Abstract
In bacteria, nutrient deprivation evokes the stringent response, which is mediated by the small intracellular signaling molecule ppGpp. In Gram negatives, the RelA enzyme synthesizes and SpoT hydrolyzes ppGpp, although the latter protein also has weak synthetase activity. DksA, a recently identified RNA polymerase binding transcription factor, acts as a coregulator along with ppGpp for controlling the stringent response. Recently, we have shown that three genes, relA, spoT, and relV, govern cellular levels of ppGpp during various starvation stresses in the Gram-negative cholera pathogen Vibrio cholerae. Here we report functional characterization of the dksA gene of V. cholerae (dksAVc), coding for the protein DksAVc. Extensive genetic analyses of the ΔdksAVc mutants suggest that DksAVc is an important component involved in the stringent response in V. cholerae. Further analysis of mutants revealed that DksAVc positively regulates various virulence-related processes, namely, motility, expression of the major secretory protease, called hemagglutinin protease (HAP), and production of cholera toxin (CT), under in vitro conditions. We found that DksAVc upregulates expression of the sigma factor FliA (σ28), a critical regulator of motility in V. cholerae. Altogether, it appears that apart from stringent-response regulation, DksAVc also has important roles in fine regulation of virulence-related phenotypes of V. cholerae.
INTRODUCTION
Expression of genes in microorganisms is a highly regulated process and often involves complex genetic circuits controlling several phenotypes for their growth and survival under various environmental conditions. This is further complicated if we consider that bacteria, including pathogens in nature, are found in complex communities. Thus, signaling mechanisms in bacteria must be robust in order for them to sustain various environmental onslaughts and to survive and grow through tremendous competition with the community microorganisms in a particular niche. As a result, bacteria have evolved with multiple gene regulatory circuits to sense and combat various environmental stresses. The most important one among such adaptive responses is the stringent response, where bacterial cells undergo rapid and complex metabolic adjustments through negative and positive regulation of gene expression during nutritional starvation. The global changes in gene expression associated with the stringent response are triggered mainly by the intracellular accumulation of two small molecules called guanosine 3′-diphosphate 5′-triphosphate (pppGpp) and guanosine 3′,5′-bis diphosphate (ppGpp), together called (p)ppGpp, and are characterized by negative regulation of rRNA transcription, positive regulation of amino acid biosynthesis, readjustment of metabolic pathways according to physiological requirements, and induction of stationary-phase genes needed for survival (10).
In Gram-negative organisms, including Escherichia coli, the products of the relA and spoT genes synthesize (p)ppGpp. However, SpoT is a bifunctional enzyme having strong hydrolyzing and weak (p)ppGpp synthetase activities (10, 32). Although the exact mechanism is not yet clearly known, it appears that (p)ppGpp binds to a site adjacent to, but not overlapping, the active site on the β and β′ subunits of the RNA polymerase (RNAP) core enzyme and affects gene transcription at the stage of initiation during open promoter complex formation (2, 31). However, recent studies indicate that (p)ppGpp alone is not involved in the process; rather, a small protein, DksA, the product of the dksA gene, acts as a coregulator to facilitate the function of (p)ppGpp during the stringent response (39–41). Like the relA and spoT genes, dksA is also conserved in Gram-negative bacteria (11, 41). From a structural point of view, DksA belongs to an unusual family of transcriptional regulators, whose members do not bind directly to the regulatory part of a gene but rather bind directly to the secondary channel of RNAP (36, 39, 41). The crystal structure of the DksA protein of E. coli indicates a globular domain and a coiled-coil structure with C4 zinc finger motif (41). When DksA binds directly to RNAP, two highly conserved aspartic acid residues present at the tip of the coiled-coil domain of the protein help to stabilize the (p)ppGpp-Mg2+-RNAP complex. Based on several reports, it appears that apart from participation in the stringent response, DksA is also involved in multiple cellular processes in different Gram-negative bacteria. Among these functions, the conspicuous ones are modulation of multiple gene expression (55), quorum sensing (QS) (8, 24), and virulence (24, 30, 33, 36, 46, 49, 55). Most recently, it has been shown that DksA along with (p)ppGpp is directly involved in regulation of transcription of E. coli flagellar genes and ribosomal protein coding genes (27, 28).
Although most of the DksA-related studies have so far been conducted in E. coli, at present our knowledge regarding how DksA modulates different gene functions in pathogens is limited. Previously we functionally characterized the relA and spoT genes of Vibrio cholerae (13, 14, 20, 38), a Gram-negative bacterium and the causative agent of the severe diarrheal disease cholera. We discovered that apart from the canonical relA and spoT genes, this pathogen also possesses a novel (p)ppGpp synthetase gene, relV (14), and thus (p)ppGpp metabolism is quite complex in this organism. However, very little information is currently available about the function of V. cholerae DksA (DksAVc). This study aims to explore further the role of DksAVc in the stringent response. Since DksA in other enteric pathogens has been reported to be involved in regulation of pathogenicity, we wanted to check this possibility in the case of V. cholerae. Regulation of virulence genes in V. cholerae is quite complex, and several positive and negative regulators are involved in the process. Among these regulators, HapR, the master regulator of QS, plays a crucial role since virulence gene expression in this pathogen is QS dependent (56). At low cell density (LCD), when the cellular HapR level is low, expression of major virulence determinants, such as cholera toxin (CT)-, toxin-coregulated pilus-, and biofilm formation-related genes, is upregulated. Furthermore, at this condition, the cellular level of cyclic diguanylic acid (c-di-GMP), the newly identified second messenger, also remains high (19, 53). In contrast, at high cell density (HCD), the intracellular HapR concentration is increased, leading to repression of the above-described processes and upregulation of expression of the hemagglutinin protease gene hapA, which codes for the major protease HAP of V. cholerae. Several reports indicate that HAP is most likely involved in V. cholerae's pathogenesis program (7, 16, 35, 47), including its role in detaching adhered V. cholerae cells on intestinal epithelial cell surfaces.
To study the function of DksAVc, the dksA gene locus ( J. Craig Venter Institute annotation no. VC0596) was identified bioinformatically using the genome sequence information of the V. cholerae O1 El Tor strain N16961 (21). The gene was cloned and manipulated further to construct chromosomally deleted nonpolar ΔdksAVc strains. As with the E. coli ΔdksA mutant (ΔdksAEc), ΔdksAVc mutant cells exhibited poor growth in M9 minimal (M9M) medium and sensitivity toward 3-amino-1,2,4-triazole (AT). However, unlike the case with E. coli, the ΔdksAVc mutant showed growth in serine-, methionine-, glycine-, and leucine (SMGL)-containing M9M medium. Furthermore, the ΔdksAVc mutant gave the following virulence-related phenotypes compared to its wild-type (Wt) strain: (i) decreased HAP production, (ii) decreased motility, and (iii) poor production of CT under in vitro conditions. The results clearly indicate that along with the stringent response, several other genes of V. cholerae, involved in pathogenicity, dissemination, and persistence in the environment, are also controlled by the circuit of the DksAVc regulome.
(Part of this work was presented at the 44th U.S.-Japan Conference on Cholera and Other Bacterial Enteric Infections, San Diego, CA, 12 to 14 October 2009; International Symposium on 50 Years of Discovery of Cholera Toxin: a Tribute to SN De, Kolkata, India, 25 to 27 October 2009; and International Symposium on Molecular and Pathophysiological Research on Enteric Infections, Kolkata, India, 27 to 29 January 2011).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Details of recombinant plasmid and strain constructions are provided in Supplement S1 in the supplemental material. Both E. coli and V. cholerae cells were routinely grown in Luria broth (LB) (Difco) at 37°C with shaking essentially as described previously (20). For plate culture, LB was used with 1.5% agar (Difco). Antibiotics (all from Sigma-Aldrich) were used at the following concentrations unless otherwise indicated: ampicillin, 100 μg/ml; streptomycin, 100 μg/ml; kanamycin, 40 μg/ml; spectinomycin, 50 μg/ml; tetracycline, 10 μg/ml for E. coli and 1 μg/ml for V. cholerae. In some experiments, bacterial cells were grown in M9M medium (Sigma-Aldrich) containing 0.4% glucose as a carbon source (13). The Wt E. coli strain CF1648 (MG1655) (Table 1) has a frameshift mutation in the RNase PH-coding gene, leading to a weak requirement of uracil for its growth in M9M (23). It has been reported that the situation was further aggravated after deletion of the dksA gene of MG1655 (strain CF9240) (Table 1), which is then unable to grow in M9M without uracil (9). Therefore, we added 20 μg/ml of uracil (SRL Pvt. Ltd., India) as a supplement in M9M agar plates for growing the ΔdksAEc strain. Bacterial strains were maintained at −70°C in LB containing 20% sterile glycerol. To avoid development of any suppressor, all the mutant strains were minimally subcultured, and before any experiment they were directly inoculated from −70°C stock. The growth of bacterial culture was monitored spectrophotometrically by measuring the optical density at 600 nm (OD600). The growth kinetic experiments were repeated at least three to five times, and their average values were plotted.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| V. cholerae | ||
| N16961 | Wild type, lacking hapR function, O1 serogroup, biotype El Tor, Smr | 14 |
| C6709 | Wild type, hapR+, O1 serogroup, biotype El Tor, Smr | 20 |
| N-DksA1 | N16961 ΔdksA∷kan; Kmr Smr | This study |
| C-DksA1 | C6709 ΔdksA∷kan; Kmr Smr | This study |
| NRVDK2 | N-DksA1 ΔrelV∷aadA1; Kmr Smr Spr | This study |
| E. coli | ||
| DH5α | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 (ϕ80dlacZΔM15) | Promega |
| SM10λpir | thi thr leu tonA lacY supE recA∷RP4-2-Tc∷Mu λpir R6K | 20 |
| S17-1λpir | thi proA hsdR recA∷RP4-2-Tc∷Mu-1 kan∷Tn7 integrant λpir R6K; Tpr Smr | Lab stock |
| CF1648 | Wild type MG1655 | 54 |
| CF9240 | CF1648 ΔdksA∷Tet; Ttr | 9 |
| Plasmids | ||
| pDrive | pUC origin, high-copy-no. cloning vector; Apr Kmr | Qiagen |
| PBluescript II KS(+) | ColE1, high-copy-no. cloning vector; Apr | Stratagene |
| pKAS32 | rpsL suicide vector with oriR6K mobRP4; Apr | Lab stock |
| pBR322 | pMB1 origin, general-purpose cloning vector; Apr Ttr | Lab stock |
| pUC4K | Source of kanamycin resistance gene cassette; Apr Kmr | Pharmacia |
| pBAD24 | pBR322 origin, l-arabinose-inducible vector; Apr | Lab stock |
| pBS20 | 2.9-kb ΔrelV∷aadA1 allele in pKAS32; Apr Spr | 14 |
| pJK537 | 2.3-kb dksA region of E. coli strain MG1655 in pBR322; Apr | 9 |
| pDDKW1 | 841-bp dksA region of V. cholerae strain N16961 containing dksA gene with its putative natural promoter cloned in pDrive; Apr Kmr | This study |
| pBSDA3.5 | 0.6-kb ΔdksA allele in pBluescript II KS(+); Apr Kmr | This study |
| pBSDA4.8 | 1.8-kb ΔdksA∷kan allele in pBluescript II KS(+); Apr Kmr | This study |
| pKDK1 | 1.9-kb ΔdksA∷kan allele in pKAS32; Apr Kmr | This study |
| pDksAVc | 841- bp dksA region of V. cholerae strain N16961 from pDDKW1 subcloned pBR322; Apr | This study |
| pDksABAD | V. cholerae dksA ORF cloned in pBAD24; Apr | This study |
| pFliABAD | V. cholerae fliA ORF cloned in pBAD24; Apr | This study |
| pRelVBAD | V. cholerae relV ORF cloned in pBAD24; Apr | This study |
Molecular biological methods.
Standard molecular biological methods (3) for chromosomal and plasmid DNA preparations, electroelution of DNA fragments, restriction enzyme digestion, DNA ligation, bacterial transformation, conjugation, agarose gel electrophoresis, etc., were followed unless stated otherwise. All restriction enzymes and nucleic acid-modifying enzymes were purchased from New England BioLabs, Inc., and were used essentially as directed by the manufacturer. Electrocompetent V. cholerae cells were prepared as described previously (13). Transformants were selected by plating transformed cells on LB agar plates containing appropriate antibiotics.
AT and SMGL tests.
Sensitivity of bacterial strains toward the histidine analogue AT or in SMGL medium was examined essentially as described previously (13, 14). When needed, the amino acid l-histidine (Sigma-Aldrich) was added (4 μg/ml) to the AT medium.
Determination of intracellular (p)ppGpp by TLC.
Intracellular accumulation of (p)ppGpp under amino acid or glucose starvation in various strains, including the ΔdksAVc mutant, was determined by the thin-layer chromatography (TLC) method essentially as described previously (13, 14, 20).
RT-PCR and qRT-PCR assays.
For reverse transcriptase PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR) assays, total cellular RNA was prepared from bacterial cells grown in LB to an OD600 of ∼1 using TRIzol reagent (Invitrogen) as described by the vendor. The purity check and quantitation of the prepared RNA were done spectrophotometrically (3). A standard RT-PCR experiment was carried out using the Qiagen One Step RT-PCR kit as directed by the manufacturer (Qiagen, Germany). The PCR-amplified product was checked by agarose gel electrophoresis using appropriate DNA size markers. To confirm absence of any contaminating DNA in prepared RNA samples, PCR assay of each sample was also done with Taq DNA polymerase (Invitrogen). Lack of amplification in the absence of RT confirmed that the desired PCR product was generated only from cDNAs.
For qRT-PCR, cDNA was prepared from 1 μg of DNase I-treated RNA using SuperScriptIII RT (Invitrogen) essentially as described by the manufacturer. The qRT-PCR was done using either Power SYBR green PCR master mix (Applied Biosystems Inc.) or the One Step SYBR PrimeScript RT-PCR kit (TaKaRa, Japan) essentially as described by the manufacturer. The primer sets FliA-F/FliA-R and HapA-F/HapA-R (see Table S1 in the supplemental material) were used for qRT-PCR analysis. Relative expression values (R) were calculated using the equation R = 2−(ΔCT target − CT reference), where CT is the fractional threshold cycle. In each experiment, as an internal control, the recA-specific primers recA-F/recA-R (see Table S1 in the supplemental material) were used. The experiments were repeated at least thrice using three different batches of prepared RNA.
Motility assay.
Motility assay of V. cholerae strains was performed on LB soft-agar plates containing 0.3% agar (Difco) at 30°C as described previously (18), and a reading was taken after 8 to 10 h of incubation. Experiments were repeated at least thrice, and the average values were used.
HAP assays.
HAP activity was studied by milk plate assay as described previously by Vance et al. (51). Each strain was examined thrice by milk plate assay, and an average was taken. HAP was also quantitated by azocasein assay as described previously (4). The amount of enzyme required for increasing 0.01 units in OD at 440 nm per hour was considered one azocasein unit.
GM1-ELISA of CT.
For detection of CT production by V. cholerae strains under in vitro conditions, the cells were grown in AKI medium (1.5% Bacto peptone [Difco], 0.4% yeast extract [Difco], 0.5% NaCl [Merck], and 0.3% sodium bicarbonate [Sigma-Aldrich]) essentially as described earlier by Iwanaga et al. (22). V. cholerae cells were initially grown statically in a test tube containing freshly prepared AKI medium at 37°C for 4 h, and then the culture was aseptically transferred to a sterile conical flask, followed by continuation of incubation overnight at 37°C with shaking (22). CT present in culture supernatant was assayed by GM1-enzyme linked immunosorbent assay (ELISA) (29, 34) using pure CT (Sigma-Aldrich) and phosphate-buffered saline (10 mM, pH 7.2) as positive and negative controls, respectively. A standard curve of known CT concentrations was plotted and used to estimate the amount of CT present in each sample.
Assessment of CT production by rabbit ileal loop assay.
In vivo CT production by V. cholerae strains, including mutants, was assayed by using the ligated rabbit ileal loop model essentially as described previously (15). Fluid accumulation (FA) in the ligated ileal loop was measured as the ratio of loop fluid volume to loop length and expressed as ml/cm, and an FA ratio of 1 or more than 1 was considered high production of CT under in vivo conditions. In all experiments, sterile 0.9% NaCl (normal saline) was used as a negative control, and as a positive control, Wt V. cholerae N16961 (Table 1) live culture was used. Each strain was tested at least thrice in three different animals. The experimental protocol used in this study was reviewed and approved by the institutional animal ethics committee of Indian Institute of Chemical Biology, Kolkata, India.
SEM.
For microscopy, the V. cholerae sample was prepared as described previously (14, 45), and bacterial cells were examined using a scanning electron microscope (SEM) (model Vega II Lsu; Tescan, Czech Republic) at 10 kV. The images in the figures are representative of what was observed in 10 random fields in each of two independent experiments.
DNA sequencing.
DNA sequencing reactions were carried out using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems Inc.) essentially as recommended by the manufacturer. The samples were run on an ABI3130 genetic analyzer using the Pop-7 polymer (Applied Biosystems Inc.). Results were analyzed using the software DNA Sequencing Analysis V5.1 (Applied Biosystems Inc.).
Computational analyses.
DNA sequence data were compiled and analyzed by using the DNASIS software program (Hitachi Corporation, Yokohama, Japan). The National Center for Biotechnology Information (NCBI) BLASTN program was used to search for homologous sequences in the database (www.ncbi.nlm.nih.gov). The open reading frames (ORFs) were subsequently subjected to a database search using the BLASTP program, version 2.2.15 (www.ncbi.nlm.nih.gov). For designing PCR and other primers, the Primer3 software program was used (http://frodo.wi.mit.edu/). Genomatix software (www.genomatix.de/cgi-bin/dialign/dialign.pl) was used for the alignment of protein sequences.
Statistical analysis.
Where needed, pairwise comparison of data for each sample was analyzed for statistical significance using Student's t test.
RESULTS
Functional analysis of dksAVc.
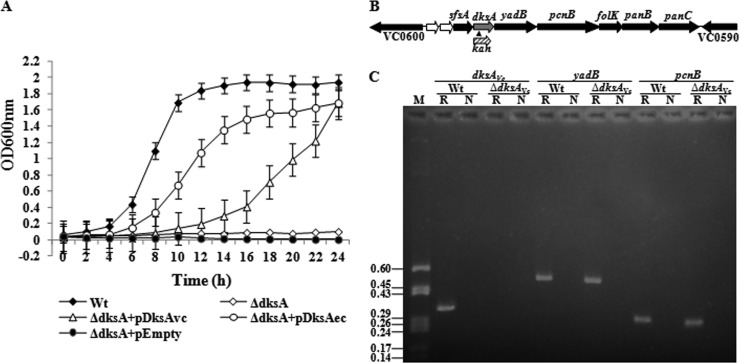
Bioinformatics analysis of the whole-genome-sequenced strain N16961 of V. cholerae (21) indicated that the large chromosome of the organism carries the dksAVc gene with an open reading frame (ORF) of 447 bp (VC0596) having 66% identity with the sequence of the E. coli dksA gene (dksAEc). While dksAEc codes for a 151-amino-acid-long DksA protein (here it will be designated DksAEc), DksAVc is composed of 148 amino acids (41), with a calculated molecular mass of about 17.2 kDa (www.jcvi.org). BLASTP (blast.ncbi.nlm.nih.gov/blast.cgi) analysis of DksAVc showed 77.8% identity and 84.7% similarity with DksAEc. For functional verification, the identified dksAVc gene of the strain N16961 (Table 1) along with its natural promoter was cloned into the plasmid pBR322 or pDrive (Table 1), and the recombinant plasmid was designated pDksAVc or pDDKW1 (Table 1), respectively. Brown et al. (9) previously reported that a ΔdksAEc strain is unable to grow in M9M medium, which is identical to the phenotype of an E. coli ΔrelA ΔspoT mutant (ppGpp0 strain). Introduction of the plasmid pDDKW1 into the ΔdksAEc strain CF9240 (Table 1) enabled growth of the strain in M9M salt solution, although at a lower rate than that of the Wt (Fig. 1A). On the other hand, CF9240 complemented with the dksAEc gene through the plasmid pJK537 (Table 1) showed better growth in M9M medium than the dksAVc-complemented strain. However, after 24 h of incubation, both CF9240(pDDKW1) and CF9240(pJK537) reached to an OD600 value similar to that of the Wt strain, CF1648 (Fig. 1A). In sharp contrast, CF9240 carrying the empty vector failed to grow in M9M salt solution (Fig. 1A). We have also verified the growth of these strains in M9M agar plates after 18 to 24 h of incubation and found that only the Wt, ΔdksAEc strain carrying the plasmid pDDKW1, pDksAVc, or pJK537 could grow (data not shown). The complementation results are consistent with those reported earlier (9). Furthermore, the experimental results support that the promoter PdksA of V. cholerae and the DksAVc protein both are functional in E. coli.
Fig 1.
(A) Growth complementation of the ΔdksAEc strain with a functional dksAVc gene in M9M medium. E. coli strains used are as follows: Wt, CF1648; ΔdksA, CF9240; ΔdksA+pDksAvc, CF9240(pDDKW1); ΔdksA+pDksAec, CF9240(pJK537); ΔdksA+pEmpty, CF9240(pDrive). Error bars indicate standard deviations. (B) Genomic arrangement of the dksA gene (gray arrow), including its flanking genetic determinants (VC0590 to VC0600), in V. cholerae. The direction of each arrow indicates the direction of transcription of a gene. VC0598 and VC0599 are two small hypothetical ORFs (white arrows). The insertion location of the kanamycin resistance gene (kan) cassette (small filled triangle) and its direction of transcription are also shown. (C) RT-PCR analysis to show that the deletion of dksAVc did not hamper transcription of downstream genes. V. cholerae strains used are the Wt (C6709) and the ΔdksAVc mutant (C-DksA1). Lanes: M, pBluescript II KS(+) plasmid DNA digested with HaeIII, used as markers; sizes (in kb) of the DNA fragments are given in the left margin; R, RT with Taq DNA polymerase; N, only Taq DNA polymerase (used as a negative control).
To define the functions of DksAVc in more detail, in-frame dksA deletion mutants of N16961 (lacking hapR function) and C6709 (hapR+) were constructed by the positive selection method using the kanamycin resistance gene (kan) as a marker (see Supplement S1 in the supplemental material), and the ΔdksAVc mutants thus constructed were designated N-DksA1 and C-DksA1 (Table 1), respectively. Since the two mutants showed almost similar phenotypes, here they will be collectively called the ΔdksAVc mutants unless mentioned otherwise. Brown et al. suggested that dksAEc could be present in an operon with the flanking genes sfsA and yadB (9). Preliminary examination of the dksAVc locus (VC0596) appears to be organized in a similar fashion, where VC0597 and VC0595 are the sfsA and yadB genes, respectively (Fig. 1B). However, BioCyc analysis (http://biocyc.org/vcho/new-image?type=gene&object=vc0596) of the locus predicted that the gene dksAVc could alone be a single transcriptional unit. Therefore, to confirm that deletion of the dksAVc gene in strain C-DksA1 had no polar effect, we examined the transcript levels of two physically linked genes, yadB (VC0595) and pcnB (VC0594), present downstream of dksAVc (VC0596) by employing the RT-PCR method using the specific primer sets VCO595-F/VC0595-R and PcnB-F/PcnB-R (see Table S1), respectively. While in both cases the desired cDNA of sizes 0.5 and 0.3 kb of yadB and pcnB, respectively, was generated, no cDNA was detectable for the dksAVc gene using the specific primers Dksint-F/Dksrt-R (see Table S1), as shown in Fig. 1C. Similar results were obtained in the case of the N-DksA1 mutant strain (data not shown), indicating a similar arrangement of the locus in both V. cholerae strains used. The results confirmed the authenticity of deletion of the dksAVc gene, and such deletion most likely had no polar effects on its downstream genes.
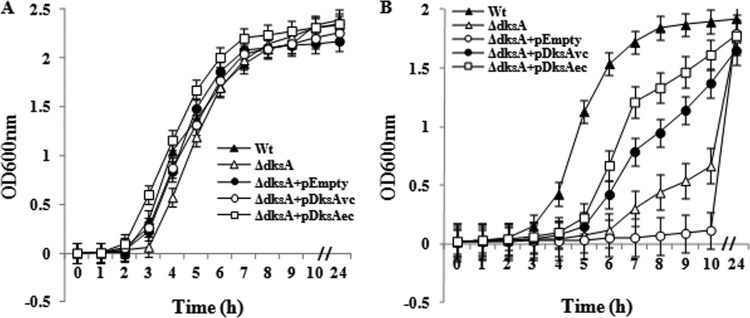
Initially, we checked cellular levels of (p)ppGpp in the ΔdksAVc mutant under amino acid- or glucose-starved conditions using the TLC method and found no significant change in the concentration of (p)ppGpp compared to that for the Wt (data not shown). This result is consistent with the report of Brown et al. (9), who found similar levels of (p)ppGpp in the ΔdksAEc and Wt strains. To analyze other phenotypes of the ΔdksAVc strain, we first compared its growth in nutritionally rich (LB) and poor (M9M) media. The ΔdksAEc mutant is unable to grow in M9M medium (Fig. 1A), but it grows as does the Wt in LB (9; this study). Although the ΔdksAVc mutant showed no growth defect in LB (Fig. 2A), there was significant growth retardation in M9M medium for ∼5 h compared to growth of the Wt (Fig. 2B). The growth defect of the ΔdksAVc strain in M9M medium was partially corrected by expressing the DksAVc or DksAEc protein in trans through the plasmid pDDKW1 or pJK537 (Table 1), respectively, but not by the empty vector (Fig. 2B). To rule out the possibility that growth of the ΔdksAVc strain and the ΔdksAVc strain carrying the empty vector after overnight incubation was due to development of any suppressor, each overnight-grown culture was reinoculated separately into fresh M9M medium and their growth was monitored spectrophotometrically. Both of them showed growth patterns with ∼5 h of an extended lag period, suggesting that they are indeed the ΔdksAVc mutant and not a suppressor (data not shown). The results support the view that the growth defect is due to a lack of the DksAVc protein and not to the polar effect of deleting the gene. This further suggests that DksAEc is functional in V. cholerae. We have already provided evidence that DksAVc is functional in E. coli, and thus, functions of the DksA protein in this respect appear to be conserved between the two species.
Fig 2.
(A) V. cholerae strains, including the dksAVc mutant, showed no growth defect in LB (nutrient-rich medium). Strains used are as follows: Wt, N16961; ΔdksA, N-DksA1; ΔdksA+pEmpty, N-DksA1(pDrive); ΔdksA+pDksAvc, N-DksA1(pDDKW1); ΔdksA+pDksAec, N-DksA1(pJK537). (B) Growth phenotypes of V. cholerae cells in M9M medium. Strains are as indicated for panel A. Error bars indicate standard deviations.
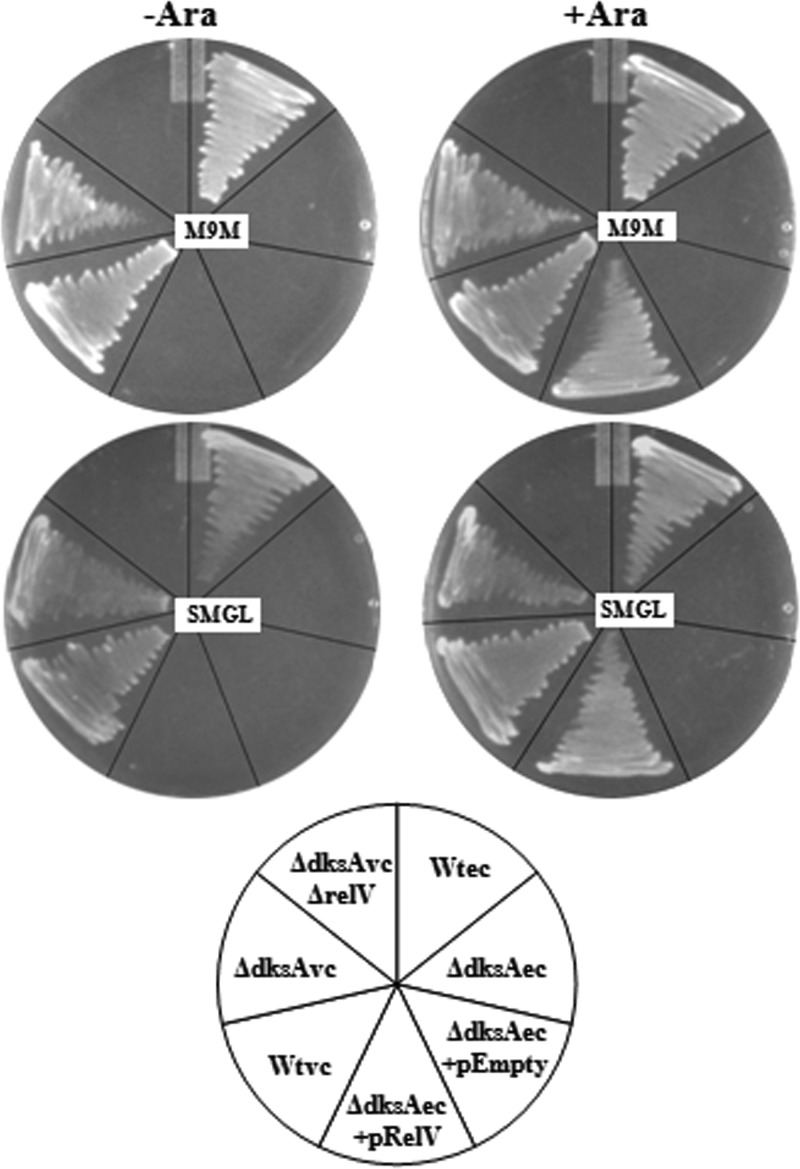
It has been reported that DksAEc is crucial for the function of (p)ppGpp and it acts as a cofactor by binding with the secondary channel of RNA polymerase, leading to positive regulation of amino acid biosynthesis operons (40). When ΔdksAVc cells were grown in M9M medium supplemented with all the amino acids, the mutant showed a growth pattern similar to that of the Wt (data not shown), which supports that a lack of DksAVc probably leads to downregulation of amino acid biosynthesis operons even when the cells are RelA+ SpoT+, i.e., cells are (p)ppGpp+. It is interesting to note that the ΔdksAEc strain CF9240 failed to grow in M9M medium even after overnight incubation at 37°C (Fig. 1A). In sharp contrast, the ΔdksAVc strain showed initiation of growth in M9M medium after ∼5 h of incubation at a similar temperature and reached saturation (OD600 > 1.8) after overnight (16 h) incubation (Fig. 2B). Similar results were obtained when the growth phenotype of the ΔdksAVc mutant was compared with that of the ΔdksAEc strain along with appropriate control strains using an M9M agar plate assay (Fig. 3). Similarly, we also checked the growth sensitivity of ΔdksAVc cells toward AT and SMGL. The principal effect of AT (a histidine analog) is blockage of biosynthesis of the amino acid histidine (44). It has been demonstrated that the ΔdksAEc strain is unable to grow on AT medium (9). Although AT sensitivity could be overcome by an adequate amount of (p)ppGpp synthesis (54), as in the case of the Wt, it should be noted that the ΔdksAEc strain is a (p)ppGpp+ strain and still failed to grow in AT medium. In SMGL agar plates the growth of a (p)ppGpp0 strain is inhibited due to increased intracellular levels of methylenetetrahydrofolate (44, 50). This inhibitory effect of methylenetetrahydrofolate could also be overcome by optimal cellular levels of (p)ppGpp. According to Paul et al. (40), DksA and (p)ppGpp act synergistically to regulate the transcription of various amino acid biosynthetic pathway genes during the stringent response. Therefore, the ΔdksAEc strain should be sensitive to AT and SMGL. When these assays were performed with appropriate controls, interestingly, ΔdksAVc cells were able to grow in SMGL-containing (Fig. 3) but not in AT-containing (Fig. 4) medium. This difference in growth phenotypes between the ΔdksAEc and ΔdksAVc strains in M9M and SMGL media could be explained as follows. It has recently been shown that the V. cholerae genome carries a novel (p)ppGpp synthetase gene, relV, apart from the canonical relA and spoT genes (14). We hypothesize that this could be due to the presence of the relV gene in V. cholerae, which probably helped ΔdksAVc cells to overcome growth defects in M9M/SMGL medium through optimal production of (p)ppGpp. In favor of this hypothesis, one piece of indirect evidence is that E. coli lacks a relV-like gene and thus accumulation of excess (p)ppGpp is not possible although the cells are RelA+ and SpoT+, and probably for this reason, the ΔdksAEc strain failed to grow in SMGL/M9M medium. If this is the case, then a V. cholerae ΔdksA ΔrelV double mutant, like the ΔdksAEc strain, will not be able to grow in SMGL/M9M medium. Therefore, we constructed a V. cholerae ΔdksA ΔrelV double mutant strain, NRVDK2 (Table 1; see also Supplement S1 in the supplemental material), which, as hypothesized, failed to grow in M9M and SMGL (Fig. 3) media. It is to be noted that like the ΔdksAVc mutant, NRVDK2 was unable to grow in AT medium (data not shown), and in this respect it behaved just like the ΔdksAEc strain. It may be argued that the supplying of functional relV in ΔdksAEc cells in trans may allow the strain to behave like a ΔdksAVc strain. When the ΔdksAEc strain CF9240 was transformed with the plasmid pRelVBAD (Table 1) carrying the relV ORF under the arabinose-inducible promoter (PBAD), as rationalized, the strain CF9240(pRelVBAD) showed growth on M9M and SMGL agar media, while CF9240 carrying the empty vector pBAD24 failed to grow (Fig. 3). On the other hand, AT sensitivity of the ΔdksAVc mutant [a (p)ppGpp+ strain] could be due to the lack of the DksAVc protein, which seems to be essential for upregulation of the his operon of V. cholerae. Therefore, we thought that supplementation of histidine in AT medium should rescue the ΔdksAVc mutant from histidine auxotrophy. In fact, the ΔdksAVc mutant showed growth in AT agar medium containing the amino acid l-histidine, as shown in Fig. 4. This is also true for E. coli, since the ΔdksAEc strain showed growth in an l-histidine-containing AT agar plate (Fig. 4).
Fig 3.
The functional relV gene confers growth on M9M agar and SMGL media in the ΔdksA background. Growth of different V. cholerae (vc) and E. coli (ec) strains on M9M and SMGL media without (−) or with (+) 0.01% l-arabinose (Ara) after 24 h is shown. Strains used are as follows: Wtvc, N16961; ΔdksAvc, N-DksA1; ΔdksAvcΔrelV, NRVDK2; Wtec, CF1648; ΔdksAec, CF9240; ΔdksAec+pEmpty, CF9240(pBAD24); ΔdksAec+pRelV, CF9240(pRelVBAD).
Fig 4.
DksA is essential to overcome histidine auxotrophy caused by AT. Growth of different V. cholerae (vc) and E. coli (ec) strains in AT medium without (−) and with (+) 4 μg/ml l-histidine (His) after 24 h. Strains used are as follows: Wtec, CF1648; ΔdksAec, CF9240; Wtvc, N16961 or C6709; ΔdksAvc, N-DksA1 or C-DksA1.
DksAVc is required for optimal production of HAP.
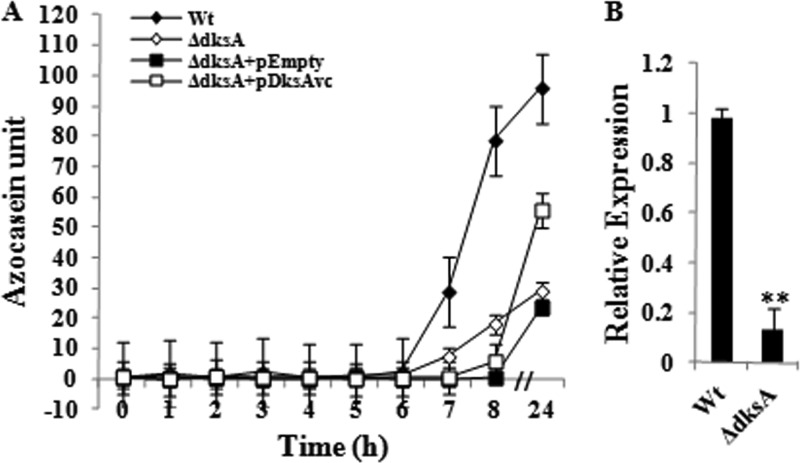
After establishing the function of DksAVc in stringent-response-related phenotypes, we wished to study further its role, if any, in virulence-related phenotypes. We found that the dksAVc-deleted strain C-DksA1 produced a 3-fold smaller amount of HAP after 24 h of growth than its parent HapR+ HapA+ DksA+ Wt strain, C6709 (Fig. 5A). Furthermore, the qRT-PCR assay with the hapA gene-specific primers HapA-F/HapA-R (see Table S1 in the supplemental material) revealed a ∼5-fold decrease in the hapA transcript level in the C-DksA1 mutant with respect to that for the Wt strain, C6709 (Fig. 5B). The result was also consistent with the direct measurement of HAP in culture supernatant by azocasein assay (Fig. 5A). It is well established that HAP is produced at stationary phase under HCD conditions (4). We found that HAP in the culture supernatant of the Wt strain is detectable after 6 h of growth and reached its maximum concentration in overnight stationary culture, as shown in Fig. 5A. However, when the C-DksA1 (ΔdksAVc) mutant strain was similarly tested, it showed a distinct shift in the timing of HAP production (from 8 h of growth), and the amount produced was also substantially less in a saturated culture than with the Wt strain, C6709 (Fig. 5A). Expression of DksAVc in trans through the plasmid pDksAVc in the C-DksA1 strain partially complemented the mutant phenotype and thus provided further evidence that the downregulation of HAP production in C-DksA1 was probably due to the lack of the DksAVc protein (Fig. 5A). This is further supported by the fact that C-DksA1 carrying the empty vector pBR322 failed to complement (Fig. 5A).
Fig 5.
DksAVc modulates HAP production in V. cholerae. (A) Kinetics of HAP production in various V. cholerae strains derived from the parental strain C6709. Error bars indicate standard deviations. Strains used are as follows: Wt, C6709; ΔdksA, C-DksA1; ΔdksA+pEmpty, C-DksA1(pBR322); ΔdksA+pDksAvc, C-DksA1(pDksAVc). (B) Quantitative measurement of hapA transcript of V. cholerae. Relative expression of hapA in the ΔdksA strain (C-DksA1) with respect to that in the Wt (C6709) was measured by qRT-PCR. Error bars indicate standard deviations. Significant differences (**, P < 0.01) in expression are indicated from multiple comparison of each mutant versus the Wt.
DksAVc positively regulates motility.
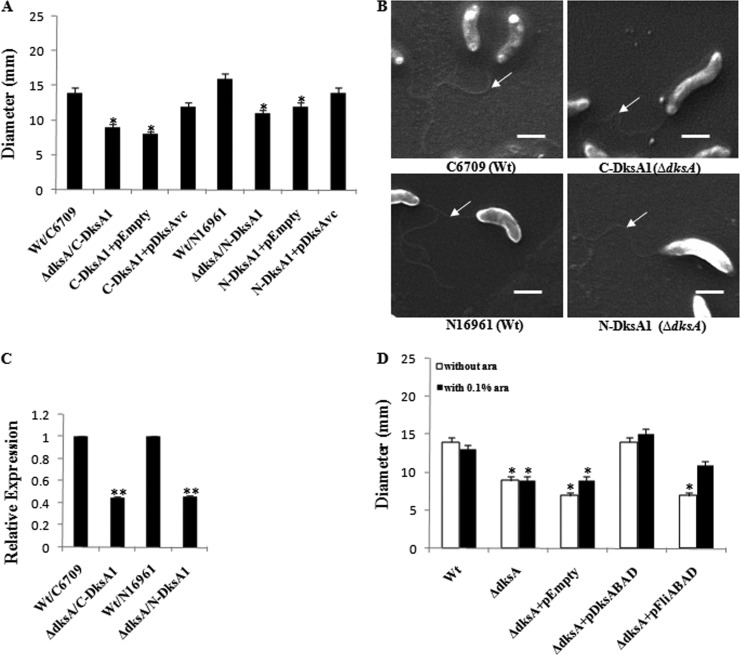
Recently Åberg et al. (1) reported that E. coli cells deficient in DksAEc are hyperflagellated, leading to hypermotility, and this observation was further supported by the work of Lemke et al. (27) in analyzing transcription of the flagellar biosynthesis genetic cascade. Österberg et al. (37) reported that dksA deletion leads to a decrease in motility in another Gram-negative bacterium, Pseudomonas putida. In this study, unlike the case with E. coli, the ΔdksAVc strain showed about a 30% reduction in motility in a soft-agar plate assay, considering the motility of a Wt strain as 100% (Fig. 6A). Since the decreased motility of the ΔdksAVc strain was complemented by expressing DksAVc in trans through the plasmid pDksAVc (Fig. 6A), it may be concluded that DksAVc is involved in regulation of motility of V. cholerae.
Fig 6.
DksAVc is involved in regulation of motility of V. cholerae. (A) Motility assay of V. cholerae strains was carried out on LB soft agar plates. Strains used are as indicated. Error bars indicate standard deviations. Significant differences (*, P < 0.05) in motility are indicated from multiple comparison of each mutant with Wt strains. (B) SEM analysis of V. cholerae cells. Strains used are as indicated. White arrows indicate flagella. Bars correspond to 1 μm. (C) Relative expression of the fliA gene of V. cholerae, determined by qRT-PCR assay. Significant differences (**, P < 0.01) in fliA transcript levels between mutant and Wt strains are indicated. Error bars indicate standard deviations. (D) Complementation of motility defect of ΔdksA mutant strain C-DksA1. Strains are as indicated. Significant differences (*, P < 0.05) in motility are indicated from multiple comparisons of mutant, gene-complementing plasmid, or empty vector (pBAD24) strains with the Wt strain, C6709.
Motility-related flagellar gene expression in V. cholerae is highly complex, and there are four distinct levels (class I to IV) of the gene regulation cascade (43). As with the QS pathway, several motility genes are transcribed with the help of sigma factor RpoN, or σ54 (25, 43). The master regulatory gene (class I category) is flrA, the product of which in the presence of σ54 RNA polymerase holoenzyme activates several class II genes, including flrC and fliA (which codes for sigma factor 28, or σ28). FlrC, with the help of the σ54 holoenzyme, promotes expression of the class III genes, including the flagellin gene flaA. Finally, the σ28 RNA polymerase holoenzyme promotes expression of the class IV genes, including the flagellar motor component genes motABY (43). It is noteworthy that unlike the V. cholerae ΔrpoN and ΔfliA mutants (both of which are nonmotile), the ΔdksAVc mutant strain showed motility (decreased from that of the Wt) as revealed by a soft agar assay (Fig. 6A). Furthermore, SEM analysis of the Wt and the ΔdksAVc mutant showed the presence of a single polar flagellum in more than 90% of cells (Fig. 6B), suggesting normal flagellation of both the strains. Interestingly, SEM analysis revealed distinct elongated morphology of ΔdksAVc cells compared to that of the Wt (Fig. 6B). Since ΔdksAVc cells were hypomotile with their intact flagella, this suggested that flagellar motor gene functions are most probably affected, instead of functions of genes related to flagellar synthesis. As mentioned above, expression of the flagellar motor component genes motABY in V. cholerae is controlled by the sigma factor fliA, or σ28 (43). In other bacteria, DksA has been shown to be involved in regulation of expression of σ28 (12, 27). Therefore, we hypothesize that DksAVc may carry out a similar function. To examine this, we performed qRT-PCR experiments by using total cellular RNA of the ΔdksAVc mutant along with the Wt as a control, and the result indicated about a 2-fold downregulation of expression of the σ28 gene in the ΔdksAVc genetic background (Fig. 6C). Thus, it seems that DksAVc is most likely needed for optimal expression of σ28 of V. cholerae. To further confirm, the fliA ORF (21) of V. cholerae was cloned under an arabinose-inducible promoter, generating the plasmid pFliABAD (Table 1) (see Supplement S1 in the supplemental material) and introduced into the ΔdksAVc strain. Controlled expression of FliA (σ28) through the plasmid pFliABAD partially complemented the motility defect of the ΔdksAVc strain, while the empty vector pBAD24 or pFliABAD without arabinose induction failed to complement (Fig. 6D). Thus, it appears that DksAVc positively regulates σ28 for its optimal expression.
DksAVc modulates CT production under in vitro conditions.
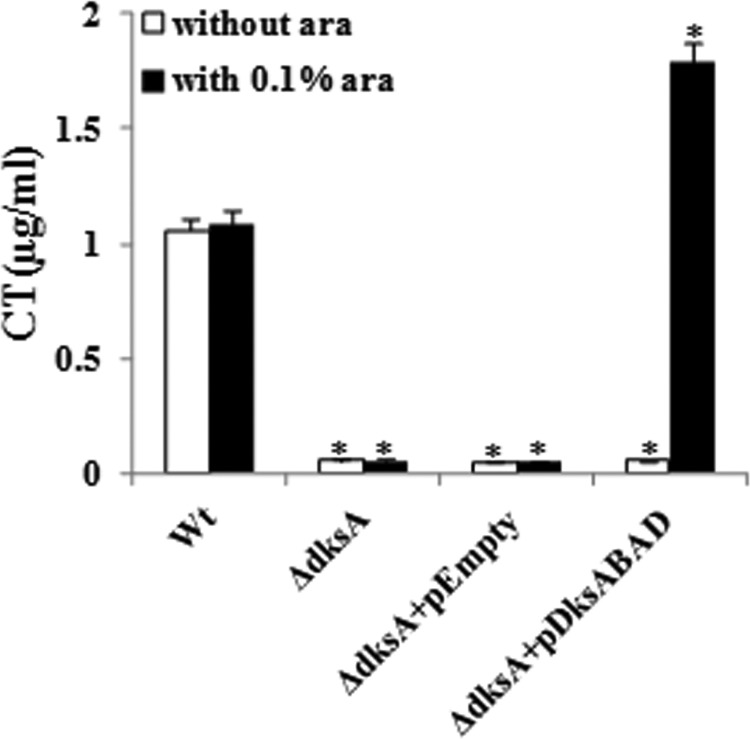
Since DksAVc appears to be involved in modulation of expression of HAP as well as regulation of motility, both of which are pathogenicity-related phenomena, we checked the status of production of CT, the principal virulence factor of V. cholerae, in ΔdksAVc cells. To do this, both ΔdksAVc and its isogenic Wt strain were grown in CT-inducing AKI medium (for details, see Materials and Methods), and the amount of CT produced was measured by GM1-ELISA. Interestingly, the ΔdksAVc strain produced significantly less CT than the Wt (Fig. 7). Since this observation is true for both the dksAVc mutants derived from C6709 (Fig. 7) and N16961 (data not shown), it appears that DksAVc most likely has a role in CT production under in vitro conditions, and this is most probably not strain specific. When the DksAVc protein was expressed in the ΔdksAVc strain C-DksA1 through the plasmid pDksABAD (Table 1) using 0.1% l-arabinose as an inducer, it complemented CT production in the mutant (Fig. 7). A similar result was obtained when N-DksA1(pDksABAD) was examined (data not shown). We also wished to know whether the in vitro defect in CT production by ΔdksAVc mutant cells is also true in an in vivo situation. Therefore, the strains were tested for CT production in rabbit ileal loops. However, the ΔdksAVc mutant and the Wt showed similar FA ratios (for details, see Materials and Methods) of about 1.2 ml/cm, indicating in vivo induction of an unknown factor(s), which could easily overcome the lack of DksAVc. These results support the view that the signaling cascade in CT production by V. cholerae differs under in vitro and in vivo conditions (26).
Fig 7.
Deletion of dksAVc affects CT production. Strains used are as follows: Wt, C6709; ΔdksA, C-DksA1; ΔdksA+pEmpty, C-DksA1(pBAD24); ΔdksA+pDksABAD, C-DksA1(pDksABAD). Error bars indicate standard deviations. Significant differences (*, P < 0.05) in CT production are indicated from multiple comparisons of each mutant with the Wt strain, C6709.
DISCUSSION
In the present study, we have for the first time functionally characterized the stringent-response-related dksA gene of the cholera pathogen V. cholerae. Our experimental results suggest that DksAVc is indeed involved in the stringent response in conjunction with (p)ppGpp. We showed that the DksAEc and DksAVc proteins are functionally similar and both of them are active in homologous and heterologous genetic backgrounds (Fig. 1 and 2). It has previously been reported from this laboratory that unlike the case with E. coli, the intracellular level of (p)ppGpp in V. cholerae is governed by three enzymes, RelA, SpoT, and RelV (14), and we have provided evidence that RelV through its (p)ppGpp synthetase activity indeed helps the ΔdksAVc mutant to grow in M9M salt solution or agar plate (Fig. 2 and 3) after a certain period of lag time (∼5 h). This is also true in the case of SMGL agar medium (Fig. 3), which is not possible for the ΔdksAEc mutant since it naturally lacks the relV gene. This is further supported by expressing the relV gene in the ΔdksAEc strain, which rescued the growth defect of the mutant in M9M and SMGL media, and the strain behaved like the ΔdksAVc mutant (Fig. 3). It should further be noted that although DksA has been proposed as an essential cofactor for the action of (p)ppGpp, our experimental results suggest that most probably the cellular concentration of (p)ppGpp is crucial for bacterial growth, which might work in a DksA-independent manner. Alternatively, the RelV protein itself may be involved in the process, which needs further investigation. We found that the ΔdksAVc mutant failed to grow in AT medium (Fig. 4). AT is a histidine analog and blocks protein synthesis by inhibiting histidine biosynthesis. It appears that DksAVc is essential to overcome this inhibitory effect of AT in V. cholerae. In fact, this may be the case, since Paul et al. (40) have shown that DksAEc is absolutely needed for upregulation of histidine biosynthesis in E. coli.
Apart from regulation of the stringent response, our experimental results also suggest for the first time that DksAVc is likely to be involved in fine regulation of important virulence-related phenotypes in clinical V. cholerae strains. We found that DksAVc positively regulates HAP production (Fig. 5). In support of our study, it may be mentioned here that mutation in the dksA gene of P. aeruginosa, a Gram-negative organism, also led to significant downregulation of expression of the secreted elastase enzyme LasB (24), a highly homologous protein of HAP. Our results also suggest that DksAVc positively controls expression of the critical motility regulator σ28 of V. cholerae, and this could be one of the reasons for the decreased-motility phenotype showed by the DksAVc-negative strain (Fig. 6). Dalebroux et al. (12) have recently reported that for flagellar morphogenesis in the Gram-negative human pathogen Legionella pneumophila, DksA is required for basal σ28 promoter activity. Furthermore, it has also been shown that deletion of the dksA gene of P. putida leads to a significant motility defect (37). Although our SEM analysis indicated normal flagellation (Fig. 6), we believe that downregulation of σ28 expression in the ΔdksAVc strain possibly affected the flagellar motor functions, which are under the control of σ28 (43). In sharp contrast, recently DksAEc has been shown to be a negative regulator of σ28 expression in E. coli (1, 27). Although the basis of these opposite activities of DksA in V. cholerae and E. coli is currently unknown, it could be due to a difference in their lifestyle.
Our study indicates that CT production under in vitro conditions is positively regulated by DksAVc (Fig. 7). CT production by V. cholerae cells is a highly regulated process and is QS dependent (56). At LCD, when cellular levels of the QS master regulator HapR are low, AphA, a positive transcriptional regulator of virulence gene expression in V. cholerae, is derepressed, leading to a series of reactions which ultimately allow V. cholerae cells to express the virulence master regulator ToxT followed by production of the principal virulence factor CT. It should also be noted that the cellular level of the newly established second messenger c-di-GMP is critical for biofilm formation, virulence factor production, and motility in V. cholerae (5, 42, 48, 53). c-di-GMP is synthesized in bacteria by the action of the GGDEF domain, containing the diguanylate cyclase enzyme, on two molecules of GTP and degraded by the EAL/HD-GYP domain, containing phosphodiesterase enzymes (17). It should be noted that the V. cholerae genome codes for a large numbers of GGDEF/EAL domain-containing proteins (6, 17), and thus, maintenance of the cellular level of c-di-GMP appears to be controlled by a complex regulatory network about which our current knowledge is limited. A high cellular level of c-di-GMP negatively regulates CT production and motility (5, 42, 48), the phenotypes of the ΔdksAVc mutant observed in this study. In a recent study, the authors have shown that c-di-GMP regulates the production of HAP in a negative manner, because an artificial increase in the cellular c-di-GMP pool through overexpression of a diguanylate cyclase caused poor expression of HAP (52). Thus, it cannot be ruled out that DksAVc possibly acts as a direct/indirect negative regulator in critically maintaining the intracellular c-di-GMP pool in V. cholerae, which needs to be addressed. Our preliminary experimental results (R.R. Pal, S. Bag, S. Dasgupta, and R.K. Bhadra, unpublished observation) suggest that overexpression of a c-di-GMP-degrading phosphodiesterase (VCA0681) in the ΔdksAVc mutant could rescue the motility defect.
One enigma of our finding is that CT production by the ΔdksAVc strain is significantly reduced only under in vitro conditions and not in the in vivo situation, where DksAVc appears to not be required (see Results), suggesting possible host-specific activation of some unknown factor(s) in overcoming the deficiency of DksAVc. The result further indicates that our knowledge regarding signaling pathways under in vitro and in vivo conditions is extremely limited. It is plausible, however, that CT production may be needed by the pathogen when it is residing in a particular environmental niche for its protection/survival, and under that condition, DksAVc may facilitate the process.
Last, it is noteworthy that the ΔdksAVc strains described in this study were not growth defective when cultivated in nutritionally rich media (Fig. 2), and our experiments conducted to assess the virulence-related phenotypes of the ΔdksAVc strains were always done in nutritionally rich medium. Therefore, we believe that phenotypes shown by the ΔdksAVc strains are most likely due to the lack of the DksAVc protein and not to any growth defect of cells. Altogether, this study suggests that different global regulators, namely, DksAVc, FliA (σ28), etc., exert positive and negative effects on various crucial genetic circuits involved in expression of virulence determinants, including motility of the cholera pathogen, and further investigations are needed for a clear understanding of how this is achieved.
Supplementary Material
ACKNOWLEDGMENTS
We thank Siddhartha Roy for his constant support and encouragement. We are grateful to M. Cashel, National Institutes of Health, Bethesda, MD, for the generous gift of E. coli strains CF1648 and CF9240 and the plasmid pJK537. We thank T. Muruganandan, Pratap C. Koyal, and Sib Prasad Sharma for their technical assistance in this work.
The work was supported by research grants from the Council of Scientific and Industrial Research (CSIR), Government of India. R.R.P. and S.D. are grateful for research fellowships from CSIR, and S.B. is grateful for a research fellowship from ICMR, New Delhi, India.
Footnotes
Published ahead of print 17 August 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J. Bacteriol. 191: 3226–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Artsimovitch I, et al. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117: 299–310 [DOI] [PubMed] [Google Scholar]
- 3. Ausubel FM, et al. 1989. Current protocols in molecular biology. John Wiley and Sons, New York, NY [Google Scholar]
- 4. Benitez JA, Silva AJ, Finkelstein RA. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 69: 6549–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188: 3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhadra RK, Shah S, Das B. 2011. Small molecule signaling systems in Vibrio cholerae, p 185–201 In Ramamurthy T, Bhattacharya SK. (ed), Epidemiological and molecular aspects on cholera. Humana Press, New York, NY [Google Scholar]
- 7. Booth BA, Boesman-Finkelstein M, Finkelstein RA. 1984. Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect. Immun. 45: 558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Branny P, et al. 2001. Inhibition of quorum sensing by a Pseudomonas aeruginosa dksA homologue. J. Bacteriol. 183: 1531–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184: 4455–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cashel M, Gentry DR, Hernandes VJ, Vinella D. 1996. The stringent response, p 1458–1496 In Neidhardt FC. (ed), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed American Society for Microbiology, Washington, DC [Google Scholar]
- 11. Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. 2010. ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74: 171–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dalebroux ZD, Yagi BF, Sahr T, Buchrieser C, Swanson MS. 2010. Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol. Microbiol. 76: 200–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Das B, Bhadra RK. 2008. Molecular characterization of Vibrio cholerae DrelA DspoT double mutants. Arch. Microbiol. 189: 227–238 [DOI] [PubMed] [Google Scholar]
- 14. Das B, Pal RR, Bag S, Bhadra RK. 2009. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol. Microbiol. 72: 380–398 [DOI] [PubMed] [Google Scholar]
- 15. Dasgupta U, Bhadra RK, Panda DK, Deb A, Das J. 1994. Recombinant derivative of a naturally occurring non-toxinogenic Vibrio cholerae 01 expressing the B subunit of cholera toxin: a potential oral vaccine strain. Vaccine 12: 359–364 [DOI] [PubMed] [Google Scholar]
- 16. Finkelstein RA, Boesman-Finkelstein M, Chang Y, Hase CC. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203: 11–21 [DOI] [PubMed] [Google Scholar]
- 18. Gardel CL, Mekalanos JJ. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64: 2246–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hammer BK, Bassler BL. 2009. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J. Bacteriol. 191: 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haralalka S, Nandi S, Bhadra RK. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 185: 4672–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heidelberg JF, et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iwanaga M, et al. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30: 1075–1083 [DOI] [PubMed] [Google Scholar]
- 23. Jensen KF. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175: 3401–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jude F, et al. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 185: 3558–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klose KE, Mekalanos JJ. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28: 501–520 [DOI] [PubMed] [Google Scholar]
- 26. Lee SH, Hava DL, Waldor MK, Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99: 625–634 [DOI] [PubMed] [Google Scholar]
- 27. Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 74: 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemke JJ, et al. 2011. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U. S. A. 108: 5712–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maiti D, et al. 2006. Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology 152: 3633–3641 [DOI] [PubMed] [Google Scholar]
- 30. Mogull SA, Runyen-Janecky LJ, Hong M, Payne SM. 2001. dksA is required for intercellular spread of Shigella flexneri via an RpoS-independent mechanism. Infect. Immun. 69: 5742–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murray HD, Gourse RL. 2004. Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol. Microbiol. 52: 1375–1387 [DOI] [PubMed] [Google Scholar]
- 32. Murray KD, Bremer H. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 259: 41–57 [DOI] [PubMed] [Google Scholar]
- 33. Nakanishi N, et al. 2006. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61: 194–205 [DOI] [PubMed] [Google Scholar]
- 34. Nandi S, Maiti D, Saha A, Bhadra RK. 2003. Genesis of variants of Vibrio cholerae O1 biotype El Tor: role of the CTXphi array and its position in the genome. Microbiology 149: 89–97 [DOI] [PubMed] [Google Scholar]
- 35. Nielsen AT, et al. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2: 933–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Osterberg S, Del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65: 37–55 [DOI] [PubMed] [Google Scholar]
- 37. Osterberg S, Skarfstad E, Shingler V. 2010. The sigma-factor FliA, ppGpp and DksA coordinate transcriptional control of the aer2 gene of Pseudomonas putida. Environ. Microbiol. 12: 1439–1451 [DOI] [PubMed] [Google Scholar]
- 38. Pal RR, Das B, Dasgupta S, Bhadra RK. 2011. Genetic components of stringent response in Vibrio cholerae. Indian J. Med. Res. 133: 212–217 [PMC free article] [PubMed] [Google Scholar]
- 39. Paul BJ, et al. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118: 311–322 [DOI] [PubMed] [Google Scholar]
- 40. Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U. S. A. 102: 7823–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perederina A, et al. 2004. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118: 297–309 [DOI] [PubMed] [Google Scholar]
- 42. Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282: 12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prouty MG, Correa NE, Klose KE. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39: 1595–1609 [DOI] [PubMed] [Google Scholar]
- 44. Rudd KE, Bochner BR, Cashel M, Roth JR. 1985. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J. Bacteriol. 163: 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shah S, Das B, Bhadra RK. 2008. Functional analysis of the essential GTP-binding-protein-coding gene cgtA of Vibrio cholerae. J. Bacteriol. 190: 4764–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma AK, Payne SM. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62: 469–479 [DOI] [PubMed] [Google Scholar]
- 47. Syngkon A, et al. 2010. Studies on a novel serine protease of a DhapADprtV Vibrio cholerae O1 strain and its role in hemorrhagic response in the rabbit ileal loop model. PLoS One 5: e13122 doi:10.1371/journal.pone.0013122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73: 5873–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Turner AK, Lovell MA, Hulme SD, Zhang-Barber L, Barrow PA. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uzan M, Danchin A. 1976. A rapid test for the relA mutation in E. coli. Biochem. Biophys. Res. Commun. 69: 751–758 [DOI] [PubMed] [Google Scholar]
- 51. Vance RE, Zhu J, Mekalanos JJ. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71: 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang H, Wu JH, Ayala JC, Benitez JA, Silva AJ. 2011. Interplay among cyclic diguanylate, HapR, and the general stress response regulator (RpoS) in the regulation of Vibrio cholerae hemagglutinin/protease. J. Bacteriol. 193: 6529–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190: 2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiao H, et al. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266: 5980–5990 [PubMed] [Google Scholar]
- 55. Yun J, et al. 2008. Role of the DksA-like protein in the pathogenesis and diverse metabolic activity of Campylobacter jejuni. J. Bacteriol. 190: 4512–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu J, et al. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.