Abstract
Integration of viral DNA into cellular DNA is an essential step in the replication cycle of HIV and other retroviruses. The first antiviral drugs that target integrase, the viral enzyme that catalyzes DNA integration, have recently been approved and more are in the pipeline. These drugs bind to an intermediate in DNA integration called the intasome, in which a pair of viral DNA ends are synapsed by a tetramer of integrase, rather than free integrase enzyme. We discuss the biochemical mechanism of integration, which is now quite well understood, and recent progress towards obtaining atomic-resolution structures of HIV intasomes in complex with inhibitors. Such structures are ultimately required to understand the detailed mechanism of inhibition and the mechanisms by which mutations in integrase confer resistance. The path from early biochemical studies to therapeutic inhibitors of integrase highlights the value of basic science in fighting human diseases.
Keywords: HIV-1, intasome, integrase, integration, retrovirus
Retroviruses integrate a DNA copy of the viral genome into host DNA as an obligatory step in the replication cycle [1, 2]. Integration can occur at essentially any location in the genome, but certain regions of chromatin are preferred [3]. The integrated viral DNA is stably maintained and replicated along with cellular DNA through cycles of cell division. This presents a challenge to the treatment of retroviral infections. Although great strides have been made in antiviral therapy for HIV, the integrated virus persists in long-lived cells and eradication is an elusive goal [4]. The key enzyme that integrates retroviral DNA into the host genome is the virally encoded integrase protein. The first clues that a viral protein mediates DNA integration came from genetic studies [5]. Mutations were identified in the viral pol gene that allowed viral DNA to be synthesized at normal levels by reverse transcription, but this DNA failed to integrate. This part of the pol gene encodes the protein we now call integrase that is cleaved from a polyprotein precursor by the viral protease. The first antiviral drugs that target integrase have been recently developed [6], and these drugs complement the established protease and reverse transcriptase inhibitors.
Retroviral DNA is synthesized by reverse transcription within the cytoplasm of the infected cell. This DNA forms part of a large nucleoprotein complex, termed the preintegration complex (PIC), that is derived from the core of the infecting virion [7]. The PIC is transported to the nucleus and the viral DNA is integrated into cellular DNA by integrase. PICs are poorly defined because their low abundance in extracts of infected cells prevents direct analysis by biophysical methods. HIV PICs have been reported to contain the viral proteins integrase, nucleocapsid, matrix, reverse transcriptase and Vpr [8–15], in addition to a number of cellular proteins. PICs efficiently integrate their viral DNA into a target DNA in vitro with all the hallmarks of integration in vivo.
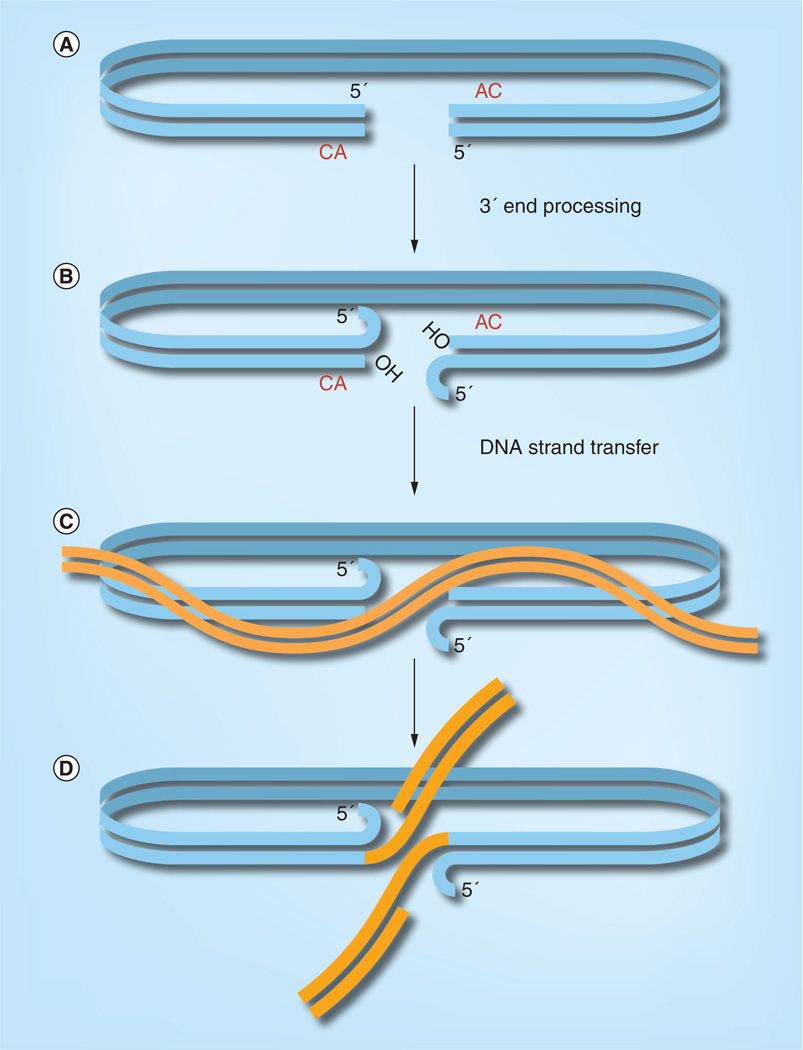
PICs derived from virus-infected cells efficiently integrate their viral DNA into a target DNA in vitro [11, 16, 17]. The DNA cutting and joining steps of retroviral DNA integration (Figure 1) were determined by analysis of the integration intermediate formed in vitro with PICs as the source of viral DNA and plasmid DNA as the target for integration [18, 19]. In the first step, 3′ end processing, two nucleotides are cleaved from each 3′ end of the initially bluntended DNA. This exposes the CAOH 3′ ends that mark the junction between viral and target DNA upon integration. In the next step, DNA strand transfer, the hydroxyl groups at the 3′ ends of the viral DNA attack a pair of phosphodiester bonds in the target DNA. The sites of attack are separated by five nucleotides on each DNA strand in the case of HIV. The result is an integration intermediate in which the 3′ ends of the viral DNA are covalently attached to target DNA. The 3′ ends of the target DNA and the 5′ ends of the viral DNA, two nucleotides of which are unpaired, are not joined. Cellular enzymes are required to complete the integration process by repairing the integration intermediate. The required steps are removal of the two unpaired nucleotides at 5′ end of the viral DNA, filling in of the single strand gaps, and ligation.
Figure 1. DNA cutting and joining steps of retroviral DNA integration.
(A) The viral DNA synthesized by reverse transcription is initially blunt ended. (B) The 3′ end processing reaction removes two nucleotides from each 3′ end. (C) Next, in the DNA strand transfer reaction, the 3′ hydroxyls at the ends of the viral DNA attack a pair of phosphodiester bonds in the target DNA; in the case of HIV, the sites of attack are separated by five nucleotides on the two target DNA strands. (D) The result is the integration intermediate, in which the 3′ ends of the viral DNA are joined to the 5′ ends of the target DNA at the site of integration. The integration intermediate is then repaired by cellular enzymes to complete the integration process.
Biochemistry of DNA integration
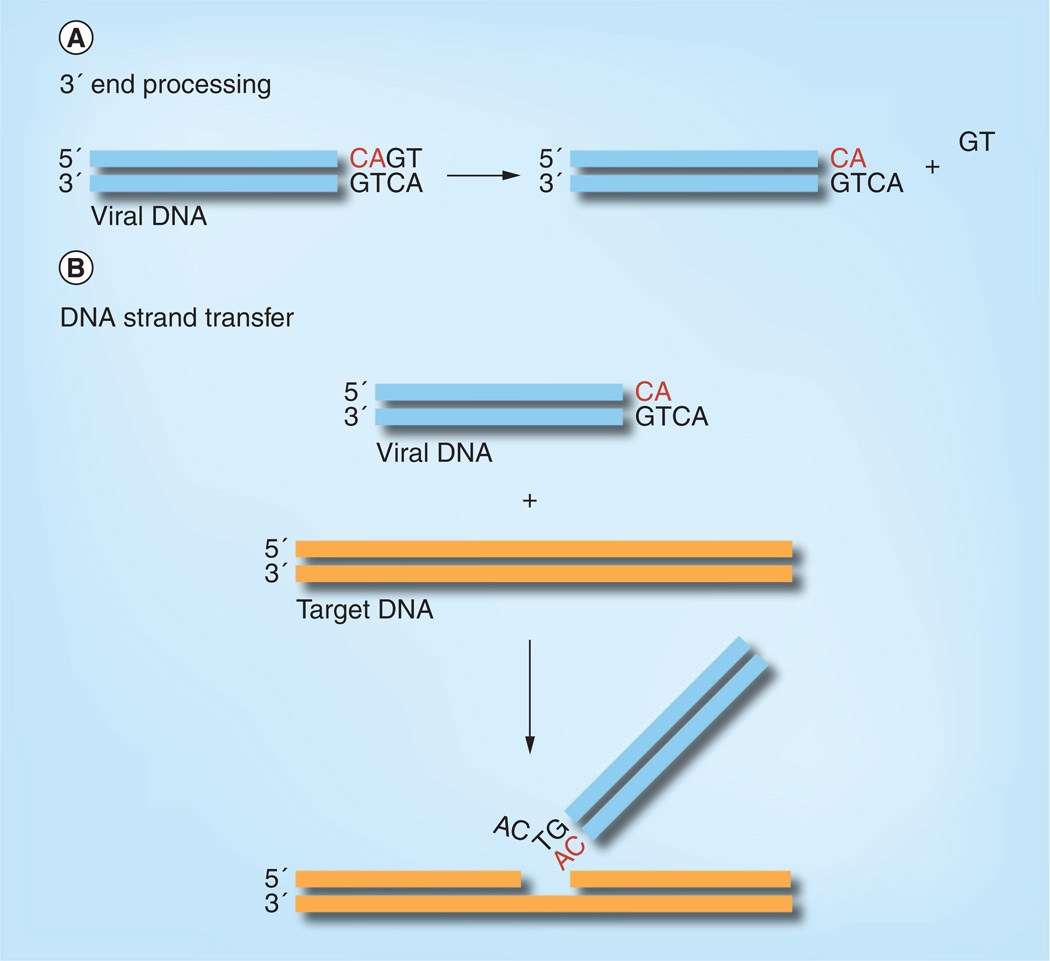
Retroviral integrases share a common biochemical mechanism of integration. Although we focus on the HIV enzyme, important advances have come from parallel studies of HIV and closely related retroviruses, notably avian sarcoma and leukosis virus, Moloney murine leukemia virus and, more recently, prototype foamy virus (PFV). Purified retroviral integrases, in the presence of a divalent metal ion, catalyze both 3′ end processing and DNA strand transfer in vitro with oligonucleotide DNA substrates that mimic the ends of the viral DNA (Figure 2) [20–22]. This simplified reaction system revealed that the chemical mechanism of DNA integration is one-step transesterification [23]; when chiral phosphorothioate is substituted for phosphate in the target DNA, the chirality is inverted in the integration product. This contrasts with DNA recombination enzymes that form a covalent intermediate between the enzyme and DNA.
Figure 2. In vitro catalytic activities of HIV integrase.
(A) Integrase catalyzes 3′ end processing on an oligonucleotide DNA substrate mimicking the ends of HIV DNA. The GT dinucleotide is released, exposing the conserved CA that is to be joined to the target DNA. (B) Integrase also catalyzes DNA strand transfer, in which the 3′ end of the viral DNA attacks a phosphodiester bond in the target DNA, covalently joining the viral to target DNA. Integration can occur at essentially any location in the target DNA. Note that, in vitro with oligonucleotide DNA substrates, most of the reaction products result from joining only one viral DNA end into one strand of target DNA, rather than concerted integration of a pair of viral DNA ends as occurs in vivo.
Mutagenesis studies of several retroviral integrases showed that mutations generally affected 3′ end processing and DNA strand transfer in parallel [24–27], suggesting a common active site for these two reactions, a hypothesis that was later confirmed by structural studies. Although these two reactions at first sight appear to be quite different, they share the same chemical mechanism. In the 3′ end processing reaction the nucleophile is water, whereas in the DNA strand transfer reaction it is the 3′ hydroxyl at the ends of the viral DNA [23].
Although integrase efficiently catalyzes the chemical reactions of integration in vitro with oligonucleotide substrates, there are limitations to the most simplified systems. Firstly, the reaction takes place in an aggregated state as evidenced by pelleting of DNA substrate, product and protein with low-speed centrifugation [28]. This is often unappreciated and there are many conclusions in the literature based on kinetic analyses that are not readily applicable to the system because unreacted substrates and reaction products form mixed heterogeneous aggregates. Secondly, the vast majority of products result from the integration of only a single viral DNA end into one strand of target DNA, rather than the concerted integration of a pair of viral DNA ends [29] (compare the concerted integration of a pair of viral DNA ends shown in Figure 1 with the insertion of a single viral DNA end shown in Figure 2B). The reaction system resembles a ‘halfsite’ reaction that carries out correct chemistry, but lacks the full fidelity of integration catalyzed by PICs in vitro and in vivo.
Recently, improved in vitro reaction systems have been developed that catalyze concerted integration of a pair of viral DNA ends [30–34]. Under these reaction conditions, specific complexes are formed between integrase and DNA substrate that mimic the tight association of integrase with viral DNA in the PIC [35–37]. A tetramer of integrase stably bridges a pair of viral DNA ends to form a stable synaptic complex or intasome. Once formed, intasomes catalyze concerted integration with an efficiency approaching 100%. 3′ end processing occurs within the intasome, and strand transfer of the two viral DNAs into the target occurs sequentially, with a significant temporal separation between the joining of the two viral DNA ends to the target DNA. The resulting integration intermediate remains tightly associated with integrase in a complex termed the strand transfer complex (STC). It is likely that cellular enzymes are required to actively disassemble the STC before it can be repaired to complete integration.
Inhibitors of HIV integrase
The in vitro reaction system with oligonucleotide DNA substrates laid the foundation for large-scale screening for inhibitors of integrase within the pharmaceutical industry. Progress was painstakingly slow and the first integrase inhibitor, raltegravir, was only approved by the US FDA in 2007 [6]. Several other integrase inhibitors are currently in late-stage clinical trials [38]. These inhibitors share the property of having low affinity for integrase alone, but high affinity for intasomes in which the active sites are engaged with a pair of viral DNA ends [39]. The implication is that DNA binding elicits a protein conformational change central to high-affinity drug binding and/or the DNA itself makes direct contacts with the inhibitor. By extension, knowledge of the structures of intasomes is required to understand the molecular mechanism of action, and structures of the protein alone are unlikely to be informative.
In addition to inhibitors of integrase that target the active site, compounds have been identified that inhibit integration in vitro by an entirely different mechanism. One promising class of compounds binds to the LEDGF/p75 (discussed below) cofactor binding site of integrase [40–42].
Structure of retroviral integrase nucleoprotein intermediates in DNA integration
Structural studies of most retroviral integrases have been frustrated by the poor solubility of the protein and the flexibility between domains. With the exception of PFV [43, 44], no structures of a full-length retroviral integrase have been determined. The structures of the catalytic domain of HIV and ASV integrase were determined almost simultaneously [45, 46]. They were essentially the same and confirmed the identity of the catalytic residues postulated based on mutagenesis studies. Otherwise, the structures were largely uninformative in retrospect. They were nearly spherical dimers with a pair of active sites diametrically opposed away from the dimer interface. The spacing is incompatible with the 5-base pairs staggered integration sites on a target DNA and suggested that a higher order multimer other than a dimer is required for the authentic integration reaction. The N-terminal domain is a helix bundle, stabilized by the coordination of a zinc ion to a pair of conserved His and Cys residues [47, 48]. Finally, the isolated C-terminal domain closely resembles an SH3 domain [49, 50]. Two-domain structures comprising the N-terminal plus catalytic domain and C-terminal plus catalytic domain have also been solved for several retroviral integrases (reviewed in [51, 52]). Strikingly, the relative positions of the domains are quite different among these structures, highlighting the flexibility of the linkers.
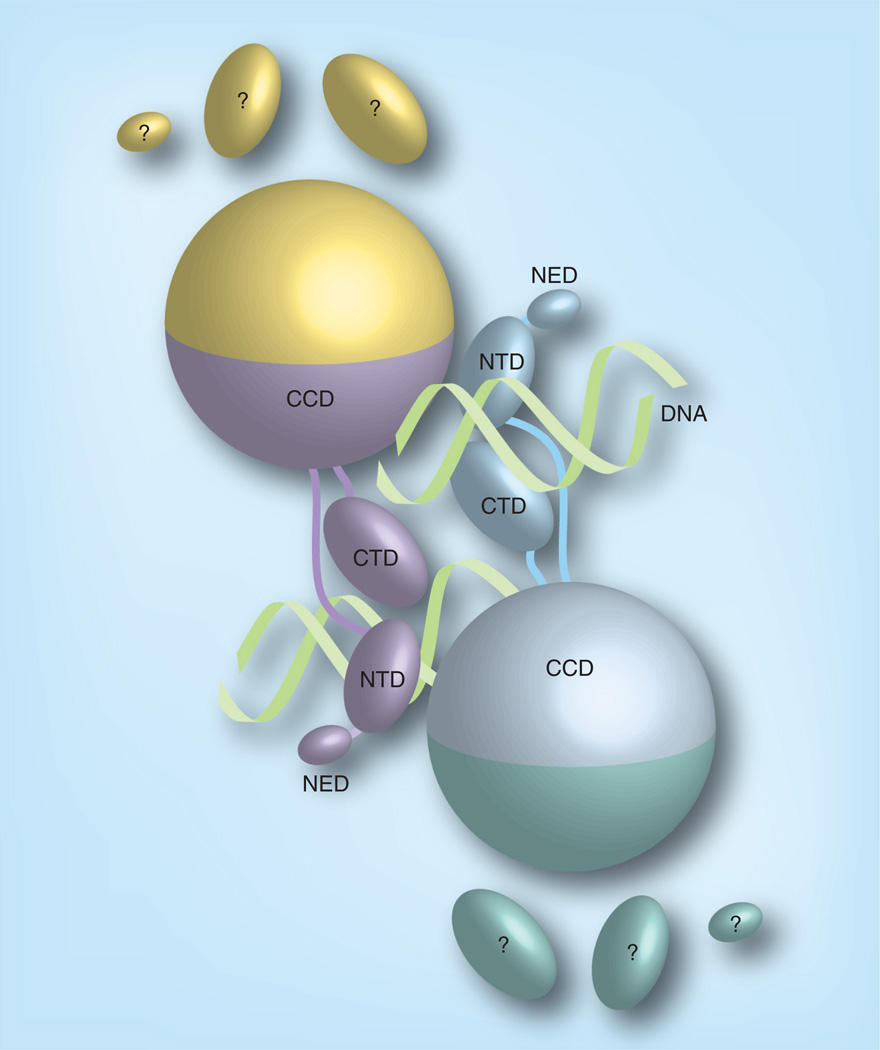
A major breakthrough in structural studies of retroviral integrase came when Cherepanov and colleagues solved the structure of the PFV intasome [43]. PFV integrase shares only limited sequence identity with HIV integrase and has an additional N-terminal domain and longer linkers between domains. Nevertheless it would be surprising if the PFV and HIV intasomes were radically different, given the functional and structural similarity of these proteins. PFV integrase is more amenable to biophysical studies than HIV integrase. First, it is more soluble, and second, unlike HIV integrase, it readily assembles intasomes on short oligonucleotide DNA substrates [53]. DNA is the glue that holds the PFV intasome together in a way that is reminiscent of the Tn5 transposase in complex with transposons [54]. It is therefore perhaps not surprising that all the models of the HIV intasome structure based on the partial structures of the protein alone were proved wrong. The organization of the PFV intasome is depicted in Figure 3. The catalytic domain dimer interface, present in all previous structures, is preserved. The integrase tetramer is a dimer of dimers, and each dimer contains an inner and outer subunit. All the DNA contacts are with the inner subunits. The N-terminal domain, C-terminal domain and the extra N-terminal extension domain of the outer PFV integrase subunits are disordered in the current structures. Structures of the PFV intasome in complex with the inhibitor raltegravir elucidate the basic inhibition mechanism. The inhibitor indeed intimately contacts the viral DNA ends, displacing the 3′ ends from the active site and making them unavailable for nucleophilic attack on the target DNA. The HIV intasome has been modeled based on the PFV structure and it is predicted to bind inhibitors in the same way [55]. The high degree of similarity in the immediate vicinity of the active site allows reliable modeling, but the proteins are too dissimilar to accurately model detailed molecular interactions further away. Structures of the HIV intasome will be required to fully understand how mutations confer resistance to drugs, because some are in residues not predicted to directly interact with the inhibitor [56]. The structure of the PFV STC has also been determined [44]. The target DNA is severely bent, explaining the preference for integration into distorted DNA [57].
Figure 3. Cartoon representation of the prototype foamy virus intasome structure.
The integrase monomers in the intasome are distinguished by their colors. The CCD, NTD, CTD and NED are distinguished by shape. The pair of viral DNA ends are represented as helices. All the contacts between integrase and viral DNA are with the inner subunits. The CTD, NTD and NED of the outer subunits are disordered.
CCD: Catalytic core domain; CTD: C-terminal domain; NED: N-terminal extension domain; NTD: N-terminal domain.
Adapted with permission from [81].
Interaction of integrase with other proteins
Although integrase is both necessary and sufficient for integration in vitromany proteins have been reported to interact with integrase, and at least some of them are functionally relevant to the integration process. Roles that have been proposed include directing the PIC to particular regions of chromatin, import of the PIC across the nuclear envelope, and blocking integration of the viral DNA into itself (autointegration). A complete list of cellular factors is outside the scope of this review, but the following are a few examples of the role of cellular factors in integration. The topic has been reviewed in depth in [57–60].
The most studied cellular factor is LEDGF/p75 (reviewed in [61, 62]). This protein was first identified as an interacting partner in affinity screens [63, 64]. Early siRNA studies were inconclusive regarding the functional relevance of the interaction, probably because trace amounts are sufficient to support HIV replication. Later experiments with more complete knockdown and knockout cell lines clearly demonstrated that LEDGF stimulates HIV replication up to approximately 100-fold in cell culture [65–67]. Strikingly, PICs isolated from LEDGF/p75-knockout cells are fully competent for integration in vitro [66], demonstrating that LEDGF/p75 does not play a role in PIC assembly or intrinsic activity. How, then, does LEDGF/p75 exert its activity on integration in cells? A convincing body of evidence indicates that LEDGF/p75 tethers PICs to chromatin prior to catalysis of integration. HIV preferentially integrates into active transcription units [68]. This correlates well with the distribution of LEDGF/p75 in chromatin, where ‘LEDGF islands’ preferentially lie in transcription units [69]. Biochemical and structural studies indicate that LEDGF/p75 can serve as a bridge between PICs and chromatin. LEDGF/p75 contains an integrasebinding domain that binds tightly to integrase [70, 71]. The structure of the catalytic domain of HIV integrase complexed with the integrase-binding domain has been determined by X-ray crystallography [72] and small molecules that mimic the LEDGF/p75 integrase interaction have defined a novel class of allosteric integrase inhibitors [42, 73]. LEDGF/p75 also contains a PWWP chromatin-binding domain and an A/T hook domain that is likely involved in DNA binding [74, 75].
Treatment of Moloney murine leukemia virus PICs with high ionic strength, followed by size exclusion chromatography to remove small liberated factors, abolishes intermolecular integration in vitro [76]. Instead, the viral DNA uses itself as a target in a self-destructive reaction termed autointegration. Intermolecular integration is restored and autointegration is abolished by addition of an extract of uninfected cells. Using this as an assay, the protein factor BAF (or BANF1) was identified [77]. It is a highly conserved DNA bridging protein that compacts DNA [78]. It was hypothesized that BAF blocks autointegration by compacting the viral DNA, making it inaccessible as a target for integration. Such compaction of DNA by BAF was later confirmed by total internal reflection fluorescence microscopy experiments [79]. It is unclear if BAF also performs a similar function for HIV PICs. Because BAF is an essential protein, it is difficult, if not impossible, to directly test the effect of knockdown or knockout in vivo because of the indirect effects of perturbing normal cellular processes. This is a conundrum that has also hindered the investigation of the role of other essential cellular proteins in DNA integration.
Ongoing work
The biochemical mechanism of DNA integration is quite well understood, but atomic-level structural details are incomplete. The structure of the HIV intasome remains elusive. The success with the PFV intasome structure is a giant step forward, but the protein is sufficiently different that the HIV intasome structure is required to properly understand the mechanism of drug resistance. Six of the domains are disordered in the PFV intasome structure. Are these domains required for function or are they dispensable? Additional structures of retroviral intasomes and biochemical studies are required to answer this question.
A major focus of ongoing research is on the role of interacting partners of HIV integrase. PICs are much too large to passively enter the nucleus, and so an active process must be involved. This field has been controversial and is not yet settled. Numerous claims have been made, some involving proteins that interact with integrase and some not. There is likely to be redundancy in the nuclear import pathways [80] and this may in part explain the apparent contradictions in the literature. The role of cellular proteins in targeting PICs to chromatin is generally accepted. The dual interaction of LEDGF/p75 with HIV integrase and chromatin is clearly a major factor in the case of HIV DNA integration. Are other proteins involved in targeting integration of HIV, and do other factors substitute for LEDGF/p75 for retroviruses with integrases that do not bind LEDGF? There is still much to learn.
Future perspective
The groundwork for studies on HIV integrase was laid with biochemical studies of closely related DNA transposition systems. It was not clear at the time that such basic science would contribute to the treatment of an emerging disease. The early biochemical studies of HIV integrase resulted in assays that were adapted by the pharmaceutical industry for high-throughput screening for integrase inhibitors. Almost two decades after initiating these efforts, the first antiviral drug that targets integrase was brought to market and more are in the pipeline. The story highlights the value of basic research to tackle future emerging diseases. It is now necessary to study the atomic details of how these drugs work and the mechanisms of drug resistance.
Executive summary.
DNA integration
-
▪
DNA integration is essential for the replication of HIV and other retroviruses and can be a target for antiretroviral drugs.
Integrase
-
▪
The viral enzyme integrase catalyzes the key DNA cutting and joining steps of integration.
Biochemistry
-
▪
A tetramer of integrase stably bridges the ends of the viral DNA in a nucleoprotein complex called the intasome. The chemical steps of integration take place within intasomes.
-
▪
Integrase cleaves two nucleotides from the 3′ ends of the viral DNA (3′ end processing).
-
▪
Integrase catalyzes nucleophilic attack of the 3′ hydroxyl group at the ends of the processed DNA on a pair of phosphodiester bonds in the target DNA (DNA strand transfer).
-
▪
Cellular enzymes complete integration by repairing the resulting integration intermediate.
Inhibitors
-
▪
The first integrase inhibitors were recently approved by the US FDA.
-
▪
These inhibitors have high affinity for intasomes, but low affinity for free integrase.
-
▪
The mechanism involves displacement of the 3′ ends of the viral DNA away from the active site, making them unavailable for nucleophilic attack on the target DNA.
Structure
-
▪
Knowledge of the structures of HIV intasomes is needed to understand the detailed mechanism of inhibitors and mutations that confer resistance.
-
▪
Only partial structures of HIV integrase without DNA have so far been determined.
-
▪
Prototype foamy virus is currently the only retrovirus for which intasome structures are available.
Future perspective
-
▪We need to:
-
–Determine high-resolution structures of HIV intasomes in complex with inhibitors.
-
–Apply these structures to understand the mechanism of inhibition at the atomic level and how mutations in integrase confer resistance.
-
–Develop new classes of integrase inhibitors.
-
–
Acknowledgements
The author thanks A Engelman and W Yang for reading the manuscript.
This work was supported by the Intramural Program of NIDDK, NIH and by the AIDS Targeted Antiviral Program of the Office of the Director of the NIH.
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Varmus H, Brown PO. Retroviruses. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, DC, USA: American Society of Microbiology; 1989. pp. 53–108. [Google Scholar]
- 2.Coffin JM, Hughes SH, Varmus HE. Retroviruses. NY, USA: Cold Spring Harbor Laboratory Press; 1977. [PubMed] [Google Scholar]
- 3.Ciuffi A. Mechanisms governing lentivirus integration site selection. Curr. Gene Ther. 2008;8(6):419–429. doi: 10.2174/156652308786848021. [DOI] [PubMed] [Google Scholar]
- 4.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 5.Goff SP. Genetics of retroviral integration. Ann. Rev. Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 6.Summa V, Petrocchi A, Bonelli F, et al. Discovery of Raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 2008;51(18):5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 7.Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 8.Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Intracytoplasmic maturation of the human immunodeficiency virus type I reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology. 2006;3:4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl Acad. Sci. USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71(7):5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnet CM, Haseltine WA. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnet CM, Bushman FD. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 13.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res. Hum. Retrovir. 1993;9(9):817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 15.Gallay P, Swingler S, Song JP, Bushman F, Trono D. HIV nuclear import Is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83(4):569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 16.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 17.Ellison V, Abrams H, Roe T, Lifson J, Brown P. Human immunodeficiency virus integration in a cell-free system. J. Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 19.Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl Acad. Sci. USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 22.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 23.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 24.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Gent DC, Groeneger AA, Plasterk RH. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl Acad. Sci. USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leavitt AD, Shiue L, Varmus HE. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 28.van Gent DC, Elgersma Y, Bolk MW, Vink C, Plasterk RH. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bushman FD, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl Acad. Sci. USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aiyar A, Hindmarsh P, Skalka AM, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: Dependence on both long terminal repeat termini. J. Virol. 1996;70(6):3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J. Virol. 2002;76(7):3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Craigie R. Processing the viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 2005;79(13):8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey KK, Bera S, Grandgenett DP. The HIV-1 Integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry. 2011;50(45):9788–9796. doi: 10.1021/bi201247f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25(6):1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M, Ivanov V, Mizuuchi M, Mizuuchi K, Craigie R. DNA requirements for assembly and stability of HIV-1 intasomes. Protein Sci. 2012;21:249–257. doi: 10.1002/pro.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J. Mol. Biol. 2010;399(3):491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beare KD, Coster MJ, Rutledge PJ. Diketoacid inhibitors of HIV-1 integrase: From L-708,906 to Raltegravir and beyond. Curr. Med. Chem. 2012;19(8):1177–1192. doi: 10.2174/092986712799320565. [DOI] [PubMed] [Google Scholar]
- 39.Espeseth AS, Felock P, Wolfe A, et al. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl Acad. Sci. USA. 2000;97(21):11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Luca L, Ferro S, Gitto R, et al. Small molecules targeting the interaction between HIV-1 integrase and LEDGF/p75 cofactor. Bioorg. Med. Chem. 2010;18(21):7515–7521. doi: 10.1016/j.bmc.2010.08.051. [DOI] [PubMed] [Google Scholar]
- 41.De Luca L, Ferro S, Morreale F, De Grazia S, Chimirri A. Inhibitors of the interactions between HIV-1 IN and the cofactor LEDGF/p75. Chem. Med. Chem. 2011;6(7):1184–1191. doi: 10.1002/cmdc.201100071. [DOI] [PubMed] [Google Scholar]
- 42.Kessl J, Jena N, Koh Y, et al. A multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 2012;287(20):16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468(7321):326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 46.Bujacz G, Jaskolski M, Alexandratos J, et al. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 47.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 48.Eijkelenboom AP, van den Ent FM, Vos A, et al. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr. Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 49.Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 50.Lodi PJ, Ernst JA, Kuszewski J, et al. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 51.Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 2004;4(9):965–977. doi: 10.2174/1568026043388547. [DOI] [PubMed] [Google Scholar]
- 52.Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009;276(11):2926–2946. doi: 10.1111/j.1742-4658.2009.07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valkov E, Gupta SS, Hare S, et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37(1):243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 55.Krishnan L, Li XA, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl Acad. Sci. USA. 2010;107(36):15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl Acad. Sci. USA. 2010;107(46):20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bushman F, Lewinski M, Ciuffi A, et al. Genome wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3(11):848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 58.Greene WC, Peterlin BM. Charting HIV’s remarkable voyage through the cell: basic science as a passport to future therapy. Nat. Med. 2002;8(7):673–680. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki Y, Craigie R. The road to chromatin – nuclear entry of retroviruses. Nat. Rev. Microbiol. 2007;5(3):187–196. doi: 10.1038/nrmicro1579. [DOI] [PubMed] [Google Scholar]
- 60.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 2006;31(2):98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 61.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4(3):e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell. Mol. Life Sci. 2008;65(9):1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cherepanov P, Maertens G, Proost P, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278(1):372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 64.Turlure F, Devroe E, Silver PA, Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 65.Llano M, Saenz DT, Meehan A, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 66.Shun MC, Raghavendra NK, Vandegraaff N, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21(14):1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandekerckhove L, Christ F, Van Maele B, et al. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 2006;80(4):1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schroder ARW, Shinn P, Chen HM, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 69.De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 2010;38(18):6135–6147. doi: 10.1093/nar/gkq410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell. Sci. 2005;118(8):1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 71.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279(47):48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 72.Cherepanov P, Ambrosio ALB, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl Acad. Sci. USA. 2005;102(48):17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Christ F, Voet A, Marchand A, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 2010;6(6):442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 74.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360(4):760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 75.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34(5):1653–1665. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee MS, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc. Natl Acad. Sci. USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl Acad. Sci. USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng RL, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl Acad. Sci. USA. 2000;97(16):8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skoko D, Li M, Huang Y, et al. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc. Natl Acad. Sci. USA. 2009;106(39):16610–16615. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee K, Ambrose Z, Martin TD, et al. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7(3):221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin Z, Craigie R. Modeling the HIV-1 intasome: a prototype view of the target of integrase inhibitors. Viruses. 2010;2(12):2777–2781. doi: 10.3390/v2122777. [DOI] [PMC free article] [PubMed] [Google Scholar]