Abstract
Containment strategies for outbreaks of invasive Neisseria meningitidis disease are informed by serogroup assays that characterize the polysaccharide capsule. We sought to uncover the genomic basis of conflicting serogroup assay results for an isolate (M16917) from a patient with acute meningococcal disease. To this end, we characterized the complete genome sequence of the M16917 isolate and performed a variety of comparative sequence analyses against N. meningitidis reference genome sequences of known serogroups. Multilocus sequence typing and whole-genome sequence comparison revealed that M16917 is a member of the ST-11 sequence group, which is most often associated with serogroup C. However, sequence similarity comparisons and phylogenetic analysis showed that the serogroup diagnostic capsule polymerase gene (synD) of M16917 belongs to serogroup B. These results suggest that a capsule-switching event occurred based on homologous recombination at or around the capsule locus of M16917. Detailed analysis of this locus uncovered the locations of recombination breakpoints in the M16917 genome sequence, which led to the introduction of an ∼2-kb serogroup B sequence cassette into the serogroup C genomic background. Since there is no currently available vaccine for serogroup B strains of N. meningitidis, this kind capsule-switching event could have public health relevance as a vaccine escape mutant.
INTRODUCTION
Neisseria meningitidis is a Gram-negative bacterium that has the potential to cause sudden and acute diseases, including meningitis and septicemia (25). N. meningitidis is protected from host-mediated, complement-dependent bacteriolysis and phagocytosis by a polysaccharide capsule (25) and is classified into 12 serogroups determined by the chemical composition of the capsule. Distinct serogroup capsule compositions differ by either their particular sugar makeup or the linkages between sugars (7, 23). Meningococci frequently take up and incorporate genomic DNA from the environment via homologous recombination (12, 15). This may produce a change in capsule genes, resulting in a change in the composition/serogroup of the capsule sometimes referred to as capsule switching (29). The most typically disease-associated serogroups are A, B, C, W-135, and Y (35). Effective vaccines are available for disease-associated serogroups A, C, W-135, and Y (3), whereas there is currently no protective vaccine specifically against the serogroup B capsule (11). Therefore, capsule switching may produce escape mutants such as C to B. Accordingly, rapid identification of N. meningitidis capsule serogroups is a critical component of public health response to meningococcal disease.
The chemical composition of the N. meningitidis polysaccharide capsule is classically inferred by using the slide agglutination serogrouping (SASG) assay (23, 24). The SASG assay reveals the capsule type with serogroup-specific antibodies that yield positive or negative reactions. Ideally, exactly one reactivity will be positive per isolate but there are occasionally multiple or no positive reactions. In these cases, the isolate is characterized as “nongroupable” (NG). Isolates that have multiple positive SASG assay results are referred to as NG-polyagglutinating. Mothershed et al. introduced the serogroup-specific real-time PCR (SGS-PCR) assays to help resolve NG SASG assay results (22). The SGS-PCR assay is a real-time PCR test that uses primers and probes specific to each serogroup. The target of these assays is typically the gene that encodes the capsule polymerase specific to each serogroup. The CDC Meningitis Laboratory and other reference labs routinely perform both SASG and SGS-PCR assays to group isolates quickly and unambiguously.
Despite the availability of these two assays for the characterization of N. meningitidis capsule serogroups, there remain instances of NG isolates and at times the SASG and SGS-PCR assay results do not agree. Examination of these NG isolates reveals three distinct classes: autoagglutinating, nonaggultinating, and polyagglutinating. Autoagglutinating isolates agglutinate in the saline control for the SASG assay via a mechanism unrelated to serogroup specificity. Nonagglutinating isolates show no evidence of capsule expression, and a number of such isolates have been genetically characterized. This work showed that nonaggultination can result from a loss of capsule expression through point mutations and/or phase variation, as well as partial or complete deletions of capsule operon sequences (5, 34). Polyagglutinating isolates are defined as showing cross-reactivity to two or more serogroup-specific antisera. At this time, the genetic basis of polyagglutination is less well understood. Polyagglutination has been hypothesized to result from the expression of multiple capsule types but has more often been attributed simply to the quality of the antiserum reagents used in the SASG assay.
In this study, we sought to characterize the genomic basis of an NG polyagglutination result for a cerebrospinal fluid (CSF) isolate (M16917) from a patient with acute meningococcal disease. To this end, we characterized and analyzed its complete genome sequence. The M16917 genome sequence was analyzed to determine its likely genomic origin, and the capsule-encoding locus was compared with capsule loci from genomes with known serogroups in order to determine its specific structure. The M16917 capsule locus was found to be chimeric, with sequences derived from both B and C serogroup-encoding regions.
Nucleotide sequence accession number.
The genome sequence data obtained in this study have been submitted to the NCBI sequence read archive and assigned accession number SRX111309.
MATERIALS AND METHODS
Isolation and characterization of M16917.
The M16917 strain of N. meningitidis was isolated from the CSF of a 31-year-old male with acute meningococcal disease. The patient presented on 15 December 2007 with acute onset (∼3 h) of mental status changes, agitation, combativeness, and diaphoresis. He had complained of cough, congestion, and body aches for several days. The patient was HIV negative and did not have any apparent underlying immunocompromising conditions. On presentation, the patient's temperature was 100°F and his blood pressure was 146/76. The patient's skin was flushed, cool, and moist with no rash. The patient's CSF had a white blood cell count of 260 cells/mm3, 95% neutrophils, a protein content of 1,570 mg/liter, a glucose level of 3 mg/dl, and a culture positive for N. meningitidis. Blood cultures were negative for meningococci. The hospital course was complicated by fever, hypoxia, and pneumonia requiring intubation. The patient was treated with vancomycin and ceftriaxone and improved. He recovered and was eventually extubated and discharged home on 28 December 2007.
The M16917 strain was initially characterized with the SASG assay by the Illinois Department of Public Health in Chicago. M16917 was subsequently characterized by the CDC Meningitis Laboratory using both the SASG and SGS-PCR assays as previously described (22, 23). The genome sequence of M16917 was characterized by 454 pyrosequencing by the CDC core facility and analyzed with the CG-Pipeline tool as previously reported (17). 454 sequencing is prone to result in errors at homopolymeric sequences, and this occurred at the adk locus, which is one of the housekeeping loci used in multilocus sequence typing (MLST) analysis. In order to resolve this homopolymeric region in the adk locus, we performed Sanger sequencing as described at http://pubmlst.org/ (13, 14).
N. meningitidis reference genome sequences.
All publicly available N. meningitidis reference complete genome sequences were downloaded from the “completed genomic sequence” portion of the NCBI Entrez Genome database entry for N. meningitidis (http://www.ncbi.nlm.nih.gov/genomes/geblast.cgi?taxid=487). These sequences correspond to four serogroups (A, B, C, and cnl) and 14 sequence types (STs) (Table 1).
Table 1.
Complete N. meningitidis reference genomes used in this study
| Strain | SGa | ST | CC | RefSeq accession no. | Geographic origin | Yr isolated | PMIDb |
|---|---|---|---|---|---|---|---|
| WUE 2594 | A | 5 | 5 | NC_003116 | Germany | 1991 | 21296965 |
| Z2491 | A | 4 | 4 | NC_013016 | Gambia | 1983 | 10761919 |
| alpha710 | B | 136 | 41/44 | NC_017512 | Germany | 2010 | 20709895 |
| G2136 | B | 8 | 8 | NC_01751 | United Kingdom | 1986 | 21368196 |
| H44/76 | B | 32 | 32 | NC_017516 | Norway | 1976 | 21368196 |
| M01-240149 | B | 41 | 41/44 | NC_017514 | United Kingdom | 2001 | 21368196 |
| M01-240355 | B | 213 | 213 | NC_017517 | United Kingdom | 2001 | 21368196 |
| M04-240196 | B | 269 | 269 | NC_017515 | United Kingdom | 2004 | 21368196 |
| MC58 | B | 74 | 32 | NC_003112 | Gloucester, United Kingdom | 1985 | 10710307 |
| NZ-05/33 | B | 42 | 41/44 | NC_017518 | New Zealand | 2005 | 21368196 |
| 8013 | C | 177 | 18 | NC_017501 | France | 1989 | 19818133 |
| 053442 | C | 4821 | 4821 | NC_010120 | Anhui Province, China | 2003 | 18031983 |
| FAM18 | C | 11 | 11 | NC_008767 | North Carolina | 1983 | 17305430 |
| alpha14 | cnl | 53 | 53 | NC_017505 | Bavaria, Germany | 1999 | 18305155 |
| M16917 | NG | 11 | 11 | N/A | Illinois | 2007 | N/A |
SG, serogroup.
PMID, PubMed identifier.
Genome-based MLST analysis.
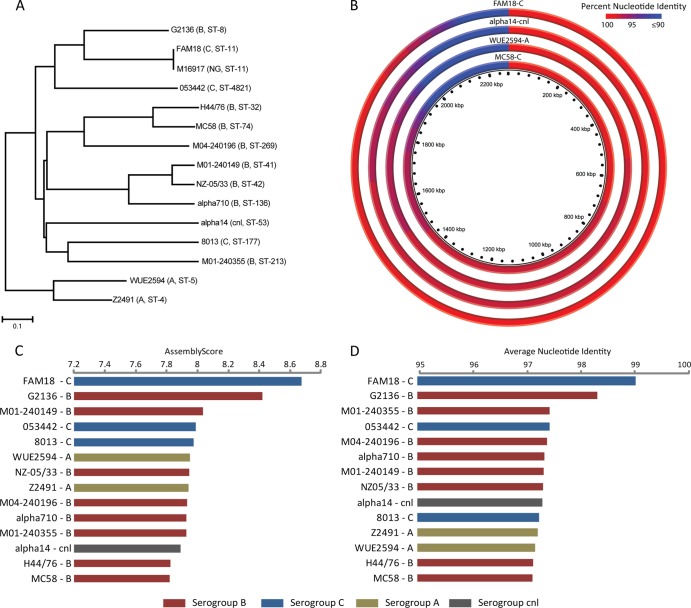
We employed an MLST analysis, which characterizes bacterial isolates based on the alleles of seven highly conserved housekeeping genes (12, 19), on the complete genome sequence of M16917. The genome sequence was assembled de novo using the CG-Pipeline (v0.2.2) software tool (17), and the assembly was compared to the PubMLST database of alleles for each of the seven housekeeping loci (13, 14) using BLAST (v2.2.17) (1). We determined the presence of an allele on the basis of whether the best hit of the BLAST analysis had 100% identity, with no gaps or mismatches, to that allele, as well as an alignment length equal to the full length of the allele. The unique combination of alleles across the seven loci yields an ST, which may belong to a family of STs called a clonal complex (CC). STs from all publicly available N. meningitidis reference complete genome sequences were determined in the same way, and the phylogenetic relationships among all STs (Fig. 1A) were determined by using the neighbor-joining algorithm implemented in the tree-drawing program of the PubMLST database (http://pubmlst.org/perl/mlstanalyse/mlstanalyse.pl?site=pubmlst&page=treedraw&referer=pubmlst.org).
Fig 1.
Whole-genome sequence comparison of M16917 with complete N. meningitidis reference genome sequences. (A) Phylogenetic analysis of M16917 and complete N. meningitidis genome sequences based on their STs. The serogroups and STs of the strains are shown. (B) Distributions of sequence similarity values (percentages of nucleotide identity) between M16917 contigs and complete genome sequences representative of four different serogroups. Percentages of nucleotide identity are color coded as shown and sorted along the genomes according to similarity levels (highest to lowest). (C and D) Assembly scores and average nucleotide identities of M16917 reference assemblies to the N. meningitidis reference genome sequences shown.
Reference-based assembly.
We performed reference-based assemblies of M16917 by using the program Newbler (20) against all publicly available N. meningitidis reference complete genome sequences (Table 2). The quality of each reference-based assembly was evaluated on the basis of a number of metrics, i.e., the number of large contigs produced, the number of bases assembled, and the N50 size. These three metrics were combined into an overall reference assembly quality score (AS) by using the equation AS = log10 (N50 × [number of bases assembled/number of large contigs]).
Table 2.
Comparison of M16917 reference assemblies against different N. meningitidis genomes
| Reference | Serogroup | No. of contigsa | Assembly sizeb | N50c | Assembly scored | % Alignmente | ANI (%)f |
|---|---|---|---|---|---|---|---|
| FAM18 | C | 143 | 2,090,956 | 32,086 | 8.67 | 95.67 | 99.05 |
| G2136 | B | 186 | 2,073,087 | 23,750 | 8.42 | 95.15 | 98.35 |
| M01-240149 | B | 275 | 1,974,903 | 15,169 | 8.04 | 90.94 | 97.35 |
| 053442 | C | 271 | 1,936,077 | 13,556 | 7.98 | 89.07 | 97.45 |
| 8013 | C | 283 | 1,976,593 | 13,400 | 7.97 | 90.38 | 97.26 |
| NZ-05/33 | B | 295 | 1,975,340 | 13,257 | 7.95 | 90.91 | 97.34 |
| WUE2594 | A | 273 | 1,937,602 | 12,529 | 7.95 | 88.65 | 97.18 |
| Z2491 | A | 279 | 1,937,199 | 12,446 | 7.94 | 88.98 | 97.23 |
| alpha710 | B | 304 | 1,975,804 | 13,069 | 7.93 | 90.71 | 97.37 |
| M01-240355 | B | 299 | 1,986,559 | 12,720 | 7.93 | 91.00 | 97.46 |
| M04-240196 | B | 287 | 1,978,914 | 12,413 | 7.93 | 91.41 | 97.41 |
| alpha14 | cnl | 298 | 1,922,701 | 11,961 | 7.89 | 87.62 | 97.32 |
| H44/76 | B | 320 | 1,954,116 | 10,919 | 7.82 | 89.84 | 97.16 |
| MC58 | B | 319 | 1,936,480 | 10,854 | 7.82 | 90.14 | 97.14 |
Total number of contigs of >500 bp (i.e., large contigs) produced in the assembly.
Total number bases assembled into large contigs.
Size of the largest contig at which 50% of the total assembled genome length is covered.
Score combining the three metrics defined in footnotes a to c as described in the Materials and Methods.
Percentage of bases from M16917 large contigs aligned against the reference genome using MUMMER.
Average percent nucleotide identity of bases aligned between M16917 large contigs and the reference genome using MUMMER.
Whole-genome profiling.
The Newbler program was used to produce a de novo assembly of M16917, and contigs from this assembly were compared against publicly available N. meningitidis reference complete genome sequences by using the programs BLASTN (1) and MUMMER (18). For the BLAST approach, M16917 contig query sequences were compared against individual genome sequences and the percent identities of the best hits were retained. Distributions of the percent identities from the best hits were used to visually compare the overall similarity levels between M16917 and reference complete genome sequences with the program CGview (9) (Fig. 1B). For the MUMMER approach, M16917 contigs were compared against complete genome references sequences using the DNAdiff application to determine average percent nucleotide identities between M16917 and the reference genomes.
Identification and analysis of the capsule locus and capsule polymerase gene.
M16917 capsule locus sequence-containing contigs were assembled de novo to avoid any biases that may be introduced by reference-based assembly to one serogroup strain or another. De novo assembled contigs were then aligned with the N. meningitidis reference genomes MC58 (serogroup B) and FAM18 (serogroup C) by using the program wgVISTA (6, 21). Contigs that were visually confirmed to align with the capsule locus in either MC58 or FAM18 were retained for subsequent analysis. The identities of specific capsule locus genes in these contigs, including the capsule polymerase gene used in the SGS-PCR assays, were confirmed using BLAST.
The capsule polymerase gene from M16917 was compared against the N. meningitidis reference complete genome sequences using BLASTN to identify orthologous sequences and their levels of similarity (bit scores; Fig. 2A). The M16917 capsule polymerase gene sequence was then aligned with all significantly similar capsule polymerase gene sequences from serogroup B (synD) and serogroup C (synE) complete genome sequences. This alignment was used to build a neighbor-joining tree of the capsule polymerase genes by using the program MEGA (30) (Fig. 2B).
Fig 2.
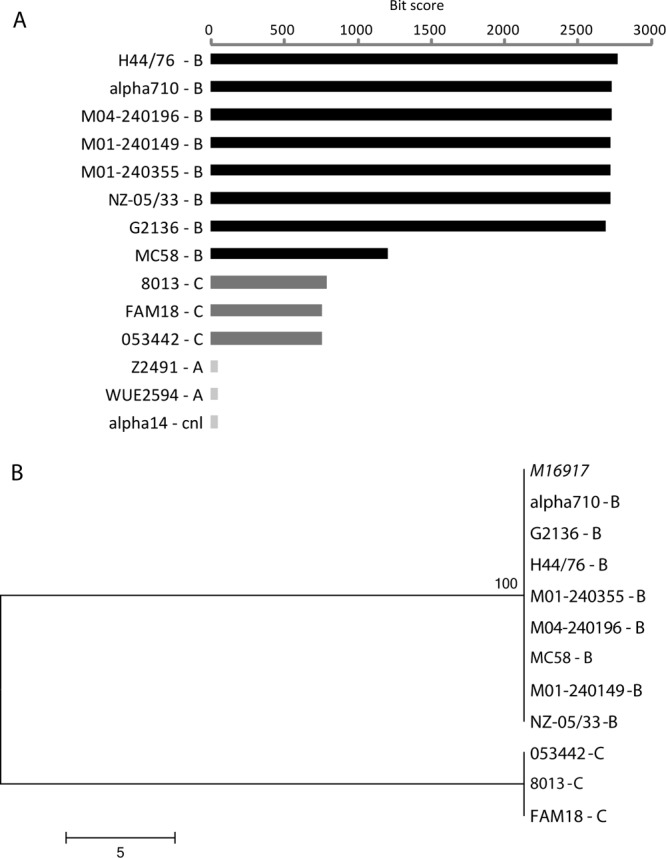
Sequence analysis of the M16917 capsule polymerase gene. (A) BLAST results showing relative sequence similarity levels, expressed as BLAST bit scores for the best hits, between the M16917 capsule polymerase gene and orthologs from N. meningitidis reference genome sequences. The serogroups of the strains are shown. (B) Phylogenetic analysis of the M16917 capsule polymerase gene sequence compared to orthologous genes from serogroup B and C genome sequences.
Identification of recombination breakpoints.
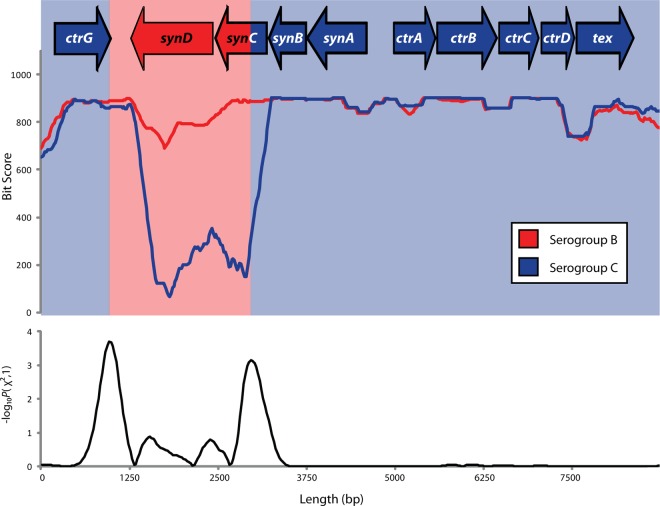
The individual contig sequence covering the capsule locus of M16917 was used in a series of BLASTN searches against reference complete genome sequences of N. meningitidis strains that have been unambiguously defined as either serogroup B or C in order to identify C/B genome sequence recombination breakpoints (Fig. 3). To do this, a sliding-window approach with a window size of 500 bp and a step size of 25 bp was used. For each window along the M16917 capsule locus, the sequence was blasted against each serogroup B or C genome and the bit score for the best hit for each genome was recorded. For each window along the capsule locus, the average bit scores for M16917-serogroup B and M16917-serogroup C comparisons were then plotted and visually compared.
Fig 3.
Recombination breakpoint analysis of the M16917 capsule loci. Average sequence levels of similarity, expressed as BLAST bit scores for overlapping 500-bp windows, between the M16917 capsule sequence and serogroup B (red) or C (blue) genome sequences are plotted along the length of the capsule locus. Statistical significance levels, expressed as −log10 P values, for the recombination breakpoint analysis along the capsule locus are plotted at the bottom. Local maxima in this plot represent the most likely recombination breakpoints, and the serogroup origins of the capsule locus and genes at the top are colored accordingly.
Recombination breakpoint analysis was then performed by using a modified version of the maximum-χ2 method (26) in order to identify the specific positions in the M16917 capsule locus that are most likely to partition adjacent genomic regions with distinct serogroup B or C origins. To do this, the cumulative M16917-serogroup B and M16917-serogroup C average bit scores (BS and CS) in adjacent upstream and downstream genomic regions were compared. Specifically, average M16917-serogroup B and M16917-serogroup C average bit scores were summed across the 10 upstream and 10 downstream windows as follows:
Average bit scores partitioned in this way were then used to populate a two-by-two contingency table as follows:
and to compute its χ2 and P values. The resulting P values were transformed as −log10 P and plotted along the M16917 reference capsule locus sequence. Local −log10 P maxima that partition adjacent regions enriched for serogroup B versus serogroup C positions were then taken as the most likely recombination breakpoints.
RESULTS AND DISCUSSION
M16917 isolation and characterization.
In 2007, the Bacteriology Laboratory at the Illinois Department of Public Health, Chicago, received an isolate of N. meningitidis cultured from the CSF of a patient with meningitis. The Illinois lab performed the SASG assay on the isolate and characterized it as NG polyagglutinating on the basis of agglutination with multiple serogroups, i.e., A, B, C, W-135, X, Y, and Z. The isolate was then sent to the CDC Meningitis Laboratory, which designated it M16917. The CDC lab redid the SASG assay and also characterized the M16917 isolate as NG polyagglutinating on the basis of agglutination with antisera to serogroups B, C, and W-135. The CDC lab also performed the SGS-PCR assay, which yielded a serogroup B result.
We characterized and analyzed the complete genome sequence of M16917 in an effort to better understand the genomic basis of the NG polyagglutination phenotype. The M16917 genome sequence assembly yielded 71 contigs >500 bp in length with a longest contig of 192,528 bp and an N50 value of 53,150 bp. The total length of the genome was found to be 2,158,678 bp, and there were 2,337 predicted protein-coding genes. These results are consistent with a number of previous N. meningitidis genome sequence projects (16, 17).
Origin of the M16917 genome.
The origin of the M16917 genome sequence was evaluated by using two approaches: a genome-based MLST analysis and comparative whole-genome profiling against N. meningitidis reference genomes from different serogroups. Genome-based MLST analysis indicated that M19617 is an ST-11 isolate. ST-11 is most commonly associated with serogroup C in the United States (10); although it has also been found to be associated less often with serogroup B and W-135 strains. ST-based phylogenetic analysis of complete genome sequences shows that strain M16917 is most closely related to serogroup C strains FAM18 and 053442, although it is also closely related to serogroup B strain G2136 (Fig. 1A). Nonetheless, the genome-based MLST result suggests the possibility that the capsule locus has a distinct origin relative to the housekeeping loci since the SGS-PCR assay resulted in a serogroup B result.
MLST is considered to be a reliable typing method in part because it utilizes seven loci spread across the N. meningitidis genome. However, the complete genome sequence should provide additional information with respect to the origin of strain M16917. To do this, contigs from the M16917 de novo assembly were blasted against complete genome sequences representative of four different serogroups. Consistent with the MLST results, the distribution of percent identities for the best hits resulting from this analysis shows that, across the entire genome, M16917 is most similar to the FAM18 ST-11 serogroup C strain and least related to the ST-74 (CC-32) serogroup B lineage (Fig. 1B).
A more in-depth whole-genome profiling of M16917 was performed using a reference-based assembly of M16917 with 14 publicly available complete reference N. meningitidis genome sequences representative of four different serogroups and 14 distinct STs (Table 1). More closely related N. meningitidis genomes will yield M16917 reference assemblies of higher quality. To evaluate the relative quality of the 14 M16917 reference assemblies performed here, a series of assembly metrics were computed for each: the number of large contigs, the assembly size, the N50 size, and an overall assembly score. Taken together, these metrics underscore the close similarity between the M16917 strain and the FAM18 ST-11 serogroup C strain (Table 2 and Fig. 1C and D). However, the next two most closely related genomes according to these scores encode serogroup B capsules. Similar results were obtained by whole-genome profiling of M16917 with the MUMMER comparative-genomics tool, which yields average percent genome sequence identities between M16917 and the 14 reference genomes.
Both the MLST and whole-genome profiling results indicate that strain M16917 is most closely related to the FAM18 serogroup C genome sequence, whereas the SGS-PCR result indicates that the capsule polymerase gene (synD) encodes a serogroup B capsule. This raises the possibility that the serogroup B synD gene, perhaps along with other genes in the capsule locus, was introduced via capsule switching. Furthermore, the close overall relationship between the serogroup B and C genomes seen here, considered together with the fact that N. meningitidis can readily take up and integrate DNA from the environment, should provide ample opportunities for sequence exchange via homologous recombination leading to capsule switching.
Origin of the M16917 capsule polymerase gene.
Whereas the SASG tests performed by the Illinois Department of Public Health and the CDC lab were inconclusive, the SGS-PCR result clearly pointed to a serogroup B-type capsule polymerase gene (synD). This serogroup B result from the SGS-PCR assay could be based on a false-positive PCR or, more intriguingly, could point to a capsule-switching event from serogroup C to B as described above. The genetic basis of capsule switching was first identified by Swartley et al., who characterized an isolate that underwent a switch from serogroup B to C based on a horizontal gene transfer and a subsequent recombination event at the capsule locus (29). This particular transfer and recombination event introduced a serogroup C-type capsule polymerase gene into a serogroup B genomic background. However, the nature of this recombination event was not defined at the genome sequence level. Subsequent studies have also implied probable capsule-switching events based on distinct genetic versus antigenic similarities of different serogroups (4, 10, 31–33).
The SGS-PCR assay is based on serogroup-specific primer sets designed to correspond to the capsule polymerase gene found in the synthesis operon of the capsule locus (22). To evaluate the origins of the M16917 capsule polymerase gene here, we isolated its sequence from the genome assembly and compared it to capsule polymerase genes from the publicly available complete N. meningitidis genome reference sequences (Table 1). To do this, the M16917 capsule polymerase gene was first blasted against the reference genomes and the best hits were recorded. The BLAST analysis shows that all serogroup B genomes encode capsule polymerase genes that are far more similar to M16917 than those of any other serogroup (Fig. 2A). Capsule polymerase genes from serogroup C genomes, while far less similar, do show statistically significant levels of similarity to the M16917 gene; best hits to genomes from other serogroups were not statistically significant. Multiple-sequence alignment and phylogenetic analysis of the homologous capsule polymerase sequences from all serogroup B and C genomes also clearly indicate that the M16917 genome encodes a serogroup B-type capsule polymerase gene (Fig. 2B). These results confirm the results of the SGS-PCR assay, and when considered together with the likely serogroup C origin of the M16917 genomic background, point to a possible C-to-B capsule-switching event. It is not immediately apparent why such a capsule-switching event would produce the particular NG polyagglutination results seen in the SASG assays.
Origin of the M16197 capsule locus.
While a number of different cases of capsule switching have previously been identified, the nature of the recombination events that underlie these switches has not been precisely characterized (2, 29, 31). This is most likely due to the relatively limited amount of sequence information available in previous studies of capsule switching. Given our characterization of the complete genome sequence of M16917, along with the availability of the complete genome sequences of numerous serogroup B and C strains, we reasoned that we would have sufficient data to (i) evaluate whether capsule switching based on recombination had in fact occurred and, if so, (ii) more precisely define the locations of recombination breakpoints in the M16917 sequence. To do this, we performed a series of BLAST searches comparing the M16917 capsule locus sequence against all serogroup B and C genomes by using query sequences from sliding windows of 500 bp along the length of the capsule locus (see Materials and Methods). For each window along the capsule locus, the average similarity of the M16917 sequence to serogroup B and C genome sequences was plotted (Fig. 3). This approach allowed us to visualize whether there are changes in the relative sequence similarity to serogroup B versus serogroup C genomes along the M16917 capsule locus. Indeed, there is a very pronounced change in the relative levels of sequence similarity just upstream of the capsule polymerase gene (synD). M16917 sequences are more similar to serogroup C genomes, on average, through the capsule transport operon and into the first three genes of the capsule synthesis operon. Within the synC gene, there is a marked change in the relative levels of sequence similarity to the two serogroups and the synD gene is clearly more related to serogroup B genomes. These data are clearly consistent with a serogroup C-to-B capsule-switching event.
In addition to this visual evidence, the distribution of average values of M16917 similarity to serogroup B and C was statistically evaluated in an effort to more precisely determine the locations of the recombination breakpoints that introduced the serogroup B-type capsule polymerase gene into the serogroup C-type genomic background (see Materials and Methods). To do this, the average serogroup B and C similarity levels were evaluated in regions immediately upstream and downstream of the individual 500-bp sequence windows used in the BLAST analysis by using a two-by-two χ2 contingency table. In this way, individual sequence windows that are flanked by the most disjointed average levels of similarity to serogroup B versus serogroup C genomes will yield the highest (and accordingly most significant) χ2 values. The locations of such windows are taken to represent the most likely positions of the recombination breakpoints. To visualize this, the P value significance levels of the χ2 values were transformed (−log10 P) and plotted along the length of the capsule locus. Local maxima in this plot, i.e., the most significant individual windows, represent the most likely recombination breakpoints. These local maxima correspond to two positions, one within the synC gene and the other downstream of the synD gene. Thus, the recombination event that led to capsule switching in this case appears to have yielded a chimeric synC gene followed by a true serogroup B synD gene. These results are consistent with capsule switching via a single recombination event, as previously assumed (2, 29, 31). Furthermore, the average size of a recombination event in N. meningitis was previously inferred to be 1.1 kb (15) and the B serogroup-specific sequence cassettes we observed is ∼2 kb in length, slightly larger but still in line with this previous result.
Capsule switching and NG polyagglutination.
Capsule switching has previously been implicated in the NG polyagglutination phenotype via changes to the capsule polymerase gene. For example, an isolate with a serogroup Y-type N. meningitidis genome was found to be NG polyagglutinating by virtue of cross-reactivity with both serogroup Y and W-135 antisera. This cross-reactivity was shown to be due to the production of both serogroup Y- and W-135-specific capsular sugars by a mutated capsule polymerase containing both Y- and W-135 characteristic amino acid residues. However, this does not appear to be the case here and it remains unclear why a C-to-B capsule-switching event would produce an NG polyagglutination phenotype with cross-reactivity to serogroup W-135 or other serogroups. It should be noted that the M16917 synD gene does not have any sequence similarity to the W-135 capsule polymerase.
The serogroup B SynD polymerase encoded by M16917 creates α2→8 linkages between sialic acid monomers to form the B capsule polysaccharide, whereas the C-type capsule polymerase gene encodes SynE, which creates a capsule with α2→9 sialic acid linkages (8, 28, 29). Steenbergen and Vimr experimentally demonstrated that the linkage specificity of identically composed Escherichia coli polysaccharide capsules could be changed from α2→8 to α2→9 by exchanging discrete regions of the polymerases that encode these specific linkages (27). Accordingly, the change between B- and C-type capsule polymerase-encoding activities may be subtle and readily achieved. Indeed, previously reported capsule-switching events involving these two serogroups were B to C (29), whereas the event reported here is C to B. Thus, interchange between these two serogroup phenotypes may involve a back-and-forth exchange that is even more dynamic and facile than previously imagined. The specific nature of the capsule-switching event uncovered here may be particularly relevant since there is no currently available vaccine to serogroup B strains, and thus, M16917 would represent a vaccine escape mutant.
ACKNOWLEDGMENTS
We thank the Illinois Department of Public Health, Chicago, Bacteriology Laboratory for donating isolate M16917 (also known as N. meningitidis 703526). This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford. We thank Nancy Messonnier for reading and providing valuable feedback on the manuscript.
This work was supported by the Alfred P. Sloan Research Fellowship in Computational and Evolutionary Molecular Biology (BR-4839 to I.K.J.), the Georgia Research Alliance (GRA.VAC09.O to I.K.J., B.H.H., J.D.T., L.W.M., and L.S.K.), and the Bioinformatics Program, Georgia Institute of Technology (to N.V.S.).
Footnotes
Published ahead of print 17 August 2012
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beddek AJ, Li MS, Kroll JS, Jordan TW, Martin DR. 2009. Evidence for capsule switching between carried and disease-causing Neisseria meningitidis strains. Infect. Immun. 77: 2989–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bilukha OO, Rosenstein N. 2005. Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 54(RR-7): 1–21 [PubMed] [Google Scholar]
- 4. Claus H, Stummeyer K, Batzilla J, Mühlenhoff M, Vogel U. 2009. Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol. Microbiol. 71: 960–971 [DOI] [PubMed] [Google Scholar]
- 5. Dolan-Livengood JM, Miller YK, Martin LE, Urwin R, Stephens DS. 2003. Genetic basis for nongroupable Neisseria meningitidis. J. Infect. Dis. 187: 1616–1628 [DOI] [PubMed] [Google Scholar]
- 6. Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frosch M, Vogel U. 2006. Structure and genetics of the meningococcal capsule, p 145–162 In Frosch M, Maiden MCJ. (ed), Handbook of meningococcal disease: infection biology, vaccination, clinical management. Wiley-VCH Verlag GmbH & Co., Weinheim, Germany [Google Scholar]
- 8. Frosch M, Weisgerber C, Meyer TF. 1989. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 86: 1669–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36: W181–W184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harrison LH, et al. 2010. Population structure and capsular switching of invasive Neisseria meningitidis isolates in the pre-meningococcal conjugate vaccine era—United States, 2000-2005. J. Infect. Dis. 201: 1208–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harrison LH, Trotter CL, Ramsay ME. 2009. Global epidemiology of meningococcal disease. Vaccine 27(Suppl 2): B51–B63 [DOI] [PubMed] [Google Scholar]
- 12. Holmes EC, Urwin R, Maiden MC. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16: 741–749 [DOI] [PubMed] [Google Scholar]
- 13. Jolley KA, Chan MS, Maiden MC. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5: 86 doi:10.1186/1471-2105-5-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolley KA, Maiden MC. 2010. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595 doi:10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC. 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22: 562–569 [DOI] [PubMed] [Google Scholar]
- 16. Katz LS, et al. 2011. Using single-nucleotide polymorphisms to discriminate disease-associated from carried genomes of Neisseria meningitidis. J. Bacteriol. 193: 3633–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kislyuk AO, et al. 2010. A computational genomics pipeline for prokaryotic sequencing projects. Bioinformatics 26: 1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5: R12 doi:10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maiden MC, et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95: 3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mayor C, et al. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047 [DOI] [PubMed] [Google Scholar]
- 22. Mothershed EA, et al. 2004. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J. Clin. Microbiol. 42: 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Popovic T, Ajello GW. 2003. Neisseria meningitidis: confirmatory identification and antimicrobial susceptibility testing. World Health Organization, Geneva, Switzerland [Google Scholar]
- 24. Popovic T, Ajello GW, Facklam R. 1999. Laboratory manual for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae. World Health Organization, Geneva, Switzerland [Google Scholar]
- 25. Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N. Engl. J. Med. 344: 1378–1388 [DOI] [PubMed] [Google Scholar]
- 26. Smith JM. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34: 126–129 [DOI] [PubMed] [Google Scholar]
- 27. Steenbergen SM, Vimr ER. 2003. Functional relationships of the sialyltransferases involved in expression of the polysialic acid capsules of Escherichia coli K1 and K92 and Neisseria meningitidis groups B or C. J. Biol. Chem. 278: 15349–15359 [DOI] [PubMed] [Google Scholar]
- 28. Swartley JS, et al. 1998. Characterization of the gene cassette required for biosynthesis of the (alpha1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J. Bacteriol. 180: 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Swartley JS, et al. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 94: 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 31. Tsang RS, et al. 2005. Potential capsule switching from serogroup Y to B: the characterization of three such Neisseria meningitidis isolates causing invasive meningococcal disease in Canada. Can. J. Infect. Dis. Med. Microbiol. 16: 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vogel U, Claus H, Frosch M. 2000. Rapid serogroup switching in Neisseria meningitidis. N. Engl. J. Med. 342: 219–220 [DOI] [PubMed] [Google Scholar]
- 33. Wang Q, et al. 2010. Genetic study of capsular switching between Neisseria meningitidis sequence type 7 serogroup A and C strains. Infect. Immun. 78: 3883–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weber MV, Claus H, Maiden MC, Frosch M, Vogel U. 2006. Genetic mechanisms for loss of encapsulation in polysialyltransferase-gene-positive meningococci isolated from healthy carriers. Int. J. Med. Microbiol. 296: 475–484 [DOI] [PubMed] [Google Scholar]
- 35. Yazdankhah SP, et al. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42: 5146–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]