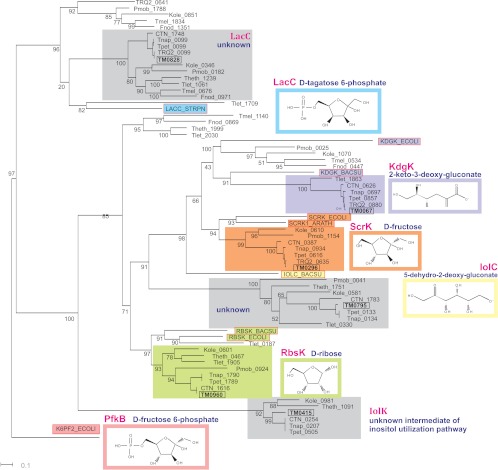
The PfkB family in T. maritima is represented by six proteins (Table 1). Orthologs of the PfkB-family kinases determined in 12 Thermotogales species (Table 2) form six distinct clusters on the phylogenetic tree, each containing 6 to 12 proteins (Fig. 3). The specificities of RbsK/TM0960, KdgK/TM0067, ScrK/TM0296, and LacC/TM0828 could initially be predicted by the significant similarity of these proteins with the kinases characterized from model organisms supported by their clustering on the phylogenetic tree. The genome context analysis confirms the homology-based functional assignments for RbsK, KdgK, and ScrK. The predicted ribulokinase gene rbsK in T. maritima is colocalized with genes encoding the ribose-specific ABC transporter (23) (Fig. 1), and this gene arrangement is conserved in other Thermotogales (see Fig. S2 in the supplemental material). The predicted 2-keto-3-deoxy-gluconate (KDG) kinase gene kdgK in five Thermotoga genomes is located within the kdgAK-uxuBA (TM0066 to TM0069) gene cluster encoding enzymes that are 27 to 40% identical to the d-glucuronate catabolic enzymes from E. coli. The predicted fructokinase scrK was found in a conserved chromosomal cluster with a gene encoding the d-mannitol dehydrogenase MtlD/TM0298 (35). Thus, the Mtl d-dependent oxidation of d-mannitol produces d-fructose, which is further phosphorylated by ScrK. The experimental assessment of T. maritima RbsK, ScrK, and KdgK kinases confirmed their stringent substrate preferences for d-ribose, d-fructose, and KDG, respectively (Table 3). Although the specific activity of ScrK on d-fructose was very low at 37°C (0.27 U/mg), it increased ∼100-fold at a higher temperature (28 U/mg at 80°C), while the enzyme remained inactive on all other sugars tested.
Fig 3.
Maximum likelihood phylogenetic tree of PfkB-family kinases in the Thermotogales. The numbers represent bootstrap values (in percent) obtained from 1,000 replicates. TRQ2, Thermotoga sp. strain RQ-2; Tpet, Thermotoga petrophila RKU-1; Tnap, Thermotoga naphthophila RKU-10; CTN, Thermotoga neapolitana DSM 4359; Tlet, Thermotoga lettingae TMO; Theth, Thermotoga thermarum DSM 5069; Tmel, Thermosipho melanesiensis BI429; Fnod, Fervidobacterium nodosum Rt17-B1; Pmob, Petrotoga mobilis SJ95; Kole, Kosmotoga olearia TBF 19.5.1.
In contrast, the putative sugar kinase lacC in T. maritima and its orthologs in other Thermotogales are located in the conserved gene cluster TM0828 to TM0838 encoding some essential cellular functions such as DNA gyrase, cell division proteins, and tRNA modification enzymes. The observed genomic context of lacC homologs and the absolute conservation of orthologous kinases in all Thermotogales were suggestive of a role in central metabolism (rather than in catabolic feeding pathways). Experimental screening for the specific activity of the TM0828 kinase with our panel of sugars showed very low activity with N-acetylmannosamine at 37°C (0.28 U/mg), which was not confirmed at the higher temperature (80°C). TM0828 is an ortholog of the d-tagatose-6-phosphate kinase LacC from Firmicutes, and it is reasonable to suggest the same enzymatic function in Thermotogales, although the metabolic pathway context for this proposed function is unknown in Thermotoga. However, this tentative assignment could not be tested since d-tagatose-6-phosphate is not commercially available and could not be easily generated in situ.
The remaining two putative PfkB-type kinases in T. maritima, TM0415 and TM0795, are only distantly related to the characterized kinases from the PfkB family (Table 1). These two kinases and their orthologs in other Thermotogales form two separate and divergent branches on the PfkB-family phylogenetic tree (Fig. 3). The genome context analysis allowed us to suggest possible involvement of the TM0415 kinase in a novel inositol catabolic pathway (Rodionova, unpublished); however, its specific substrate specificity is yet unknown.
Other families of sugar kinases.
Glk/TM1469 is a single representative of the ROK kinase family in T. maritima which was previously characterized as ATP-dependent glucokinase with stringent substrate specificity on d-glucose and, to some extent, on 2-deoxyglucose (13). Our study confirmed the glucokinase activity of Glk/TM1469 but also revealed its rather broad substrate specificity, which also includes N-acetyl-d-glucosamine, d-glucosamine, d-mannose, and d-mannosamine. The specific enzymatic activity of Glk on these substrates was 2- to 20-fold higher at 80°C than at 37°C, while it showed essentially the same preference profile (Table 4).
The determined broad substrate specificity of the Glk kinase in T. maritime is corroborated by the genomic reconstruction of the sugar utilization metabolic pathways. Thus, the pathway for utilization of chitobiose, a disaccharide of N-acetyl-d-glucosamine, which is encoded by the TM0808 to TM814 gene cluster, lacks N-acetyl-d-glucosamine-specific kinase (Fig. 1B). Similarly, d-mannose-specific kinase is absent from the reconstructed pathway for utilization of mannan and mannosides, which is encoded by the TM1224 to TM1227 gene cluster. Therefore, the respective activities of TM1469 may potentially fill in the missing steps in these two pathways (Fig. 1B).
The GalK/TM1190 protein belongs to the GHMP family and is similar to galactokinase from Pyrococcus furiosus (14). GalK is encoded in the lactose-induced TM1201 to TM1190 gene cluster in T. maritima (25), which also contains galactose-1-phosphate uridylyltransferase (GalT/TM1191), α-galactosidase (GalA/TM1192), two β-galactosidases (LacZ/TM1193 and LacA/TM1195), secreted arabinogalactan endo-1,4-β-galactosidase (GanA/TM1201), and a predicted ABC-type galactoside transporter (LtpEFGK/TM1194 to TM1199). Orthologs of the galK gene are present in all Thermotogales species, where they are always colocalized with the galT genes (Table 2). Substrate screening with the purified GalK protein confirmed its strong preference for d-galactose and determined d-galactosamine to be a secondary possible substrate with 14-fold lower activity (Table 3).
The TM0209 and TM0289 kinases belong to the PfkA family, which includes solely 6-phosphofructose-specific kinases from various microorganisms. Both PfkA-type kinases in T. maritima were previously characterized as 6-phosphofructokinases that are involved in the glycolysis pathway (9, 12). Here we screened TM0209 on the panel of 45 different sugars and showed its strong substrate preference for d-fructose-6-phosphate (Table 3). The TM0209 gene is highly conserved in all Thermotogales and clusters on the chromosome with another central metabolic enzyme, the pyruvate kinase gene pyk/TM0208.
The hypothetical sugar kinase TM1280 belongs to the COG2971 protein family, which is represented by the characterized N-acetyl-d-glucosamine-specific kinases NagK from both bacteria (e.g., from Shewanella oneidensis [41]) and eukaryotes (e.g., from human and rat [15]). However, the sequence similarity between different NagK proteins and TM1280 is weak (20 to 22% identity), suggesting a possible functional divergence. The TM1280 orthologs were found in six other Thermotoga spp., where they were always located in a putative operon with downstream gene TM1281, encoding the 6-phospho-β-glucosidase BglT (38). The genomic context suggests that TM1280 may be involved in the phosphorylation of β-glucosides, thus providing 6-phospho-β-glucosides for the BglT hydrolase. This kinase was therefore tentatively named BglK. However, the screening for specific kinase activity of BglK with three β-glucosides, namely, cellobiose, gentibiose, and chitobiose, did not confirm this prediction. Instead, the TM1280 kinase was found to be active on d-glucosamine and d-gluconate (Table 3), although the physiological implication of these observed activities for Thermotoga metabolism is not clear.
Conclusions.
The marine hyperthermophilic bacterium T. maritima has extensive and highly diversified carbohydrate utilization machinery. In this study, we applied the integrated approach to infer and experimentally assess biochemical functions of sugar kinases involved in a variety of carbohydrate utilization pathways in this bacterium. Using genome context analysis, we were able to tentatively assign biological roles in reconstructed pathways and infer substrate preferences for a substantial fraction of the analyzed sugar kinome (Table 1). For 5 of these enzymes, namely, TM0116/XylB, TM0284/AraB, TM1073/RhaB, TM0952/DrlK, and TM0415/IolK, the bioinformatic analysis predicted their biochemical functions for the first time in this study. Biochemical assays with purified recombinant proteins identified the kinase activity with a panel of 45 diagnostic mono- and disaccharides. Remarkably, nearly all of the 14 experimentally characterized enzymes displayed a strong preference toward a single substrate corresponding to their respective biological functions. The scope of experimental analysis was limited to primary screening, which was performed under suboptimal temperature conditions for T. maritima enzymes (37°C). However, our detailed analysis of one enzyme, the hexokinase Glk/TM1469, suggested a full correlation of substrate relative preference between two compared temperatures (37°C and 80°C), which is also consistent with other reports (39, 42).
Overall, the integrative sugar kinome analysis combining the subsystem-based bioinformatic approach, phylogenetic analysis, and experimental substrate screening revealed a high level of consistency between the physiological roles of the T. maritima sugar kinases and their in vitro substrate preferences. The results of this analysis confirm that sugar kinases may indeed be used as signature genes/enzymes for the inference of respective catabolic pathways from genomic data.
Supplementary Material
ACKNOWLEDGMENTS
This research was partially supported by the U.S. Department of Energy, Office of Science (Biological and Environmental Research), as part of Genomic Science Program contracts DE-FG02-08ER64686 and DE-SC0004999 with the Sanford-Burnham Medical Research Institute. Additional funding was provided by the Russian Foundation for Basic Research (10-04-01768 to D.A.R.), the Russian Academy of Sciences under the program Molecular and Cellular Biology (to D.A.R.), the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-G-5 to C.Y.), and the Towsley Foundation (Midland, MI) through the Towsley Research Scholar program at Hope College (to A.A.B.).
Footnotes
Published ahead of print 10 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andreassi JL, II, Leyh TS. 2004. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry 43:14594–14601 [DOI] [PubMed] [Google Scholar]
- 2.Benson DA, et al. 2012. GenBank. Nucleic Acids Res. 40:D48–D53 doi:10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner CA, et al. 2008. Cohesion group approach for evolutionary analysis of TyrA, a protein family with wide-ranging substrate specificities. Microbiol. Mol. Biol. Rev. 72:13–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, Sander C, Valencia A. 1993. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 2:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheek S, Ginalski K, Zhang H, Grishin NV. 2005. A comprehensive update of the sequence and structure classification of kinases. BMC Struct. Biol. 5:6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheek S, Zhang H, Grishin NV. 2002. Sequence and structure classification of kinases. J. Mol. Biol. 320:855–881 [DOI] [PubMed] [Google Scholar]
- 7.Conners SB, et al. 2006. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 30:872–905 [DOI] [PubMed] [Google Scholar]
- 8.Dehal PS, et al. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38:D396–D400 doi:10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding YR, Ronimus RS, Morgan HW. 2001. Thermotoga maritima phosphofructokinases: expression and characterization of two unique enzymes. J. Bacteriol. 183:791–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frock AD, Notey JS, Kelly RM. 2010. The genus Thermotoga: recent developments. Environ. Technol. 31:1169–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 12.Hansen T, Musfeldt M, Schonheit P. 2002. ATP-dependent 6-phosphofructokinase from the hyperthermophilic bacterium Thermotoga maritima: characterization of an extremely thermophilic, allosterically regulated enzyme. Arch. Microbiol. 177:401–409 [DOI] [PubMed] [Google Scholar]
- 13.Hansen T, Schonheit P. 2003. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS Microbiol. Lett. 226:405–411 [DOI] [PubMed] [Google Scholar]
- 14.Hartley A, et al. 2004. Substrate specificity and mechanism from the structure of Pyrococcus furiosus galactokinase. J. Mol. Biol. 337:387–398 [DOI] [PubMed] [Google Scholar]
- 15.Hinderlich S, Berger M, Schwarzkopf M, Effertz K, Reutter W. 2000. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 267:3301–3308 [DOI] [PubMed] [Google Scholar]
- 16.Huson DH, et al. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460 doi:10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnsen U, Hansen T, Schonheit P. 2003. Comparative analysis of pyruvate kinases from the hyperthermophilic archaea Archaeoglobus fulgidus, Aeropyrum pernix, and Pyrobaculum aerophilum and the hyperthermophilic bacterium Thermotoga maritima: unusual regulatory properties in hyperthermophilic archaea. J. Biol. Chem. 278:25417–25427 [DOI] [PubMed] [Google Scholar]
- 18.Kim BC, et al. 2002. Cloning, expression and characterization of l-arabinose isomerase from Thermotoga neapolitana: bioconversion of d-galactose to d-tagatose using the enzyme. FEMS Microbiol. Lett. 212:121–126 [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa M, et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 20.Lesley SA, et al. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. U. S. A. 99:11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller BG, Raines RT. 2004. Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry 43:6387–6392 [DOI] [PubMed] [Google Scholar]
- 22.Miller BG, Raines RT. 2005. Reconstitution of a defunct glycolytic pathway via recruitment of ambiguous sugar kinases. Biochemistry 44:10776–10783 [DOI] [PubMed] [Google Scholar]
- 23.Nanavati DM, Thirangoon K, Noll KM. 2006. Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars. Appl. Environ. Microbiol. 72:1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson KE, et al. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329 [DOI] [PubMed] [Google Scholar]
- 25.Nguyen TN, et al. 2004. Whole-genome expression profiling of Thermotoga maritima in response to growth on sugars in a chemostat. J. Bacteriol. 186:4824–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novichkov PS, et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 doi:10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novichkov PS, et al. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38:W299–W307 doi:10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omelchenko MV, Galperin MY, Wolf YI, Koonin EV. 2010. Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 5:31 doi:10.1186/1745-6150-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterman A, Grishin NV, Kinch LN, Phillips MA. 1994. Formation of functional cross-species heterodimers of ornithine decarboxylase. Biochemistry 33:13662–13667 [DOI] [PubMed] [Google Scholar]
- 30.Osterman AL, et al. 1995. Domain organization and a protease-sensitive loop in eukaryotic ornithine decarboxylase. Biochemistry 34:13431–13436 [DOI] [PubMed] [Google Scholar]
- 31.Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park CS, Yeom SJ, Lim YR, Kim YS, Oh DK. 2010. Characterization of a recombinant thermostable l: -rhamnose isomerase from Thermotoga maritima ATCC 43589 and its application in the production of l-lyxose and l-mannose. Biotechnol. Lett. 32:1947–1953 [DOI] [PubMed] [Google Scholar]
- 33.Rodionov DA, et al. 2010. Genomic encyclopedia of sugar utilization pathways in the Shewanella genus. BMC Genomics 11:494 doi:10.1186/1471-2164-11-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schurig H, et al. 1995. Phosphoglycerate kinase and triosephosphate isomerase from the hyperthermophilic bacterium Thermotoga maritima form a covalent bifunctional enzyme complex. EMBO J. 14:442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song SH, Ahluwalia N, Leduc Y, Delbaere LT, Vieille C. 2008. Thermotoga maritima TM0298 is a highly thermostable mannitol dehydrogenase. Appl. Microbiol. Biotechnol. 81:485–495 [DOI] [PubMed] [Google Scholar]
- 36.Swithers KS, et al. 2011. Genome sequence of Thermotoga sp. strain RQ2, a hyperthermophilic bacterium isolated from a geothermally heated region of the seafloor near Ribeira Quente, the Azores. J. Bacteriol. 193:5869–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Titgemeyer F, Reizer J, Reizer A, Saier MH., Jr 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140(Pt 9):2349–2354 [DOI] [PubMed] [Google Scholar]
- 38.Varrot A, et al. 2005. NAD+ and metal-ion dependent hydrolysis by family 4 glycosidases: structural insight into specificity for phospho-beta-d-glucosides. J. Mol. Biol. 346:423–435 [DOI] [PubMed] [Google Scholar]
- 39.Vieille C, Hess JM, Kelly RM, Zeikus JG. 1995. xylA cloning and sequencing and biochemical characterization of xylose isomerase from Thermotoga neapolitana. Appl. Environ. Microbiol. 61:1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu LF, et al. 1991. Nucleotide sequence of the Rhodobacter capsulatus fruK gene, which encodes fructose-1-phosphate kinase: evidence for a kinase superfamily including both phosphofructokinases of Escherichia coli. J. Bacteriol. 173:3117–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C, et al. 2006. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 281:29872–29885 [DOI] [PubMed] [Google Scholar]
- 42.Yang C, Rodionov DA, Rodionova IA, Li X, Osterman AL. 2008. Glycerate 2-kinase of Thermotoga maritima and genomic reconstruction of related metabolic pathways. J. Bacteriol. 190:1773–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye J, McGinnis S, Madden TL. 2006. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 34:W6–W9 doi:10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. 2009. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science 325:1544–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Zagnitko O, Rodionova I, Osterman A, Godzik A. 2011. The FGGY carbohydrate kinase family: insights into the evolution of functional specificities. PLoS Comput. Biol. 7:e1002318 doi:10.1371/journal.pcbi.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhaxybayeva O, et al. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc. Natl. Acad. Sci. U. S. A. 106:5865–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.