Abstract
The melibiose permease of Salmonella enterica serovar Typhimurium (MelBSt) catalyzes symport of melibiose with Na+, Li+, or H+. Bioinformatics and mutational analyses indicate that a conserved Gly117 (helix IV) is a component of the Na+-binding site. In this study, Gly117 was mutated to Ser, Asn, or Cys. All three mutations increase the maximum rate (Vmax) for melibiose transport in Escherichia coli DW2 and greatly decrease Na+ affinity, indicating that intracellular release of Na+ is facilitated. Rapid melibiose transport, particularly by the G117N mutant, triggers osmotic lysis in the lag phase of growth. The findings support the previous conclusion that Gly117 plays an important role in cation binding and translocation. Furthermore, a spontaneous second-site mutation (P148L between loop4-5 and helix V) in the G117C mutant prevents cell lysis. This mutation significantly decreases Vmax with little effect on cosubstrate binding in G117C, G117S, and G117N mutants. Thus, the P148L mutation specifically inhibits transport velocity and thereby blocks the lethal effect of elevated melibiose transport in the Gly117 mutants.
INTRODUCTION
The melibiose permease of Salmonella enterica serovar Typhimurium (MelBSt) shares 86% identity and 96% similarity to the primary sequence of its Escherichia coli orthologue (MelBEc) (45, 15). Like MelBEc (1, 8, 12–14, 23–25, 27, 29, 35), MeBSt catalyzes symport of galactoside with Na+, Li+, or H+ (15, 18), utilizing the free energy from the downhill translocation of one cosubstrate to drive uphill translocation of the other (3, 39, 40, 42, 44), and all three cations compete for a common binding pocket (6, 18, 26, 31). A threading model of MelB (45) based on the crystal structure of LacY (2, 16, 17, 30) suggests that MelB is a member of the major facilitator superfamily; thus, the protein is likely organized into two pseudosymmetrical six-helix bundles connected by a long middle loop surrounding an internal cavity facing the cytoplasm. Both cosubstrate-binding sites have been proposed to lie within the internal cavity (Fig. 1). This model is consistent with numerous (9–11, 14, 15, 18, 21, 28, 29, 34, 36, 46, 47) biochemical and biophysical results, as well as with low-resolution electron microscopy (EM) structures of MelBEc (20, 37).
Fig 1.
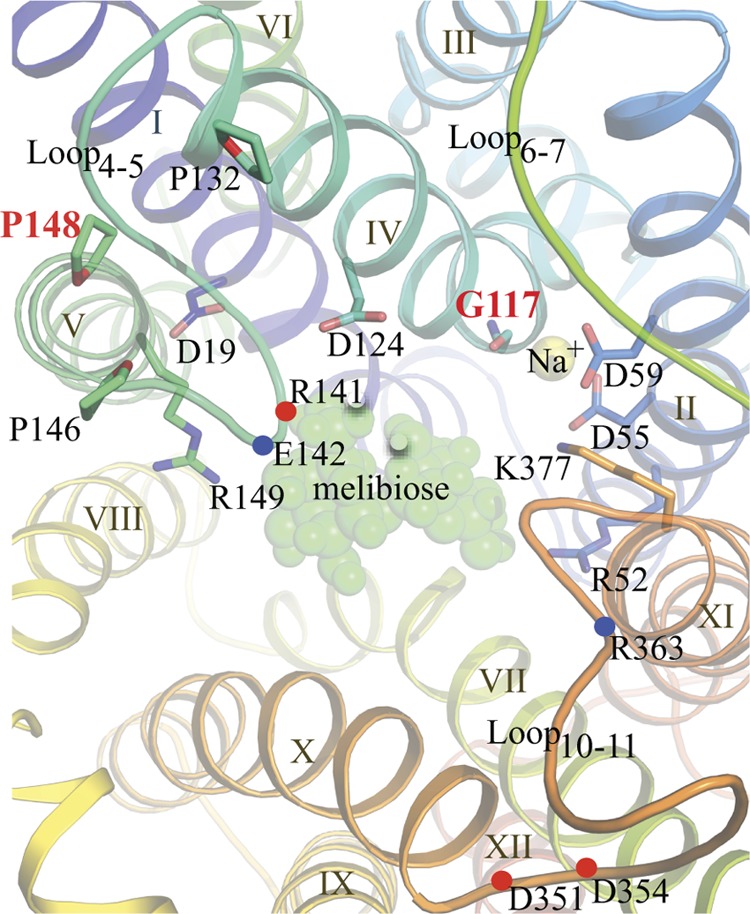
Putative cosubstrate-binding sites of MelB viewed from the cytoplasmic side. The helices are colored with the colors of the rainbow from N (blue) to C termini (red) and are numbered with Roman numerals. Side chains essential (D55 and D59) for Na+ binding and important for melibiose binding/transport (D19, D124, R52, R149, and K377) are shown as sticks. Gly117 is shown as a backbone. Three cytoplasmic loops are labeled as Loop4-5, Loop6-7, and Loop10-11. Pro132, Pro146, and Pro148 are shown as sticks. Positions for Arg141 and Glu142 in loop4-5 and Asp351, Asp 354, and Arg363 in loop10-11 are indicated by blue or red dots. A melibiose molecule and a sodium ion are shown as green and yellow spheres, respectively (45).
The proposed Na+-binding site lies between helices II and IV, and the carboxyl groups of conserved Asp55 and Asp59 (helix II) (14, 21, 28, 34, 36, 46, 47) and the carbonyl oxygen of Gly117 (helix IV) may participate in Na+ coordination (Fig. 1) (11, 15, 43, 45). Helix IV is in the center of a charge/H-bond network involved in the binding of the two cosubstrates (4, 36, 45, 47). Furthermore, two cytoplasmic loops (loop4-5 in the N-terminal domain and loop10-11 in the C-terminal domain) contain highly conserved charged and polar residues (45), some of which are functionally important (1, 7, 29). It has been postulated that rearrangements of loop4-5 and loop10-11 play an important role(s) in ligand recognition and/or conformational switching between functional states during the turnover (45).
Gly117 in MelBSt has been mutated previously to Ala, Pro, Trp, or Arg (15), and the effects of these mutations on cosubstrate binding and transport depend on the physical and chemical properties of the side chain. Compared to wild-type (WT) MelBSt, the G117A mutant exhibits little difference in either cosubstrate binding or Na+- or Li+-coupled melibiose transport; the other three mutations reduce melibiose active transport and decrease the apparent affinity for cations, with a stronger effect on Na+. Among these mutations, a bulky Trp at position 117 causes the greatest inhibition of melibiose binding. Remarkably, the G117R mutant catalyzes melibiose exchange in the presence of Na+ or Li+ but does not catalyze translocation reactions that involve net flux of the coupling cation. The data support a kinetic model in which melibiose is released prior to release of the coupling cation. The findings also support the conclusion that Gly117 plays an important role in cation binding and translocation. Further mutational analyses of Gly117 are reported in this communication.
MATERIALS AND METHODS
Materials.
[1-3H]melibiose was custom synthesized by PerkinElmer (Boston, MA). 2′-(N-Dansyl)minoalkyl-1-thio-β-d-galactopyranoside (D2G) was kindly provided by H. Ronald Kaback and Gérard Leblanc. Oligodeoxynucleotides were synthesized by Integrated DNA Technologies. MacConkey agar medium (lactose free) was from Difco. All other materials were reagent grade and obtained from commercial sources.
Bacterial strains and plasmids.
E. coli strain DW2 (melA+ ΔmelB ΔlacZY) was used for the functional characterization. E. coli XL1-Blue cells were used for DNA manipulations. The expression plasmid pK95 ΔAH/MelBSt/CHis10 (18, 35), which encodes the full-length MelBSt with L5→M and a His10 tag at the C terminus (termed the wild type), was used as the template. All mutants were constructed by a QuikChange site-directed mutagenesis kit from Stratagene and confirmed by DNA sequencing.
Protein overexpression.
E. coli DW2 cells containing a given plasmid were grown in Luria-Bertani (LB) broth (5 g yeast extract and 10 g tryptone per liter with 171 mM NaCl) with 100 mg/liter of ampicillin in a 37°C shaker. The overnight cultures were diluted by 5% with LB broth supplemented with 0.5% glycerol (LB-G) and 100 mg/liter of ampicillin, and constitutive overexpression was obtained by shaking at 30°C for another 5 h.
Preparation of crude membranes, SDS-12% PAGE, and Western blotting.
The 5-h cultures with the expressed MelBSt were washed with 20 mM Tris-HCl (pH 7.5). The preparation of crude membranes was carried out as described previously (15). After a protein assay using a Micro-bicinchoninic acid protein assay kit (Pierce), 25 μg of crude membranes was loaded onto each well of an SDS-12% PAGE plate. After transfer onto a polyvinylidene difluoride (PVDF) membrane by the Trans-Blot Turbo transfer system (Bio-Rad), the PVDF membrane was reacted with the penta-His horseradish peroxidase conjugate (Qiagen). MelBSt proteins were detected using the SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) by the ImageQuant LAS 4000 Biomolecular Imager (GE Health Care Life Science).
Melibiose effect on cell growth.
Overnight cultures in the absence of melibiose were diluted by 5% with LB-G or NaCl-removed LB-G broth in the absence or presence of melibiose at 0.4, 10, or 30 mM, respectively, and shaken at 37°C. Cell optical density was monitored hourly using the A600 for 10 h.
Preparation of RSO membrane vesicles.
Right-side-out (RSO) membrane vesicles were prepared from E. coli DW2 cells by osmotic lysis (18, 22, 38), extensively washed, and resuspended in 100 mM KPi, pH 7.5, and 10 mM MgSO4 at a protein concentration of 25 to 30 mg/ml, frozen in liquid N2, and stored at −80°C.
Melibiose fermentation and acidification.
The DW2 cells were transformed with a given plasmid, plated on MacConkey agar supplemented with melibiose at a range of concentrations between 0.01 and 30 mM (the sole carbohydrate source) and 100 mg/liter ampicillin, and incubated at 37°C (15). After 18 h, the plates were viewed and photographed immediately.
[1-3H]melibiose transport assay.
E. coli DW2 cells expressing MelBSt were washed with 100 mM KPi (pH 7.5; so-called Na+-free buffer) as described previously (18). The cell pellets were resuspended with 100 mM KPi, pH 7.5, 10 mM MgSO4 and adjusted to an A420 of 10 (∼0.7 mg protein/ml). Intracellular melibiose was assayed by fast filtration as described previously (18).
Kinetics of melibiose transport.
Initial rates of melibiose transport at a range of melibiose concentrations between 0.05 and 2.5 mM were obtained by a linear fitting of the melibiose uptake at 0, 3, 4, 6, 8, and 10 s, corrected by the rates obtained from nontransformed DW2 cells, and plotted as a function of melibiose concentration. The melibiose concentration yielding a half-maximum rate of melibiose transport (Km) and the maximum rate of melibiose transport (Vmax) were determined by fitting a hyperbolic function to the data (OriginPro 8.6).
K0.5Na+ for D2G fluorescence resonance energy transfer (FRET).
Steady-state measurements were performed with an AMINCO-Bowman series 2 spectrometer with RSO membrane vesicles at a protein concentration of ∼ 0.5 mg/ml in 100 mM KPi, pH 7.5 (15). With an excitation wavelength at 290 nm, the emission intensity was recorded at 500 nm. After the addition of 10 μM D2G (the KD [equilibrium dissociation constant] for the WT), NaCl was consecutively added until no change in fluorescence emission occurred. An identical volume of water was used for the control. Increase in intensity (ΔI, the difference before [I0] and after addition of NaCl) was expressed as the percentage of the I0, corrected by a dilution effect, and then plotted as a function of Na+ concentration. The apparent Na+ stimulation constant (K0.5Na+) value was determined by fitting a hyperbolic function to the data (OriginPro 8.6).
Melibiose concentration for the half-maximal displacement of bound D2G (IC50).
Applying the same experimental setup, melibiose was added stepwise to the samples containing the RSO vesicles supplemented with D2G (10 μM) and NaCl (20 or 200 mM) until no change in fluorescence emission occurred. An identical volume of water was added as a negative control. The decrease in intensity after each addition of melibiose (ΔF) was corrected by the dilution effect and plotted as a function of melibiose concentration. The 50% inhibitory concentration (IC50) was determined by fitting a hyperbolic function to the data (OriginPro 8.6).
RESULTS
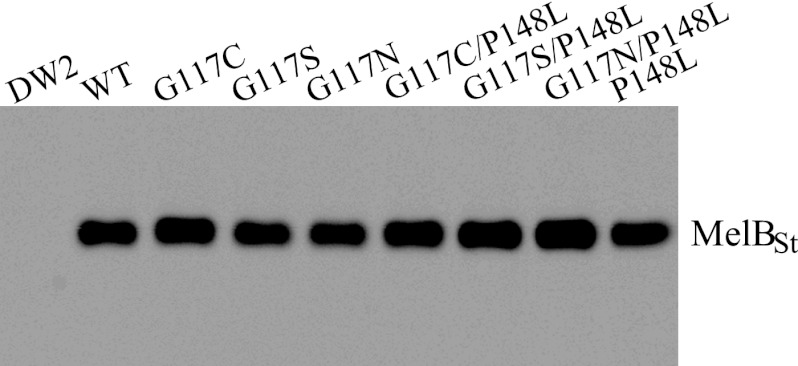
Gly117 in MelBSt was mutated to Cys, Ser, and Asn by site-directed mutagenesis. The mutations have little effect on membrane expression as detected by Western blotting with anti-His antibody (Fig. 2).
Fig 2.
Western blotting. Twenty-five μg of crude membranes was loaded onto each well for SDS-12% PAGE. After being transferred onto a PVDF membrane, MelBst proteins were detected by anti-His tag antibody.
Melibiose fermentation.
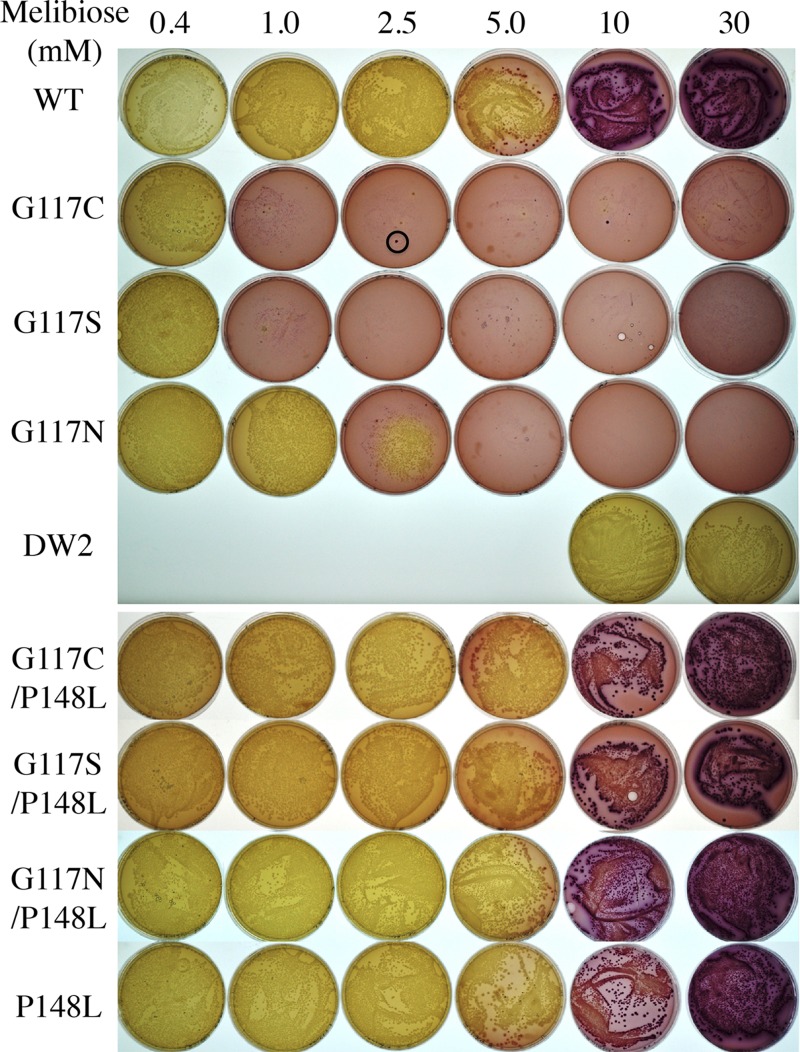
After entry into the cell, melibiose is hydrolyzed into glucose and galactose by α-galactosidase, followed by glycolysis with acidification of the surroundings, which is detected by dark red colonies when the cells are grown on MacConkey agar containing melibiose at 10 mM or higher. The rate-limiting step is the entry of melibiose (41). DW2 cells (melA+ ΔmelB) overexpressing WT MelBSt form dark red colonies when the melibiose concentration is 10 mM or greater (Fig. 3), indicating that melibiose is transported into the cell and metabolized. At decreasing concentrations of melibiose, the colonies change to lighter shades, implying weaker acidification. Nontransformed E. coli DW2 cells form pale/white colonies, denoting no melibiose transport. Although too small to see in Fig. 3, tiny red colonies are found for G117C and G117S mutants on plates with 1 mM melibiose or higher or for the G117N mutant with 2.5 mM melibiose or higher. In addition, a few dark red colonies (0 to 5) of normal size are seen, but the DNA sequences of the mutants are unchanged. Notably, G117C, G117S, and G117N mutants form pale/white colonies of normal size on the plates containing melibiose at a concentration of 0.4 mM or lower (plates containing 0.01, 0.05, or 0.1 mM melibiose not shown).
Fig 3.
Melibiose fermentation. E. coli DW2 (ΔlacY ΔlacZ melA+ ΔmelB) cells were transformed with a plasmid encoding WT or mutant MelBSt, plated on MacConkey agar (lactose free) containing melibiose at 0.4 to 30 mM, and incubated at 37°C for 18 h before photography. The black circle indicates the dark red colony of normal size with no change in the melB gene, as described in the text.
Second-site revertants.
A red colony was found with the G117C mutant on the plate containing 0.01 mM melibiose after 10 days, and DNA sequencing analysis revealed a Pro148→Leu mutation with the G117C mutation unchanged. DW2 cells transformed with the plasmid (G117C/P148L) form colonies of normal size independent of melibiose concentration and ferment melibiose similarly to WT MelBSt. Accordingly, G117C/P148L, G117S/P148L, and G117N/P148L double mutants, as well as the P148L mutant, were generated, and all show membrane expression similar to that of the WT (Fig. 2), form normal-size colonies, and ferment melibiose indistinguishably from the WT (Fig. 3).
Effect of melibiose on the lag phase of cell growth.
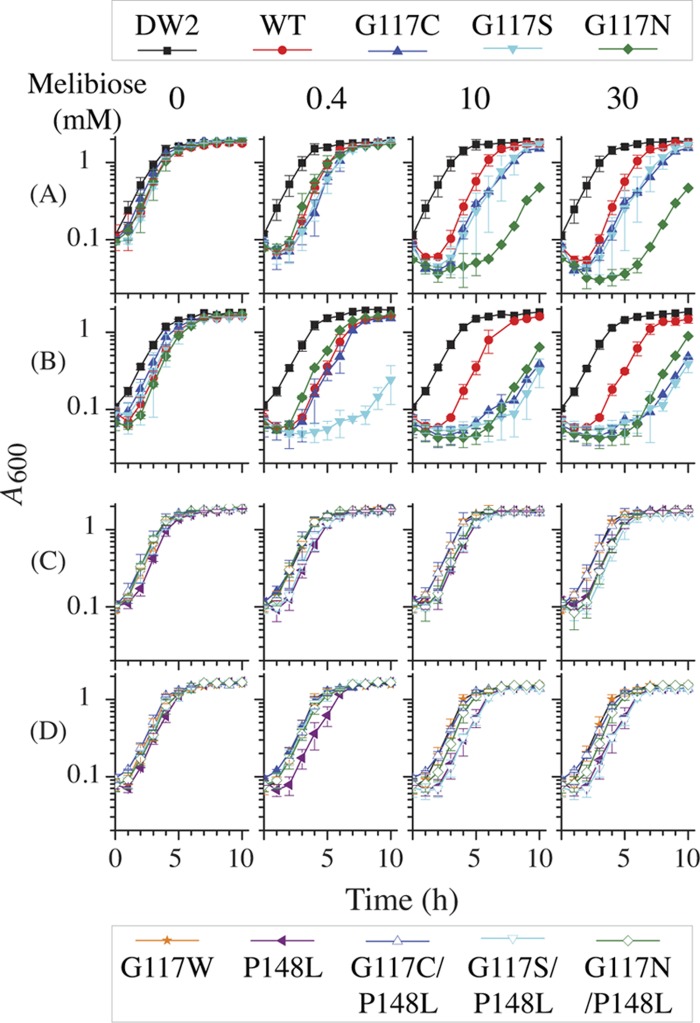
Growing in the LB-G broth, the nontransformed E. coli DW2 cells reach stationary phase in 6 h, and the growth was not affected by addition of melibiose to the medium (Fig. 4A). With cells containing the WT or G117C, G117S, or G117N mutant, a clear lag phase prior to log phase with declining cell densities is observed when diluted into melibiose-containing fresh medium. The lag phase is prolonged with up to 10 mM melibiose; however, there is no further change when the melibiose concentration is increased to 30 mM. G117C and G117S mutants manifest lag phases that are about 1 h longer than those observed with the WT at a melibiose concentration of 10 mM or higher. The G117N mutant exhibits a growth rate similar to that of the WT with 0.4 mM melibiose but shows a significantly longer lag phase of 7 h with melibiose at 10 mM or higher. A decrease in optical density occurred within a few minutes of mixing the melibiose-free overnight cultures with fresh medium containing melibiose. Viable cells during the lag phase were dramatically decreased, as indicated by CFU assay (data not shown). It is noteworthy that the rate of growth during log phase under all conditions is similar.
Fig 4.
Growth curves. E. coli DW2 cells with or without a given expression plasmid were incubated in LB broth at 37°C. Overnight cultures were diluted by 5% with LB-G broth (A and C) or NaCl-removed LB-G (B and D) in the absence or presence of melibiose at a concentration of 0.4, 10, or 30 mM and shaken at 37°C. Cell optical density was monitored hourly at A600 and averaged from 2 to 5 tests; error bars represent standard deviations.
To test osmotic effects, overnight cultures in LB-G medium were diluted by 5% to low-osmolarity LB-G broth, where 171 mM NaCl was removed. Nontransformed DW2 cells exhibit a slightly reduced growth rate in the lag phase with no effect on log phase; again, there is no melibiose effect on growth (Fig. 4B). The WT shows a lag phase that is 1 h longer in the NaCl-removed LB-G medium at each melibiose concentration. Strikingly, G117C and G117S mutants grown in the presence of melibiose at 10 mM or higher show a lag phase significantly longer than that in the LB-G media, approaching a 7-h delay that is similar to that observed in the G117N mutant. Even at a lower concentration (0.4 mM), the G117S mutant has a 7-h lag phase, which is drastically different from its growth in LB-G media.
For all three double mutants (C117C/P148L, C117S/P148L, and C117N/P148L), as well as the P148L mutant, melibiose has little or no effect on cell growth in the LB-G (Fig. 4C) or the low-osmolarity LB-G broth (Fig. 4D). Moreover, the G117W mutant (15) that neither binds nor transports melibiose (Fig. 5, bottom) behaves like the nontransformed DW2 cells (Fig. 4).
Fig 5.
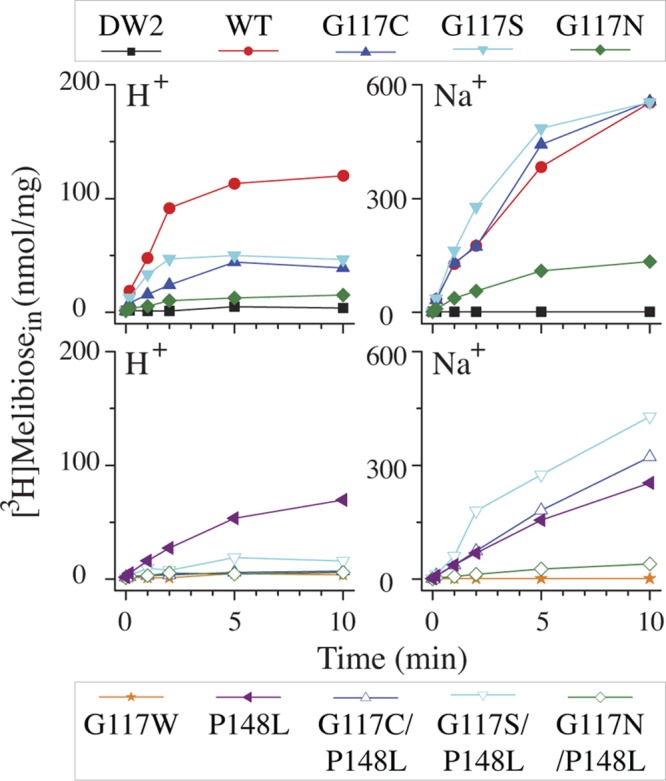
Melibiose transport in intact cells. E. coli DW2 cells were washed and resuspended with 100 mM KPi, pH 7.5, 10 mM MgSO4 and adjusted to 0.7 mg/ml of protein. Transport was initiated by adding melibiose (0.4 mM, 10 mCi/mmol) in the absence or presence of 20 mM NaCl. Intracellular melibiose is plotted as a function of time.
Melibiose transport in intact cells.
In a nominally Na+-free buffer, the WT catalyzes H+-coupled melibiose accumulation at 0.4 mM to a steady state of about 110 nmol/mg in 5 min (Fig. 5, upper); Na+ significantly increases melibiose transport (18, 32). Previously, we demonstrated that all of the accumulated [1-3H]melibiose molecules are completely exchanged with extracellular melibiose within 10 min, indicating there is little or no hydrolysis or chemical alteration of the accumulated intracellular melibiose (15). G117C and G117S mutants catalyze H+-coupled melibiose transport at a significantly reduced level; however, Na+-coupled uptake is indistinguishable from that observed in the WT, with about a 10-fold increase. The G117N mutant catalyzes H+- and Na+-coupled melibiose accumulations at less than 25% of the WT level, but the Na+ activation increases about 8-fold, similar to that observed in the G117C and G117S mutants. The P148L mutant catalyzes H+- and Na+-coupled transport at 43 to 58% of the WT level. All three double mutants containing the P148L mutation show a 3- to 5-fold inhibition compared to the parents (Fig. 5, bottom).
Melibiose transport kinetics.
In the presence of 20 mM NaCl, melibiose transport kinetics were determined with intact cells (Fig. 6A and Table 1). WT MelBSt exhibits a hyperbolic curve with a Km of 0.44 mM and Vmax of 262 nmol/mg/min. The G117C and G117S mutants show an increased Vmax to 580 nmol/mg/min with little change in Km, whereas the G117N mutant exhibits 16- and 3-fold increases in Km and Vmax, respectively. The single-site P148L mutant shows a 2-fold decrease in both Km and Vmax. All three of the G117C/P148L, G117S/P148L, and G117N/P148L double mutants show little change in Km but dramatically decreased Vmax values to levels less than 180 nmol/mg/min, with 4-, 3-, and 8-fold changes compared to the parents, respectively.
Fig 6.
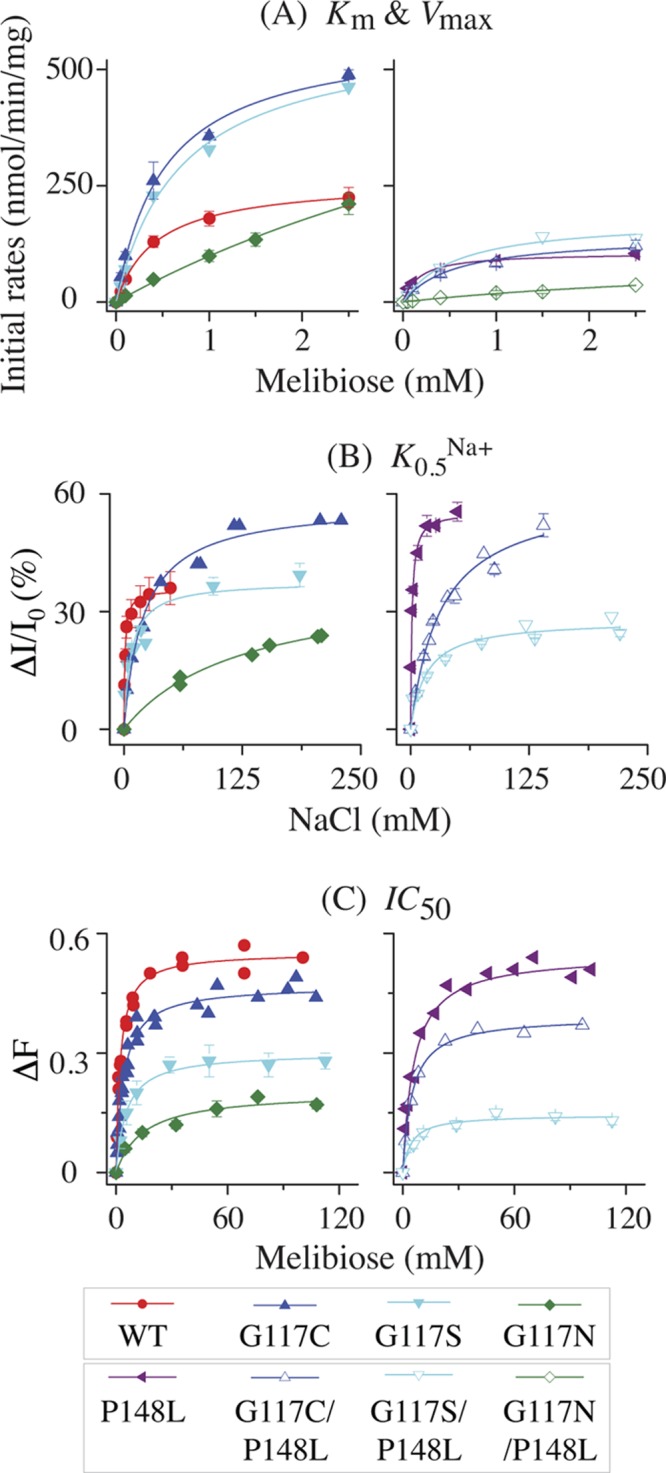
Transport kinetics and cosubstrate-binding affinity. (A) Km and Vmax for melibiose transport. Cell preparation and transport assay were performed as described in Materials and Methods. Fitted initial rates of melibiose transport at a given melibiose concentration between 0.05 and 2.5 mM (specific activity, 3.2 to 10 mCi/mmol) were corrected by the rates obtained from nontransformed DW2 cells. Means (± standard errors [SE]) from 2 to 5 tests were plotted as a function of melibiose concentration. (B) Apparent affinity for Na+ binding. Determination of K0.5Na+ for the D2G FRET was carried out with RSO vesicles (0.5 mg/ml; 100 mM KPi, pH 7.5) containing the WT or a MelBSt mutant at excitation and emission wavelengths of 290 and 500 nm, respectively. The mean values of ΔI/I0 (%) with SE and/or the values from 2 to 3 tests were plotted as a function of Na+ concentration. (C) Apparent affinity for melibiose binding. Determination of IC50 for melibiose displacement of bound D2G was performed with RSO vesicles containing 10 μM D2G and 200 mM NaCl. The mean values of the melibiose-induced change in intensity (ΔF) with SE and/or data from 2 to 3 tests were plotted as a function of melibiose concentration. The values for Km, Vmax, K0.5Na+, and IC50 were determined by fitting a hyperbolic function to the data (OriginPro 8.6).
Table 1.
Melibiose transport kinetics and cosubstrate affinitya
| Strain | Km (mM) | Vmax (nmol/mg/min) | K0.5Na+ (mM) | IC50b (mM) |
|---|---|---|---|---|
| WT MelBSt | 0.44 ± 0.03 | 262.68 ± 5.77 | 1.11 ± 0.12 | 2.22 ± 0.18 (3.53 ± 0.56) |
| G117C | 0.52 ± 0.07 | 576.48 ± 25.65 | 20.62 ± 2.47 | 3.92 ± 0.34 (4.0 ± 0.86) |
| G117S | 0.70 ± 0.08 | 583.86 ± 26.38 | 7.43 ± 2.39 | 5.43 ± 0.6 (10.9 ± 2.64) |
| G117N | 7.22 ± 1.80 | 811.22 ± 160.22 | 125.1 ± 19.8 | 14.66 ± 4.42 |
| G117C/P148L | 0.55 ± 0.12 | 142.17 ± 10.50 | 31.83 ± 4.30 | 4.87 ± 0.37 (14.70 ± 2.35) |
| G117S/P148L | 0.56 ± 0.13 | 178.01 ± 13.02 | 15.99 ± 4.02 | 4.47 ± 1.12 |
| G117N/P148L | 4.53 ± 2.01 | 99.86 ± 31.51 | –c | – |
| P148L | 0.17 ± 0.03 | 105.68 ± 4.93 | 1.35 ± 0.11 | 5.92 ± 0.57 (8.63 ± 1.91) |
The Km and Vmax values for melibiose transport were determined with intact DW2 cells. The Na+ stimulation constant for D2G FRET (K0.5Na+) and the melibiose concentration for the half-maximal displacement of bound D2G (IC50) were determined with RSO membrane vesicles prepared from DW2 cells. Methods are described in Materials and Methods and the legend to Fig. 6. Data are means ± standard errors.
The IC50s were measured in 200 mM NaCl; IC50s given in parentheses were measured in 20 mM NaCl.
–, the FRET signal is insufficient for the determination.
Affinity of Na+ and melibiose.
The quantitative measurement of binding affinity for cosubstrates (Na+ and melibiose) using FRET from endogenous Trp residues to a fluorescent sugar substrate, D2G, has been well documented (5, 15, 18, 27). With the WT, the Na+ stimulation constant (K0.5Na+) for D2G FRET (Fig. 6B) and the IC50 for melibiose displacement of bound D2G (Fig. 6C) are about 1 and 2 to 3 mM, respectively (Table 1). The G117C, G117S, and G117N mutants exhibit a K0.5Na+ value of about 20, 7, and 125 mM (19-, 7-, and 114-fold increase), respectively. The IC50 is little affected in G117C and G117S mutants but increased 7-fold in the G117N mutant.
The P148L mutant alone exhibits less than a 3-fold increase in K0.5Na+ and IC50, and G117C/P148L and G117S/P148L double mutants show little change in K0.5Na+ and IC50 relative to the parents, but the FRET intensity is significantly reduced. The FRET signal with the G117N/P148L mutant is insufficient for the determination of these constants.
DISCUSSION
The G117A mutation of MelBSt has little effect, but a bulky Trp placed at position Gly117 significantly inhibits binding for melibiose and Na+, as well as their coupled symport (15). In this study, polar residues with small side chains, including Ser, Asn, and Cys, were individually placed at position Gly117. Cells carrying these mutants form tiny red colonies on MacConkey agar supplemented with melibiose at a concentration of 2.5 mM or higher. Furthermore, cell lysis occurs after a dilution of the overnight melibiose-free cultures into fresh media containing melibiose at a concentration of 0.4 mM or greater. Each of the three mutants has an elevated Vmax for melibiose transport (Table 1) with little change in protein expression (Fig. 2). The higher the Vmax, the more severe the cell lysis and the longer the lag phase. Moreover, the osmotic stress in the NaCl-removed LB-G broth prolongs the cell lysis, specifically for G117C and G117S mutants that have Vmax values smaller than that observed with the G117N mutant. The results imply that a fast accumulation of melibiose and Na+ causes osmotic lysis in the lag phase.
Growth curves with melibiose at a concentration of 10 and 30 mM are indistinguishable in all cases. It is less likely that melibiose is depleted during the log phase; otherwise, the lag phase should last longer at 30 mM. The data suggest that the recovered cells are no longer affected by the presence of melibiose. Consistent with this interpretation, the growth rates during log phase for all conditions are similar. It is possible that one or more mechanosensor channel(s) can be activated, releasing the accumulated melibiose and Na+. The red, normal-size colonies formed in 18 h of incubation by the G117C mutant might be those adapted cells (Fig. 3, circle).
Consistent with these findings, the inactive G117W mutant shows growth curves identical to those of the nontransformed DW2 cells in the absence or presence of melibiose (Fig. 4 and 5); likewise, P148L, G117C/P148L, G117S/P148L, and G117N/P148L mutants show melibiose-independent growth and have a Vmax value of less than 180 nmol/mg/min. Although it is not clear why the spontaneous G117C/P148L mutant was identified on a plate with a melibiose concentration of 0.01 mM, the P148L mutation inhibits transport Vmax and thereby blocks the lethal effect of elevated melibiose transport in the Gly117 mutants. It is interesting that the Vmax value of WT MelBSt approaches a lethal level. In addition, the G117N mutant has higher Km and Vmax values, which explain why the melibiose uptake at a concentration of 0.4 mM is low and why a higher concentration of melibiose is required to trigger cell lysis.
All three G117C, G117S, and G117N mutants have a significantly decreased K0.5Na+ for D2G FRET; thus, it is likely that the increased Vmax for melibiose transport results from the decreased Na+ affinity. It has been documented that Na+-coupled melibiose transport in MelBEc is limited by intracellular release of Na+ (3, 33). Introduction of a polar group(s) near the Na+-binding site in these mutants may destabilize bound Na+ and thereby facilitate its intracellular release. The results support the previous conclusion that Gly117 is a component of the cation-binding site (15, 45).
Pro148 is located between loop4-5 and the N-terminal end of the important helix V (Fig. 1). Helix V contains a likely sugar-binding residue, Arg149 (adjacent to P148) (1, 45), and loop4-5 bears two residues important for transport, Arg141 and Glu142 (29). Although the role of loop4-5 has not been studied in MelBSt, a sugar-induced conformational change in loop4-5 of MelBEc (19, 29) has been well recognized; moreover, it was also proposed to be important for coordinating the two cosubstrate-binding sites (1). Our three-dimensional model also suggests that both loop4-5 and loop10-11 participate in opening and closing the cytoplasmic cavity that is probably essential for the alternating access mechanism (45). Pro146 and Pro148 (Fig. 1) may form kinks or hinges important for the transport-required rearrangement or conformational change in loop4-5. Compared to the parents, the mutation of Pro148 to Leu has little or no change in the affinity for cosubstrate binding but specifically decreases the transport velocity. The kinetic effect is consistent with the notion that loop4-5 involves a large conformational rearrangement during melibiose transport. The P148L mutation may restrain the movement of this loop, forming a rate-limiting step that significantly decreases the turnover of the permease.
ACKNOWLEDGMENTS
Alexey A. Hodkoff participated in this project at an earlier stage. S.V.J. is a master's student intern of the Center for Biotechnology and Genomics at TTU.
This work was supported by Texas Norman Hackerman Advanced Research Program 010674-0034-2009 and National Science Foundation MCB-1158085 to L.G.
Footnotes
Published ahead of print 3 August 2012
REFERENCES
- 1. Abdel-Dayem M, Basquin C, Pourcher T, Cordat E, Leblanc G. 2003. Cytoplasmic loop connecting helices IV and V of the melibiose permease from Escherichia coli is involved in the process of Na+-coupled sugar translocation. J. Biol. Chem. 278:1518–1524 [DOI] [PubMed] [Google Scholar]
- 2. Abramson J, et al. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610–615 [DOI] [PubMed] [Google Scholar]
- 3. Bassilana M, Pourcher T, Leblanc G. 1987. Facilitated diffusion properties of melibiose permease in Escherichia coli membrane vesicles. Release of co-substrates is rate limiting for permease cycling. J. Biol. Chem. 262:16865–16870 [PubMed] [Google Scholar]
- 4. Cordat E, Leblanc G, Mus-Veteau I. 2000. Evidence for a role of helix IV in connecting cation- and sugar-binding sites of Escherichia coli melibiose permease. Biochemistry 39:4493–4499 [DOI] [PubMed] [Google Scholar]
- 5. Cordat E, Mus-Veteau I, Leblanc G. 1998. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. II. Identification of the tryptophan residues acting as energy donors. J. Biol. Chem. 273:33198–33202 [DOI] [PubMed] [Google Scholar]
- 6. Damiano-Forano E, Bassilana M, Leblanc G. 1986. Sugar binding properties of the melibiose permease in Escherichia coli membrane vesicles. Effects of Na+ and H+ concentrations. J. Biol. Chem. 261:6893–6899 [PubMed] [Google Scholar]
- 7. Ding PZ. 2004. Loop X/XI, the largest cytoplasmic loop in the membrane-bound melibiose carrier of Escherichia coli, is a functional re-entrant loop. Biochim. Biophys. Acta 1660:106–117 [DOI] [PubMed] [Google Scholar]
- 8. Ding PZ, Wilson TH. 2001. The effect of modifications of the charged residues in the transmembrane helices on the transport activity of the melibiose carrier of Escherichia coli. Biochem. Biophys. Res. Commun. 285:348–354 [DOI] [PubMed] [Google Scholar]
- 9. Franco PJ, Jena AB, Wilson TH. 2001. Physiological evidence for an interaction between helices II and XI in the melibiose carrier of Escherichia coli. Biochim. Biophys. Acta 1510:231–242 [DOI] [PubMed] [Google Scholar]
- 10. Franco PJ, Wilson TH. 1999. Arg-52 in the melibiose carrier of Escherichia coli is important for cation-coupled sugar transport and participates in an intrahelical salt bridge. J. Bacteriol. 181:6377–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ganea C, et al. 2011. G117C MelB, a mutant melibiose permease with a changed conformational equilibrium. Biochim. Biophys. Acta 1808:2508–2516 [DOI] [PubMed] [Google Scholar]
- 12. Ganea C, Pourcher T, Leblanc G, Fendler K. 2001. Evidence for intraprotein charge transfer during the transport activity of the melibiose permease from Escherichia coli. Biochemistry 40:13744–13752 [DOI] [PubMed] [Google Scholar]
- 13. Garcia-Celma JJ, et al. 2008. Rapid activation of the melibiose permease MelB immobilized on a solid-supported membrane. Langmuir 24:8119–8126 [DOI] [PubMed] [Google Scholar]
- 14. Granell M, Leon X, Leblanc G, Padros E, Lorenz-Fonfria VA. 2010. Structural insights into the activation mechanism of melibiose permease by sodium binding. Proc. Natl. Acad. Sci. U. S. A. 107:22078–22083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guan L, Jakkula SV, Hodkoff AA, Su Y. 2012. Role of Gly117 in the cation/melibiose symport of MelB of Salmonella typhimurium. Biochemistry 51:2950–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guan L, Kaback HR. 2006. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35:67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guan L, Mirza O, Verner G, Iwata S, Kaback HR. 2007. Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. U. S. A. 104:15294–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan L, Nurva S, Ankeshwarapu SP. 2011. Mechanism of melibiose/cation symport of the melibiose permease of Salmonella typhimurium. J. Biol. Chem. 286:6367–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gwizdek C, Leblanc G, Bassilana M. 1997. Proteolytic mapping and substrate protection of the Escherichia coli melibiose permease. Biochemistry 36:8522–8529 [DOI] [PubMed] [Google Scholar]
- 20. Hacksell I, et al. 2002. Projection structure at 8 A resolution of the melibiose permease, an Na-sugar co-transporter from Escherichia coli. EMBO J. 21:3569–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hama H, Wilson TH. 1994. Replacement of alanine 58 by asparagine enables the melibiose carrier of Klebsiella pneumoniae to couple sugar transport to Na+. J. Biol. Chem. 269:1063–1067 [PubMed] [Google Scholar]
- 22. Kaback HR. 1971. Bacterial membranes, p 99–120 In Kaplan NP, Jakoby WB, Colowick NP. (ed), Methods in enzymology, vol XXII Elsevier, New York, NY [Google Scholar]
- 23. Leon X, Leblanc G, Padros E. 2009. Alteration of sugar-induced conformational changes of the melibiose permease by mutating Arg141 in loop 4-5. Biophys. J. 96:4877–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leon X, Lemonnier R, Leblanc G, Padros E. 2006. Changes in secondary structures and acidic side chains of melibiose permease upon cosubstrates binding. Biophys. J. 91:4440–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leon X, Lorenz-Fonfria VA, Lemonnier R, Leblanc G, Padros E. 2005. Substrate-induced conformational changes of melibiose permease from Escherichia coli studied by infrared difference spectroscopy. Biochemistry 44:3506–3514 [DOI] [PubMed] [Google Scholar]
- 26. Lopilato J, Tsuchiya T, Wilson TH. 1978. Role of Na+ and Li+ in thiomethylgalactoside transport by the melibiose transport system of Escherichia coli. J. Bacteriol. 134:147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maehrel C, Cordat E, Mus-Veteau I, Leblanc G. 1998. Structural studies of the melibiose permease of Escherichia coli by fluorescence resonance energy transfer. I. Evidence for ion-induced conformational change. J. Biol. Chem. 273:33192–33197 [DOI] [PubMed] [Google Scholar]
- 28. Matsuzaki S, Weissborn AC, Tamai E, Tsuchiya T, Wilson TH. 1999. Melibiose carrier of Escherichia coli: use of cysteine mutagenesis to identify the amino acids on the hydrophilic face of transmembrane helix 2. Biochim. Biophys. Acta 1420:63–72 [DOI] [PubMed] [Google Scholar]
- 29. Meyer-Lipp K, et al. 2006. The inner interhelix loop 4-5 of the melibiose permease from Escherichia coli takes part in conformational changes after sugar binding. J. Biol. Chem. 281:25882–25892 [DOI] [PubMed] [Google Scholar]
- 30. Mirza O, Guan L, Verner G, Iwata S, Kaback HR. 2006. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 25:1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mus-Veteau I, Pourcher T, Leblanc G. 1995. Melibiose permease of Escherichia coli: substrate-induced conformational changes monitored by tryptophan fluorescence spectroscopy. Biochemistry 34:6775–6783 [DOI] [PubMed] [Google Scholar]
- 32. Niiya S, Moriyama Y, Futai M, Tsuchiya T. 1980. Cation coupling to melibiose transport in Salmonella typhimurium. J. Bacteriol. 144:192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pourcher T, Bassilana M, Sarkar HK, Kaback HR, Leblanc G. 1990. The melibiose/Na+ symporter of Escherichia coli: kinetic and molecular properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326:411–423 [DOI] [PubMed] [Google Scholar]
- 34. Pourcher T, Deckert M, Bassilana M, Leblanc G. 1991. Melibiose permease of Escherichia coli: mutation of aspartic acid 55 in putative helix II abolishes activation of sugar binding by Na+ ions. Biochem. Biophys. Res. Commun. 178:1176–1181 [DOI] [PubMed] [Google Scholar]
- 35. Pourcher T, Leclercq S, Brandolin G, Leblanc G. 1995. Melibiose permease of Escherichia coli: large scale purification and evidence that H+, Na+, and Li+ sugar symport is catalyzed by a single polypeptide. Biochemistry 34:4412–4420 [DOI] [PubMed] [Google Scholar]
- 36. Pourcher T, Zani ML, Leblanc G. 1993. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na(+)-dependent transport and binding properties. J. Biol. Chem. 268:3209–3215 [PubMed] [Google Scholar]
- 37. Purhonen P, Lundback AK, Lemonnier R, Leblanc G, Hebert H. 2005. Three-dimensional structure of the sugar symporter melibiose permease from cryo-electron microscopy. J. Struct. Biol. 152:76–83 [DOI] [PubMed] [Google Scholar]
- 38. Short SA, Kaback HR, Kohn LD. 1974. d-Lactate dehydrogenase binding in Escherichia coli dld− membrane vesicles reconstituted for active transport. Proc. Natl. Acad. Sci. U. S. A. 71:1461–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tokuda H, Kaback HR. 1978. Sodium-dependent binding of p-nitrophenyl alpha-D-galactopyranoside to membrane vesicles isolated from Salmonella typhimurium. Biochemistry 17:698–705 [DOI] [PubMed] [Google Scholar]
- 40. Tokuda H, Kaback HR. 1977. Sodium-dependent methyl 1-thio-β-D-galactopyranoside transport in membrane vesicles isolated from Salmonella typhimurium. Biochemistry 16:2130–2136 [DOI] [PubMed] [Google Scholar]
- 41. Tsuchiya T, Lopilato J, Wilson TH. 1978. Effect of lithium ion on melibiose transport in Escherichia coli. J. Membr. Biol. 42:45–59 [DOI] [PubMed] [Google Scholar]
- 42. Tsuchiya T, Raven J, Wilson TH. 1977. Co-transport of Na+ and methul-beta-D-thiogalactopyranoside mediated by the melibiose transport system of Escherichia coli. Biochem. Biophys. Res. Commun. 76:26–31 [DOI] [PubMed] [Google Scholar]
- 43. Wilson DM, Hama H, Wilson TH. 1995. GLY113→ASP can restore activity to the ASP51→SER mutant in the melibiose carrier of Escherichia coli. Biochem. Biophys. Res. Commun. 209:242–249 [DOI] [PubMed] [Google Scholar]
- 44. Wilson DM, Wilson TH. 1987. Cation specificity for sugar substrates of the melibiose carrier in Escherichia coli. Biochim. Biophys. Acta 904:191–200 [DOI] [PubMed] [Google Scholar]
- 45. Yousef MS, Guan L. 2009. A 3D structure model of the melibiose permease of Escherichia coli represents a distinctive fold for Na+ symporters. Proc. Natl. Acad. Sci. U. S. A. 106:15291–15296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zani ML, Pourcher T, Leblanc G. 1993. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. II. Effect on cationic selectivity and coupling properties. J. Biol. Chem. 268:3216–3221 [PubMed] [Google Scholar]
- 47. Zani ML, Pourcher T, Leblanc G. 1994. Mutation of polar and charged residues in the hydrophobic NH2-terminal domains of the melibiose permease of Escherichia coli. J. Biol. Chem. 269:24883–24889 [PubMed] [Google Scholar]