Abstract
As part of a comprehensive postgenomic investigation of the model archaeon Halobacterium sp. strain NRC-1, we used whole-genome DNA microarrays to compare transcriptional profiles of cells grown under anaerobic or aerobic conditions. When anaerobic growth supported by arginine fermentation was compared to aerobic growth, genes for arginine fermentation (arc) and anaerobic respiration (dms), using trimethylamine N-oxide (TMAO) as the terminal electron acceptor, were highly upregulated, as was the bop gene, required for phototrophic growth. When arginine fermentation was compared to anaerobic respiration with TMAO, the arc and dms genes were both induced with arginine, while TMAO induced the bop gene and major gas vesicle protein (gvpAC) genes specifying buoyant gas vesicles. Anaerobic conditions with either TMAO or arginine also upregulated the cba genes, encoding one of three cytochrome oxidases. In-frame deletion of two COG3413 family regulatory genes, bat and dmsR, showed downregulation of the bop gene cluster and loss of purple membrane synthesis and downregulation of the dms operon and loss of anaerobic respiration capability, respectively. Bioinformatic analysis identified additional regulatory and sensor genes that are likely involved in the full range of cellular responses to oxygen limitation. Our results show that the Halobacterium sp. has evolved a carefully orchestrated set of responses to oxygen limitation. As conditions become more reducing, cells progressively increase buoyancy, as well as capabilities for phototrophy, scavenging of molecular oxygen, anaerobic respiration, and fermentation.
INTRODUCTION
The physiological versatility of halophilic Archaea (Haloarchaea) has been of interest since the recognition of the three domains of life (15). Unlike most other Archaea, which are strict anaerobes, Haloarchaea grow best aerobically yet can generate metabolic energy also via anaerobic respiration, fermentation, and photophosphorylation. Among the Haloarchaea, Halobacterium sp. strain NRC-1 is a genetically tractable model organism with a 2.0-Mb chromosome and two dynamic megaplasmids or minichromosomes, pNRC100 (191 kb) and pNRC200 (365 kb) (11, 24, 25). Analysis of the genome sequence identified ∼2,500 genes, and postgenomic studies showed many to be responsive to fluctuations in environmental conditions (12). Halobacterium sp. NRC-1 is highly responsive to various oxygen levels, including microaerobic and anaerobic conditions, fluctuating levels of salinity to near saturation, and a wide range of temperatures from −20°C to 60°C (9). The organism has also been shown to be tolerant of high levels of UV and ionizing radiation, which is an adaptation to the high solar radiance and desiccating conditions in its environment (16, 21). As a result, Halobacterium sp. NRC-1 has been valuable for studies of multiple stress responses in the Archaea (30).
Although Halobacterium sp. NRC-1 grows fastest under aerobic conditions, it has been shown to have the capacity for anaerobic growth via substrate-level phosphorylation using arginine, by anaerobic respiration using dimethyl sulfoxide (DMSO) or trimethylamine N-oxide (TMAO), as well as phototrophic growth using the light-driven proton pump in its purple membrane (1, 11, 23, 28, 31). Anaerobic fermentation occurs via the arginine deiminase (ADI) pathway, which is encoded by the arcRACB genes (25, 28), while anaerobic respiration requires a DMSO/TMAO reductase and a chaperone, encoded by the dmsREABCD operon. Light-driven proton pumping utilizes bacteriorhodopsin, a retinal chromoprotein that forms a two-dimensional array in the cell membrane and that is encoded by the bop gene cluster. In contrast to the arc genes, which are located on pNRC200, both the dms operon and bop gene cluster are encoded on the Halobacterium chromosome (1, 23, 25).
A combination of pre- and postgenomic studies has established basic genetic features of purple membrane biosynthesis and phototrophy in Halobacterium sp. NRC-1. The protein component, bacterio-opsin, is the product of the bop gene, while the chromophore retinal requires crtB1 and brp, which specify the first and last committed steps in the retinal biosynthetic pathway, respectively (1, 27). These genes are clustered together with six additional genes: (i) a sensor regulator gene, bat; (ii and iii) two small genes, brz and brb, thought to be involved in regulation; (iv) bac, encoding a putative AAA+ ATPase chaperone; (v) bap, encoding a putative membrane protein of unknown function downstream of bop; and (vi) blp, encoding a soluble protein of unknown function (14, 25, 27, 29, 32, 33). The bat gene codes for a multidomain protein with a putative redox-sensing PAS-PAC or LOV (COG2202) domain and a light-sensing GAF domain (COG2203), as well as a C-terminal helix-turn-helix (HTH) DNA binding domain (COG3413) (1, 26). Extensive analysis of Halobacterium mutants lacking purple membrane implicated the bat gene product in regulation of several genes in the bop gene cluster, and saturation mutagenesis of the bop promoter region identified an upstream activator sequence (UAS) as the regulatory site of action (1, 4). Bioinformatic analysis also revealed the presence of a UAS upstream of the brp, crtB1, and blp genes, in addition to the bop gene, and these genes were shown to be induced under limiting oxygen conditions and to support phototrophic growth (1, 36). Coordinate induction of the bop gene and the major gas vesicle protein gene, gvpA, under microaerobic conditions was also reported (36). Therefore, gas vesicle production is also induced, increasing cell buoyancy and enhancing the availability of light and oxygen for photophosphorylation and oxidative phosphorylation.
In another postgenomic investigation, a custom Halobacterium sp. NRC-1 oligonucleotide microarray was used for transcriptional profiling to study the response of this model organism to anaerobic growth with either DMSO or TMAO (23). When cell growth was promoted by anaerobic respiration using either compound as the sole terminal electron acceptor, the dmsREABCD operon was found to be highly induced and essential. Deletion of the putative regulatory gene, dmsR, showed that utilization of these alternate terminal electron acceptors is dependent on this gene, consistent with its action as a transcriptional activator. The C-terminal region of the dmsR gene product was shown to contain an HTH DNA binding motif (COG3413) similar to that of the bop gene activator protein, Bat, although no LOV or GAF domain was found to be present. The cbaDBA genes, encoding a ba3-type cytochrome oxidase, were also found to be inducible when grown with TMAO (23). The cbaA and cbaB genes code for the cytochrome oxidase subunits, likely functioning at low oxygen partial pressure, while cbaD codes for an additional small hydrophobic protein (20).
In the present study, we have used transcriptomic, mutational, and bioinformatic analyses to advance our understanding of the response of Halobacterium sp. NRC-1 to anaerobic conditions. Our results show that a complex genetic program involving multiple regulatory genes orchestrates the response of Halobacterium to changes in oxygen availability and that cells stay primed for aerobic respiration even under anaerobic conditions.
MATERIALS AND METHODS
Culturing.
Halobacterium sp. NRC-1 (Table 1) was grown aerobically or anaerobically with TMAO as described previously (13, 23). For fermentative growth, arginine was added to a final concentration of 0.5% (wt/vol) to modified complex (CM+) medium containing 0.001 g/liter peptone and stored inside an anaerobic glove box (Coy Laboratory Products) for 1 day prior to inoculation. Anaerobic conditions during fermentation were ascertained with resazurin as described previously (23). The cultures were grown in the dark at 37°C and 100 rpm for up to 8 days.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source (reference) |
|---|---|---|
| Escherichia coli DH5α | Laboratory collection (10) | |
| Halobacterium spp. | ||
| NRC-1 | Wild type | Laboratory collection (2) |
| BB400 | NRC-1 Δura3 | Laboratory collection (2) |
| JAM101 | BB400 ΔdmsR | Laboratory collection (23) |
| PAD101 | BB400 Δbat | This study |
| Plasmids | ||
| pUC19 | Cloning vector, lacZα bla | New England Biolabs |
| pBB400 | pUC19 containing NRC-1 ura3 | Laboratory collection (2) |
| pPAD101 | pBB400 containing truncated bat with flanking region | This study |
Deletion construction.
The bat mutant was generated using the standard Halobacterium sp. NRC-1 deletion method (2), as reported previously for the dmsR mutant (23) (Table 1 and 2). Briefly, a suicide plasmid vector was created by triple ligation of ∼500 bp of PCR-amplified flanking regions 5′ and 3′ of the bat gene in Halobacterium sp. NRC-1 into the multiple-cloning site of pBB400 (2), which contains a wild-type copy of the Halobacterium sp. NRC-1 ura3 gene plus its native promoter, and transformed into Escherichia coli DH5α (10). Cells were plated onto LB plates containing ampicillin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Ten colonies were picked, and plasmid preparations were done using the previously described method (2). The plasmids were checked using HindIII-XhoI double digests to release a 1-kb insert. One plasmid was used to transform Halobacterium sp. NRC-1 Δura3 (Table 1) using the polyethylene glycol-EDTA method (2). Cells were plated on agar plates containing uracil dropout medium (HURA+) and grown at 42°C. DNA from individual colonies, purified as previously described (2, 15), was used as the template in PCRs to screen for suicide plasmid integration into genomic DNA. Two independent isolates were plated onto CM+ medium containing 250 μg/ml of 5-fluoroorotic acid (5-FOA) for counterselection. Candidate colonies were picked and grown in CM+ 5-FOA liquid medium. DNA was isolated and PCR amplified using flanking primers (Table 2). Two potential deletants were identified out of 17 candidates and further verified by PCR using both internal and flanking gene primers and by Sanger sequencing.
Table 2.
Oligonucleotides used for bat deletion construction
| Name | Sequence | Description |
|---|---|---|
| batK1F | CTT AAG CTT CCC ACG CCA CCA TCA TCC | Outside primer, flanking bat gene on 3′ end, HindIII site |
| batK1R | CTT ACT AGT CTC GTC GGC GCG TTC TTC | Inside primer, flanking bat gene on 3′ end, SpeI site |
| batK2F | CTT ACT AGT CTC GGA CTC GGT GTT CTG GAC | Inside primer, flanking bat gene on 5′ end, SpeI site |
| batK2R | CTT CTC GAG CGG TGA CAT CGC AAC CCG | Outside primer, flanking bat gene on 5′ end, XhoI site |
| BB1464_For | ATG ACG AGC GTC CAG AAC AC | bat gene start primer |
| BB1464_Rev | CGT CAC TCC TCG AAG AAC GC | bat gene stop primer |
| BLABBF | ATG AGT ATT CAA CAT TTC C | Forward primer for β-lactamase gene on pBB400 |
| BLABBR | TTA CCA ATG CTT AAT CAG T | Reverse primer for β-lactamase gene on pBB400 |
DNA microarrays.
Whole-genome custom DNA oligonucleotide microarrays were fabricated by Agilent Corporation and hybridized using standard Halobacterium sp. NRC-1 methods (2, 23). Cultures were harvested at an optical density at 600 nm (OD600) of 0.49 for the arginine and TMAO experiments, while the OD for the Bat experiment was 1.0 (35). RNA was isolated using spin columns (Agilent), after cell lysis and DNase (Invitrogen) treatment. Synthesis of cDNA was performed with SuperScript II RNase H reverse transcriptase (Invitrogen) using 500 ng of RNA and 2 pmol of primers from three cultures grown under identical conditions. RNA from 3 parallel cultures was pooled to minimize biological noise, and fluorescently labeled cDNA was prepared as previously described (23). The labeled cDNA targets were combined and hybridized in duplicate for 17 h at 65°C in the dark, washed, and scanned for the Cy3 and Cy5 fluorescent signals on an Agilent DNA microarray scanner (model G2565BA). Image processing was carried out using Agilent Feature Extraction software. Array data have been deposited in the GEO database (see below).
Bioinformatic analysis.
Agilent microarray data were analyzed using GeneSpring software, version 11.5.1. Green and red signal intensities of the probes were imported using custom-created software. Quantile normalization (5) was performed, and baseline transformation to the median of all samples was applied to each probe to normalize the signal intensities of each probe across all samples to its median. Green processed signal intensities were grouped and labeled Cy3, while red processed signal intensities were grouped and labeled Cy5. A new gene-level experiment was created in which quantile normalization and baseline transformation were performed. Interpretation was created by grouping the data sets into two groups: Cy3 and Cy5. Fold change analysis was performed by selecting pairs of conditions with the pairing option defining Cy3 and Cy5 as two conditions. To determine the differentially expressed genes that were statistically significant, a P value of <0.05 and a fold change cutoff threshold of ≥2 were used. Local clusters of proteins developed using reciprocal blasting (7, 8) as well as the COG cluster at NCBI were utilized to identify conserved proteins and domains.
Membrane analysis.
Purple and red membrane preparations were isolated using sucrose cushion gradients as previously described (13). Briefly, cells were grown in 1 liter medium for 1 to 2 days into stationary phase (OD600 = 1.0) and spun down in a Sorvall RC-5B centrifuge at 6,000 rpm and 4°C for 10 min. The supernatant was removed, and cells were resuspended in 5 ml basal salts solution. The cell lysate was transferred into dialysis tubing and dialyzed against 5 liters of water at 4°C with 3 changes. The cell paste was then treated with 50 μl DNase I (10 μg/μl) and incubated at 37°C for 1 h with shaking at 180 rpm. This step was repeated, when necessary, to reduce viscosity. The sucrose gradient was prepared in 40 ml nitrocellulose centrifuge tubes by adding filter-sterilized sucrose dissolved in water to the bottom of the tube in the order 11 ml 30% sucrose, 11 ml 40% sucrose, 11 ml 50% sucrose, and 2 ml 60% sucrose; and 5 ml lysate was layered on top of the gradient. The gradient was spun using an SW28 swinging-bucket rotor in a Beckman L5 ultracentrifuge for 15.5 h at 24,000 rpm under vacuum at 18°C.
Microarray data accession number.
Array data were deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under series accession number GSE38374 (18).
RESULTS
Anaerobic growth on arginine versus aerobic growth.
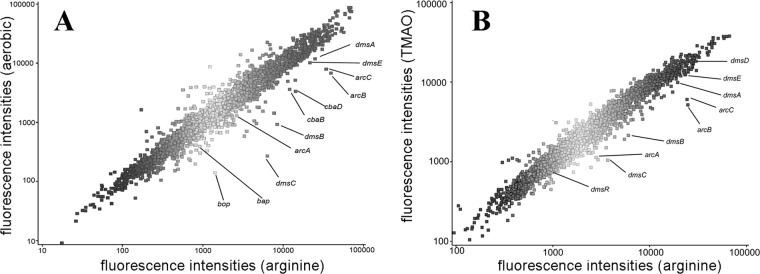
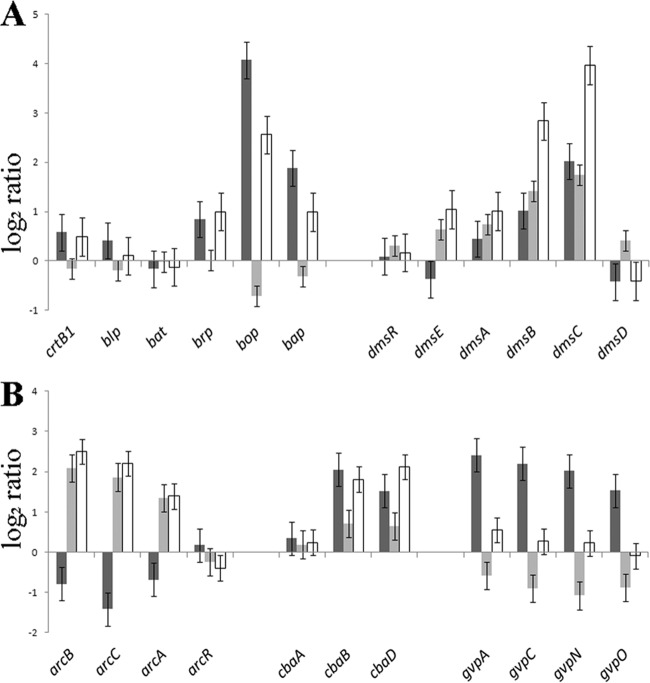
Halobacterium sp. NRC-1 was grown either aerobically in standard shaken cultures or anaerobically with 0.5% arginine. RNA was prepared from both aerobically and anaerobically grown cells, and comparative transcriptional profiling was conducted using whole-genome oligonucleotide microarrays. Several genes involved in generation of metabolic energy had significant changes in transcript levels under anaerobic versus aerobic conditions (Fig. 1A and 2, white bars). Surprisingly, the dms genes involved in DMSO/TMAO respiration were the most highly induced during arginine fermentation, although neither substrate for the DMSO/TMAO reductase was present. The arcACB genes, which are involved in arginine fermentation via the ADI pathway, were also strongly induced under anaerobic growth on arginine. During growth on arginine, the third catabolic gene, arcA, was somewhat less induced than arcC and arcB, similar to a previous report (28). The bacterio-opsin-encoding gene bop was also strongly upregulated, while the downstream transcriptionally linked gene bap was also induced, but less strongly. The other genes in the bop gene cluster, brp and crtB1, were only slightly changed, while blp and bat were essentially unchanged with arginine. Interestingly, among the cytochrome oxidase genes, the cbaD and cbaB gene transcripts were significantly upregulated under arginine fermentation almost as highly as the arc genes, while the cbaA gene was slightly upregulated. Among the gas vesicle gene cluster, only the major gas vesicle protein genes gvpA and gypC were upregulated, but not highly (Fig. 2). These results suggest that progressively more reducing conditions result in increasing transcriptional induction, in a gradient from gvp and bop to the cba, dms, and arc genes.
Fig 1.
Genome-wide transcriptomic data comparing Halobacterium sp. NRC-1 grown (A) aerobically (y axis) versus anaerobically with arginine (x axis) or (B) anaerobically with TMAO (y axis) versus arginine (x axis). DNA microarray data (fluorescence units) were generated using cDNA probes fluorescence labeled with Cy3 or Cy5 and analyzed employing Agilent's GeneSpring program (see Materials and Methods). Metabolic genes with high transcript levels during arginine fermentation are labeled with gene names (see the text).
Fig 2.
Gene expression changes in Halobacterium sp. NRC-1 grown under different conditions. Log2 ratios (y axis) in gene expression are plotted for bop gene cluster and dms genes (A) and arc genes, cba genes, and gvp genes (B) (with the genes indicated below). Dark gray bars, anaerobic growth with TMAO versus aerobic growth; light gray bars, anaerobic growth with arginine versus anaerobic growth with TMAO; white bars, anaerobic growth with arginine versus aerobic growth. Error bars indicate standard errors.
Anaerobic growth on arginine versus TMAO.
Transcriptomic analysis was carried out for Halobacterium sp. NRC-1 grown anaerobically with arginine fermentation versus anaerobic respiration using TMAO (Fig. 1B and 2, light gray bars). Among genes involved in metabolic energy generation, the dmsEABC and arcACB genes were induced during anaerobic growth on arginine compared to anaerobic respiration with TMAO. Compared to the differential expression of arginine fermentation versus aerobic growth, the degree of induction was nearly the same for arc genes but significantly less for dms genes. Like the dms genes, the cba genes also showed significantly greater induction with arginine than TMAO. In contrast, the bop gene, encoding the bacterio-opsin involved in phototrophic growth, and the transcriptionally linked bap gene, of unknown function, were downregulated under TMAO compared to their regulation under more reducing conditions in arginine. The other genes in the bop gene cluster, crtB1, blp, brp, and bat, were essentially unchanged in arginine versus TMAO. Like the bop and bat genes, the gas vesicle genes gvpACNO were downregulated in arginine compared to their regulation in TMAO. These results are consistent with previous findings comparing transcription during anaerobic respiration with TMAO to aerobic growth (Fig. 2, dark gray bars), showing coordinate induction of both purple membrane and gas vesicle synthesis under TMAO-reducing conditions. However, the finding of lower upregulation of the dms operon during TMAO respiration than with arginine fermentation was unexpected (Fig. 2). These results are consistent with a gradient of transcriptional effects for metabolic genes. Under increasingly reducing conditions, Halobacterium sp. NRC-1 progressively induces the gvp, bop, cba, dms, and arc genes.
Regulatory mutants.
In order to better understand the regulatory networks responding to oxygen limitation in Halobacterium sp. NRC-1, we targeted two positive regulatory genes of Halobacterium sp. NRC-1 for deletions, bat and dmsR, using the ura3-based deletion method (2). For the bat deletion mutant, we observed a slight difference in colony color for the mutant strain (pink) compared to that for the parent (more purple) on agar plates (Fig. 3A). Upon membrane extraction, purple membrane was detectable in the parent strain but was not detectable in the bat deletion mutant, while there was no observed difference in the red membrane fraction (Fig. 3B). DNA microarray analysis comparing the bat deletion mutant to the parent strain showed that expression of members of the bat gene cluster, including crtB1, blp, brp, bop, and bap, was also significantly lower in the mutant (Fig. 4, gray bars). These results are consistent with the purple membrane-deficient phenotype of the bat deletion mutant and confirmed the function of the bat gene in transcriptional activation of most of the genes near bop.
Fig 3.
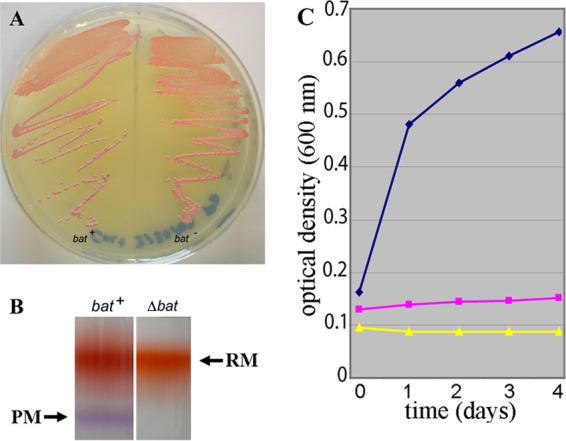
Phenotypes of regulatory mutants. (A) Halobacterium sp. NRC-1 bat wild-type (bat+) and bat deletion mutant (Δbat) strains are shown on an agar plate. A subtle phenotypic difference is visible, with the wild-type bat strain appearing purple (left) and the bat mutant strain (right) appearing pink. (B) Halobacterium sp. NRC-1 wild-type bat (left) and bat mutant (right) red membrane (RM) and purple membrane (PM) fractionated by sucrose gradient. (C) Growth of Halobacterium sp. NRC-1 wild-type dmsR and ΔdmsR strains in anaerobic liquid cultures shown by plotting optical density (at 600 nm) versus time (in days). Shown are growth curves (average of 3 cultures) for the wild-type dmsR strain in medium with TMAO (blue) or without TMAO (yellow) and the ΔdmsR strain with TMAO (pink).
Fig 4.
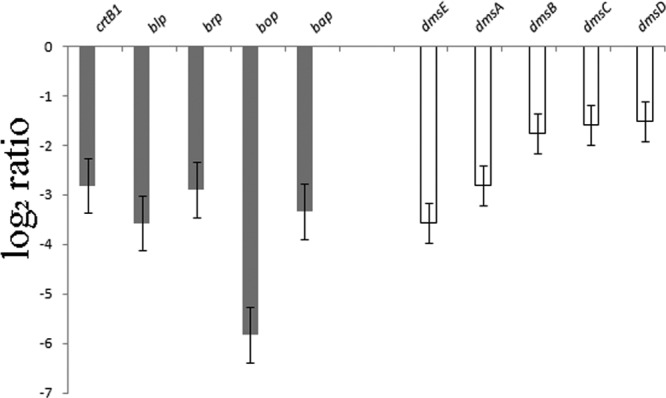
Gene expression in Halobacterium sp. NRC-1 Δbat and ΔdmsR regulatory mutants. Log2 ratios (y axis) between the Δbat mutant (gray bars) or ΔdmsR mutant (white bars) and NRC-1 are shown for genes of the bop gene cluster and the dms operon (genes are indicated above). Error bars indicate standard errors.
Previously, we had constructed a dmsR deletion strain and compared its anaerobic growth in the presence or absence of TMAO (Fig. 3C) (23). The results showed that while the wild-type dmsR strain can grow in medium with TMAO (Fig. 3C, blue curve) but not in the absence of TMAO (yellow curve), the ΔdmsR strain is unable to grow with TMAO (pink curve). We used DNA microarrays to compare the dmsR deletion strain to the parent strain grown aerobically and found that genes in the dmsR-regulated operon, dmsEABCD, were significantly reduced in the dmsR deletion strain compared to the parental strain (Fig. 4, white bars). These results confirmed that the dmsR gene is a positive regulator of transcription of the dms operon.
Bioinformatic analysis of regulators.
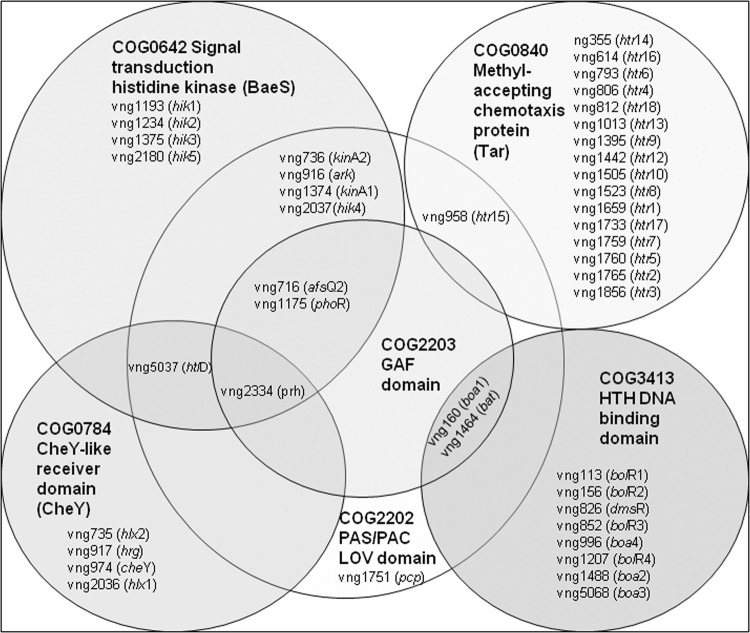
In order to discover other regulatory genes which may play a role in the response of Halobacterium sp. NRC-1 to oxygen-limiting conditions, we bioinformatically screened the Halobacterium sp. NRC-1 genome for genes encoding protein domains similar to those in the characterized regulatory genes bat (COG2202, COG2203, and COG3413) and dmsR (COG3413) (Fig. 5) (7, 8). Interestingly, we observed 10 predicted genes coding COG3413 HTH domains, one of which (boa1, a bacterio-opsin activator-like gene) also contained both COG2202 (LOV) and COG2203 (GAF) domains, like bat. In total, 12 LOV domain-containing genes were found, of which 5 contained GAF domains representing all of the members of the COG2203 GAF family. Several members of the COG2202 LOV family also contained signaling or sensor domains, including 8 out of 12 predicted histidine kinase domains (COG0642) in the Halobacterium sp. NRC-1 genome, 2 out of 6 with CheY receiver domains (COG0784), and 1 out of 17 with Tar methyl-accepting domains (COG0840). The arcR gene, encoding the regulator for the putative arc operon on pNRC200, was found to be unique, with a bacterium-type HTH domain (COG1414) in the N-terminal region (22). These bioinformatic results indicate that in addition to bat and dmsR, additional predicted regulatory genes likely participate in the anaerobic and microaerobic sensing and regulatory network for Halobacterium sp. NRC-1 (Fig. 5).
Fig 5.
Venn diagram showing conserved protein family relationships for regulatory and signaling proteins in Halobacterium sp. NRC-1. COG designations are in bold for each family, and individual members of the family in Halobacterium sp. NRC-1 are indicated by vng numbers and gene names (in parentheses) (see reference 25).
DISCUSSION
We have examined the transcriptional responses of the model halophilic archaeon Halobacterium sp. NRC-1 under different growth conditions comparing anaerobic growth with arginine fermentation to either TMAO respiration or aerobic growth in rich medium. This work expands upon our previous study examining the transcriptional response of the same organism under TMAO-respiring conditions versus aerobic growth (23). In our studies, we also constructed in-frame deletions of two regulatory genes, bat and dmsR, which are linked to genes involved in the cellular response to anaerobic conditions, bop and dms, and determined the resulting phenotypic and transcriptional changes. Together, our work now provides a comprehensive genome-wide view of the response of a model archaeon to a common perturbation in environmental growth conditions, with transcriptional effects on genes encoding gas vesicles, purple membrane, an alternate cytochrome oxidase, DMSO/TMAO oxidoreductase, and ADI pathway detected.
A gradient of changes in transcript levels was observed in the order bop, gvp, cba, dms, and arc, genes which are involved in generation of metabolic energy under hypoxic and anoxic conditions, including fermentation (arginine) and anaerobic respiration (TMAO) (Fig. 2). The arc genes were most highly induced during arginine fermentation. Apparently, the determining factor driving the expression of the arc genes is a high concentration of arginine, since anaerobiosis alone does not result in differential expression of those genes (23). Surprisingly, the dms genes were most induced during growth via arginine fermentation rather than TMAO respiration. Assuming that all components of the electron transport chain are still present in the cell during arginine fermentation (see below), this regulatory scheme is consistent with the hypothesis that the redox state of cells governs expression of the dms genes in Halobacterium sp. NRC-1. The cba ba3-type cytochrome oxidase genes were similarly induced during arginine fermentation and TMAO respiration in comparison to aerobic growth. The bop gene was also induced under anaerobic conditions, but not as dramatically in arginine as in TMAO. The gvp genes displayed a pattern similar to that for the bop gene (less induction with arginine and more with TMAO). These findings showed that cells in a partially reducing state (TMAO) induce the bop, gvp, cba, and dms genes in a gradient from more strongly to more weakly, with full induction of arc genes occurring in the presence of arginine.
Our whole-genome transcriptomic analysis also provided a broader picture of metabolic gene expression patterns under different growth conditions. Interestingly, the chromosomal cytochrome c oxidase genes (cox) were largely unchanged under the various growth conditions, in contrast to the cytochrome ba3 oxidase (cba) and, to a lesser extent, the cytochrome d oxidase (cyd), coded on the inverted repeats of pNRC100 and pNRC200, which were induced during TMAO respiration. Among the other metabolic energy systems, electron transport chain components were largely unchanged. However, the ATP synthase genes were induced with oxygen when the growth rate was higher, as were nearly all of the genes of the tricarboxylic acid cycle. Similarly, the cobalamin biosynthesis genes were upregulated at the higher growth rate with oxygen, a likely reflection of the metabolic cost of cofactor synthesis (34). Not surprisingly, both superoxide dismutase (sod) genes involved in oxidative repair of DNA damage were upregulated as well in the presence of oxygen (6). Interestingly, an increase in the abundance of buoyant gas vesicle gene transcripts (gvpACNO) under oxygen-limiting conditions may enable flotation of cells to oxic zones in the water column, with the observed regulatory scheme for cytochrome ba3 oxidase allowing scavenging of molecular oxygen whenever it becomes available (19, 23, 36).
Genetic and transcriptomic analyses of bat and dmsR confirmed and extended our understanding of the role of these regulatory genes in the anaerobic/redox response. The results clearly showed that bat and dmsR are each specific regulators of the nearby bop regulon and dms operon genes (Fig. 4). Transcriptomic results for the dmsR deletion mutant are consistent with previous findings and indicate the likely role in activation of the dmsEABCD operon (23). In contrast, the findings for the specific effect of the bat deletion on purple membrane genes are distinct from those for the spontaneous bat insertion mutation used in earlier studies, in which the mutants were pleiotropic and which resulted in the loss of both purple membrane and red bacterioruberin pigments. Previously, spontaneous bat insertion mutations resulted in a colorless or white phenotype, in contrast to the more restricted purple membrane-deficient pink phenotype for mutants with the constructed in-frame bat deletion (3, 35). A recent report also indicated that deletion of the bop gene significantly reduces bacterioruberin production (17). This disparity likely resulted from a strain-specific difference, such as the bat frameshift mutation reported in Halobacterium sp. strain S9 (1), in contrast to the wild-type background for strain NRC-1 in the present study. Expression of the retinal biosynthetic genes in the bop gene cluster (crtB1 and brp) was also strongly downregulated in the bat deletion strain, but expression changes in these genes due to growth conditions were much less pronounced. A recently described gene located downstream from bop, named bap, coding a putative membrane protein (29), was also found to be downregulated in the strain with a bat gene deletion.
While the bop, dms, and arc structural genes were highly responsive to different growth conditions, expression of the nearby regulatory genes bat, dmsR, and arcR (the putative arc operon regulator) remained essentially unchanged. These findings show that expression of transcriptional regulators does not need to be altered under different growth conditions and that their ability to affect gene expression likely occurs via changes in their protein structure and/or function. The LOV and GAF domains, which are predicted to sense redox and light, respectively, are presumably involved in Bat function (1, 17). For DmsR, where no LOV or GAF domains were identified, the N-terminal cysteine-rich region may play a role in redox sensing (23). For ArcR, which is a member of the IclR bacterium-type regulator family (COG1414) found in bacterial glycerol and acetate operons, no sensing domain has been identified (22).
Interestingly, the Bat and DmsR COG3413 HTH motif is primarily found in archaeon-type DNA binding proteins and may be quite prevalent, with 10 members in Halobacterium sp. NRC-1. Proteins in the COG3413 family are also found in Sulfolobus and Thermoplasma spp. (7, 8), which, like haloarchaea, are facultative anaerobes that can grow aerobically using organic compounds. These observations suggest that the COG3413 family may be involved in oxygen/redox regulation in other clades of facultative anaerobic Archaea. However, the COG3413 family does not appear to be exclusively used since other DNA binding domain proteins, like the N-terminal HTH-type domain in ArcR, are also involved in regulation of anaerobically induced genes.
The transcriptomic and genetic analyses in our studies have provided a detailed view of physiological changes in a model Haloarchaeon in response to environmental changes in oxygen and redox conditions. Together with studies of salinity, desiccation, radiation, and other environmental factors, we have gained a deeper understanding of how an extremophilic microorganism in the archaeal domain of life can be successful in a highly challenging and dynamic environment. Further detailed studies aimed at deciphering the regulatory networks operating in Halobacterium sp. NRC-1 are clearly necessary for a comprehensive understanding of the microbial physiology of this model organism.
ACKNOWLEDGMENTS
This work was funded by National Science Foundation grant MCB-029617, National Aeronautics and Space Administration grant NNX10AP47G, Henry M. Jackson Foundation grant HU0001-09-1-0002-660883, and Bill and Melinda Gates Foundation grant OPP1061509.
We thank Melinda D. Capes, Jong-Myoung Kim, Andrea L. Jalickee, and Susan R. Steyert for valuable assistance.
Footnotes
Published ahead of print 3 August 2012
REFERENCES
- 1. Baliga NS, Kennedy SP, Ng WV, Hood L, DasSarma S. 2001. Genomic and genetic dissection of an archaeal regulon. Proc. Natl. Acad. Sci. U. S. A. 98:2521–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berquist BR, Müller JA, DasSarma S. 2006. Genetic systems for halophilic Archaea. Methods Microbiol. 35:637–668. [Google Scholar]
- 3. Betlach M, Friedman J, Boyer HW, Pfeifer F. 1984. Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res. 12:7949–7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Betlach MC, Shand RF, Leong DM. 1989. Regulation of the bacterio-opsin gene of a halophilic archaebacterium. Can. J. Microbiol. 35:134–140 [DOI] [PubMed] [Google Scholar]
- 5. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 6. Brown-Peterson NJ, Begonia GB, Salin ML. 1995. Alterations in oxidative activity and superoxide dismutase in Halobacterium halobium in response to aerobic respiratory inhibitors. Free Radic. Biol. Med. 18:249–256 [DOI] [PubMed] [Google Scholar]
- 7. Capes MD, et al. 2011. The information transfer system of halophilic archaea. Plasmid 65:77–101 [DOI] [PubMed] [Google Scholar]
- 8. Capes MD, DasSarma P, DasSarma S. 2012. The core and unique proteins of haloarchaea. BMC Genomics 13:39 doi:10.1186/1471-2164-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coker JA, DasSarma P, Kumar J, Müller JA, DasSarma S. 2007. Transcriptional profiling of the model Archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature. Saline Systems 3:6 doi:10.1186/1746-1448-3-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dagert M, Ehrlich S. 1979. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23–28 [DOI] [PubMed] [Google Scholar]
- 11. DasSarma S. 2004. Genome sequence of an extremely halophilic archaeon, p 383–399 In Fraser CM, Read T, Nelson KE. (ed), Microbial genomes. Humana Press Inc., Totowa, NJ [Google Scholar]
- 12. DasSarma S, Berquist BR, Coker JA, DasSarma P, Müller JA. 2006. Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1. Saline Systems 2:3 doi:10.1186/1746-1448-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DasSarma S, Fleischmann EM. 1995. Archaea, a laboratory manual—halophiles. p 55–58, 179–184. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 14. DasSarma S, RajBhandary UL, Khorana HG. 1984. Bacterio-opsin mRNA in wild-type and bacterio-opsin deficient Halobacterium halobium strains. Proc. Natl. Acad. Sci. U. S. A. 81:125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DasSarma S, Coker JA, DasSarma P. 2010. Archaea—overview, p 118–139 In Schaechter M. (ed), Desk encyclopedia of microbiology, 2nd ed Academic Press, San Diego, CA [Google Scholar]
- 16. DeVeaux LC, et al. 2007. Extremely radiation-resistant mutants of a halophilic archaeon with increased single-stranded DNA binding protein (RPA) gene expression. Radiat. Res. 168: 507–514 [DOI] [PubMed] [Google Scholar]
- 17. Dummer AM, et al. 2011. Bacterioopsin-mediated regulation of bacterioruberin biosynthesis in Halobacterium salinarum. J. Bacteriol. 193:5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hechler T, Pfeifer F. 2009. Anaerobiosis inhibits gas vesicle formation in halophilic Archaea. Mol. Microbiol. 71:132–145 [DOI] [PubMed] [Google Scholar]
- 20. Mattar S, Engelhard M. 1997. Cytochrome ba3 from Natronobacterium pharaonis—an archaeal four-subunit cytochrome-c-type oxidase. Eur. J. Biochem. 250:332–341 [DOI] [PubMed] [Google Scholar]
- 21. McCready S, et al. 2005. UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Systems 1:3 doi:10.1186/1746-1448-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Molina-Henares AJ, Krell T, Eugenia Guazzaroni M, Segura A, Ramos JL. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30:157–186 [DOI] [PubMed] [Google Scholar]
- 23. Müller JA, DasSarma S. 2005. Genomic analysis of anaerobic respiration of the archaeon Halobacterium sp. strain NRC-1: dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors. J. Bacteriol. 187:1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ng W-L, et al. 1998. Snapshot of a large dynamic replicon from a halophilic Archaeon: megaplasmid or minichromosome? Genome Res. 8:1131–1141 [DOI] [PubMed] [Google Scholar]
- 25. Ng WV, et al. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. U. S. A. 97:12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pathak GP, Losi A, Gärtner W. 2012. Metagenome-based screening reveals worldwide distribution of LOV-domain proteins. Photochem. Photobiol. 88:107–118 [DOI] [PubMed] [Google Scholar]
- 27. Peck RF, et al. 2001. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J. Biol. Chem. 276:5739–5744 [DOI] [PubMed] [Google Scholar]
- 28. Ruepp A, Soppa J. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178:4942–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma AK, et al. 2007. Evolution of rhodopsin ion pumps in haloarchaea. BMC Evol. Biol. 7:79. doi:10.1186/1471-2148-7-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slonczewski JL, Coker JA, DasSarma S. 2010. Microbial growth with multiple stressors. Microbe 5: 110–116 [Google Scholar]
- 31. Sumper M, Reimeier H, Oesterhelt D. 1976. Biosynthesis of the purple membrane of halobacteria. Angew. Chem. Int. Ed. Engl. 16:187–194 [DOI] [PubMed] [Google Scholar]
- 32. Tarasov VY, et al. 2008. A small protein from the bop-brp intergenic region of Halobacterium salinarum contains a zinc finger motif and regulates bop and crtB1 transcription. Mol. Microbiol. 67:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tarasov V, Schwaiger R, Furtwangler K, Dyall-Smith M, Oesterhelt D. 2011. A small basic protein from the brz-brb operon is involved in regulation of bop transcription in Halobacterium salinarum. BMC Mol. Biol. 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Woodson JD, Reynolds AA, Escalante-Semerena JC. 2005. ABC transporter for corrinoids in Halobacterium sp. strain NRC-1. J. Bacteriol. 187:5901–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang CF, DasSarma S. 1990. Transcriptional induction of purple membrane and gas vesicle synthesis in the archaebacterium Halobacterium halobium is blocked by a DNA gyrase inhibitor. J. Bacteriol. 172:4118–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang C-F, Kim J-M, Molinari E, DasSarma S. 1996. Genetic and topological analyses of the bop promoter of Halobacterium halobium: stimulation by DNA supercoiling and non-B-DNA structure. J. Bacteriol. 178:840–845 [DOI] [PMC free article] [PubMed] [Google Scholar]