Abstract
Ionizing radiation produces clustered lesions in DNA. Since the orientation of bi-stranded lesions affects their recognition by DNA repair enzymes, clustered damages are more difficult to process, and thus more toxic, than single oxidative lesions. In order to understand the structural determinants that leads to differential recognition, we use NMR spectroscopy and restrained molecular dynamics to solve the structure of two DNA duplexes, each containing two stable abasic site analogs positioned on opposite strands of the duplex and staggered in the 3’ (−1 duplex, (AP)2−1 duplex) or 5’ (+1 duplex, (AP)2+1 duplex) direction. Cross-peak connectivities observed in the non-exchangeable NOESY spectra indicate compression of the helix at the lesion site of the duplexes, resulting in the formation of two abasic bulges. The exchangeable proton spectra show the AP site partner nucleotides forming inter-strand hydrogen bonds that are characteristic of a Watson-Crick G•C base pairs, confirming the extra helical nature of the AP residues. Restrained molecular dynamics simulations generate a set of converging structures in full agreement with the spectroscopic data. In the (AP)2−1 duplex, the extra helical abasic site residues reside in the minor groove of the helix, while they appear in the major groove in the (AP)2+1 duplex. These structural differences are consistent with the differential recognition of bi-stranded abasic site lesions by human AP endonuclease.
Keywords: DNA damage, DNA structure, AP lesions, clustered abasic sites, ionizing radiation
Ionizing radiation causes oxidative DNA damage in the cells producing various types of base damages, strand breaks and abasic sites (AP sites). In an aqueous environment, DNA damages are indirect resulting from the reaction with hydroxyl free radicals induced by radiation (1–3). Since a single energy deposition event can form several hydroxyl radicals, ionizing radiation has the potential to produce clustered DNA damages, defined as two or more lesions within one turn of the DNA helix (1, 4–5). Repair of clustered lesions can have different cellular outcomes including the generation of toxic double strand breaks (DSBs), lesions that form the basis for the utilization of ionizing radiation in cancer therapy. On the other hand, they are highly mutagenic when left unrepaired.
Base Excision Repair (BER) of AP lesions starts when a DNA glycosylase with β–δ elimination activity removes the abasic nucleotide, or after an AP endonuclease incises the phosphodiester bond at the 5’-side of the lesion (6–8). However, the repair of clustered AP lesions, as well as that of multiple DNA damage sites in general, are more complex and can elicit different outcomes. Deletion mutants for the three major DNA glycosylases in E. coli, which leaves bacteria unable of removing oxidized bases, show greater survival rates and a lower number of DSBs after exposure to ionizing radiation than wild type cells (9). Human hematopoietic cells can efficiently join DSBs within 24 h after irradiation but the number of bi-stranded abasic sites clusters decreases very slowly over a 14 days period (10).
In vitro, inhibition of DNA incision occurs with both bacterial and mammalian proteins when the bi-stranded lesions are up to five base pairs away (11–13). E. coli formamidopyrimidine DNA glycosylase (Fpg) can remove an 8-oxo-dG lesion or an AP site positioned 3 or 6 base pairs away from a single strand break in the opposing strand. However, incision is less efficient when the clustered lesions are staggered in the +1 or −1 orientation (13) (see Figure 1 for the sign convention used throughout this manuscript). Clustered lesions also affect the efficiency of mammalian BER proteins (10, 14–18). Human AP endonuclease 1 (hApe1) incises +1 and +3 bi-stranded (AP)2 substrates less efficiently than a single AP duplex but still reasonably well. In contrast, it shows greatly reduced activity processing −1 and −3 (AP)2 clusters (11) and is only 30% active with two directly opposed AP lesions (18). Furthermore, human cell extracts from mononuclear blood cells can efficiently cleave bi-stranded abasic lesions separated by five base pairs or more, irrespective of their orientation, but respond somewhat poorly with closely spaced lesions, especially on clusters staggered in the 5’ direction (10). This differential recognition fades with increase separation between the lesions and bi-stranded (AP)2 sites located six or more nucleotides apart behave as single AP lesions. The structural determinants that affect BER recognition of bi-stranded (AP)2 sites are not understood. Many research groups have investigated the structures of short DNA duplexes having a single of abasic site (natural, oxidized, reduced or the stable THF analog) placed opposite to a purine (apyrimidinic site), a pyrimidine (apurinic site) or in the context of a single base deletion. (19–33). In all cases, the helix remains right-handed with the AP residue stacked inside the helix or looped out depending to the temperature, sequence context and the nature of the opposing base. When the orphan base is a purine, both the AP site rand its partner residue remained stacked inside the helix with backbone perturbations occurring on the lesion site and adjacent base pairs (19–21, 25–31). However, when the abasic site is located in an A-tract in which case the disturbance extends over several base pairs at either side of the lesion (25). On the other hand, apurinic sites and sometimes the pyrimidine counter residue can be extra helical, depending on the temperature and duplex sequence context (21, 23, 26–27).
Figure 1.
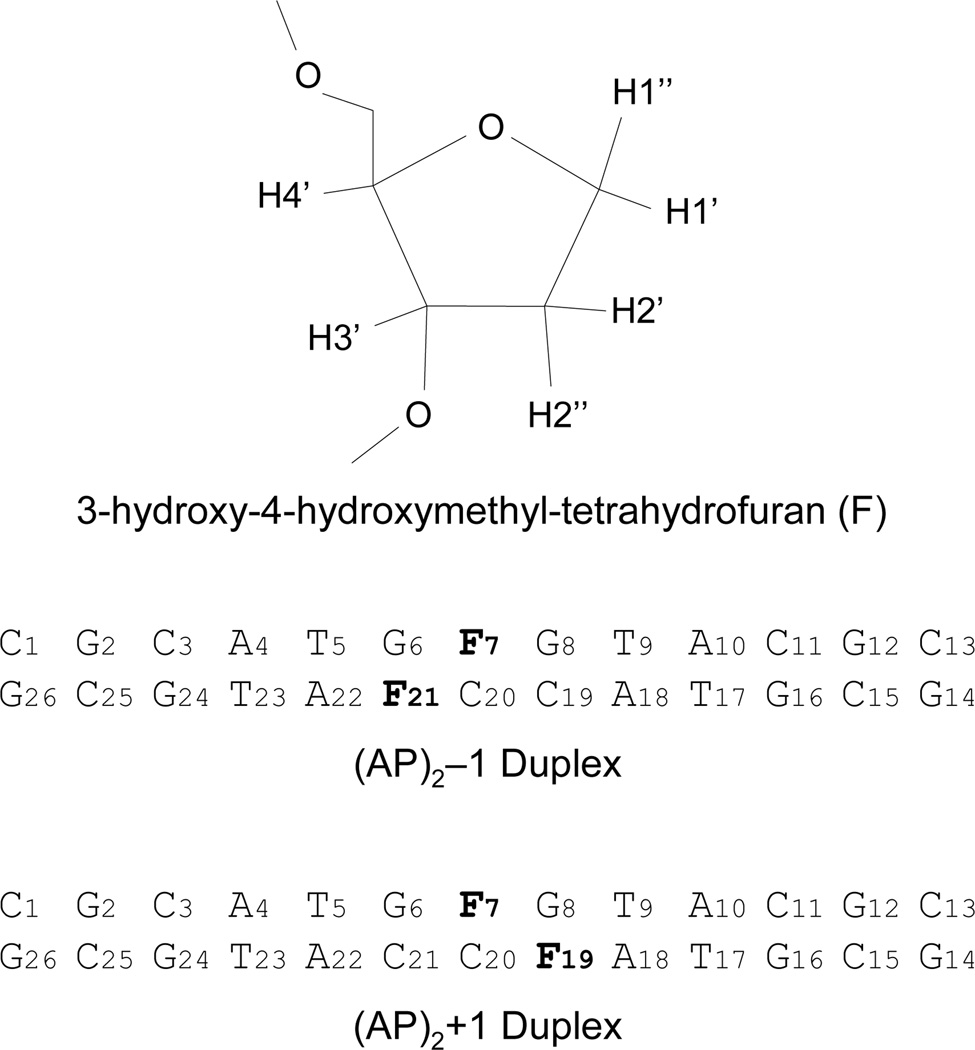
Chemical structure of the tetrahydrofuran AP residue and DNA sequences of the (AP)2−1 and the (AP)2+1 duplexes. Signs indicate the relative orientation of the second AP site being negative when positioned towards the 5’-side of the first (reference) lesion.
Previously, we reported the solution structure of DNA containing a (AP)2−1 or (AP)2+1 cluster observing the extrusion of the AP residues in both clusters, with the formation of a novel G•A mismatch only in the −1 orientation (34). In order to address the role of duplex sequence, we established the solution structure of two DNA duplexes containing a bi-stranded (AP)2−1 or (AP)2+1 cluster, with dC and dG pairing the abasic residues, as determined by NMR spectroscopy and restrained molecular dynamics simulations. Figure 1 shows the sequence of (AP)2 duplexes and the chemical structure of tetrahydrofuran, the abasic site analogue used in this study.
MATERIALS AND METHODS
Oligonucleotide Synthesis and Duplex Preparation
The synthesis of AP containing duplexes was done using standard phosphoramidite chemistry procedures with commercial reagents purchased from Glen Research Corporation. Samples purification was accomplished by two runs of reverse phase HPLC, using a C18 column as previously reported (34). Conversion of the samples to their sodium salt was achieved by running then through a Sephadex G-25 column followed by a Dowex 50W ion exchange resin. The concentration of oligomeric DNA was determined by UV260 absorption using extension coefficients calculated by Generunner (V3.0, Hasting Software, Inc.). Complementary strands were annealed by heating the samples to 80°C and slowly cooling to room temperature. Samples were then lyophilized and dissolved in 0.7 ml of 25 mM phosphate buffer at pH 6.8, containing 50 mM NaCl and 0.5 mM EDTA in 99.96% D2O (D2O buffer) or in 90% H2O/10% D2O (H2O buffer). DNA concentration of samples used for NMR spectroscopy was about 2 mM.
Duplex samples used for the hApe1 incision studies were prepared as described above. The samples consisted of 36-mer duplexes containing the bi-stranded (AP)2−1 (or (AP)2+1) cluster in the same sequence context than that used for the NMR study.
NMR Methods
One- and two-dimensional NMR spectra were recorded in Varian (INOVA) spectrometers at 600 or 500 MHz field strengths. Proton spectra were collected from samples dissolved in D2O buffer at 25°C and in H2O buffer at 1°C. Proton chemical shifts were referenced to external 3-(trimethylsilyl)-propionate-2,2,3,3,-d4 at 0 parts per million (ppm). Phase-sensitive NOESY (35), COSY, COSY45, DQF-COSY, and TOCSY spectra were collected for proton assignment and to aid the determination of 2’-deoxyribose conformations. Inter-proton distances were based on NOESY (50, 120, 190, 260, and 300 ms mixing time) spectra recorded in D2O buffer using a repetition delay of 1.4 s. The residual water sample was suppressed by saturation pulse. TOCSY spectra were collected in D2O buffer using isotropic mixing times of 60 and 120 ms. NOESY spectra in H2O buffer were recorded using mixing times of 120 and 220 ms and a jump-return reading pulse (36). Two-dimensional data sets consisted of 2048 by 300 complex points in the t2 and t1 dimensions, respectively. COSY45 spectra were acquired with doubled number of complex points in both dimensions. The temperature dependence of imino protons was investigated over the 0–55°C temperature range using 5°C intervals. NMR spectra were processed an analyzed on Silicon Graphics workstations using FELIX98 (Accelrys, San Deigo, CA). NOESY and TOCSY time domain data were multiplied by 90°-shifted sinebell window functions. COSY, COSY45 and DQF-COSY data sets were multiplied by a sine bell function and an exponential function, using 4 Hz of line broadening. The water signal on spectra collected with the sample dissolved in 10% D2O buffer was suppressed further by subtraction of a convolution function before Fourier transformation.
Computational Methods
Restrained Molecular Dynamics (rMD) simulations were run on Silicon Graphics computers using XPLOR 3.1 (37). Structures were visualized with InsightII (Accelrys, San Deigo, CA) and Midas Plus (UCSF, Computer Graphics Laboratory) and helical parameters were computed using Curves 5.1 (38, 39). Molecular dynamics were done in vacuum using an all atom force field derived from CHARM (40). Partial atomic charges on the phosphates were not reduced, leaving deoxynucleotide residues with a net charge of −1. Inter-proton distances were computed using cross-peak volume intensities measured on the 50, 120, 190, and 260 ms mixing times NOESY spectra. Each of the four NOE peak volume files was input, separately, in a ‘relaxation’ protocol in XPLOR, which minimized the structure of model duplexes using only a potential energy function proportional to the difference between experimental and back calculated peak NOE intensities. Initial structures of the (AP)2−1 and the (AP)2+1 clusters were constructed in InsightII from canonical B-form 13-mer duplexes, containing central G•C base pairs and deleting a purine and a pyrimidine base from each strand to create the bi-stranded (AP)2 lesions. During the ‘relaxation’ protocol, duplex protons were energy minimized to fit the set of experimental NOE peak intensities that were given error bounds of 0.5%. Inter-proton distances were extracted from the last atom coordinates of the minimization, resulting in four inter-proton distance sets for each (AP)2 duplex. Distance files were averaged to produce a unique inter-proton distance set. Inter-proton distances fixed by covalent-geometry, such as cytosine H5–H6 or sugar H2’-H2”, were systematically underestimated by this approach and, as a result, a correction of 20% was applied to all inter-proton distances. A total of 436 and 441 experimental inter-proton distances for the (AP)2−1 and (AP)2+1 duplexes, respectively, were enforced during restrained molecular dynamics simulations by using square-well potential energy functions. Distance boundaries were set to ± 0.6 Å of the average. Distances obtained from overlapped peaks had boundaries of ± 0.8 Å. Following the analysis of NOESY spectra in H2O buffer, Watson-Crick (WC) alignments on all canonical base pairs were enforced by distance restraints with bounds of ± 0.1 Å from the values determined by X-ray crystallography. Similarly, the central G•C base pair at the lesion site was also restrained in both duplexes. Following the analysis of COSY45 spectra (41, 42), sugar conformations of the 4 terminal residues of the duplexes were restrained within the C2’-endo range using empirical dihedral angle square-well potential energy functions. Sugar conformations at the center of the (AP)2 duplexes were restrained within the C3’-exo/C2’-endo/C1’-exo range. The SHAKE algorithm was used to maintain the length of covalent bonds involving protons (43). Initial models were energy minimized before beginning molecular dynamics.
Our molecular dynamics protocol consisted in slowly raising in 70ps the temperature of the simulation from one of four starting temperatures (150, 200, 250 and 300K) to 500K, the high temperature value, while the scale of the penalty function enforcing distance restraints was gradually increased from 50 to 300 kcal/mol/Å2. The system was equilibrated at 500K for periods of 42, 44, 46, 48 and 50ps, after which the temperature of the simulation was slowly cooled to 300K in 40ps and the simulations continued by an additional 120ps at this temperature. The four initial temperature values and the five variations in the length of the high-temperature step produced a set of 20 distance refined structures for each (AP)2 duplex. Final atom coordinates underwent 1000 steps of energy minimization, generating the distance-refined models. Five refined model for each duplex showed no NOE violations >0.1 Å and exhibited pair wise root mean square deviations (RMSD) <1.5 Å in the atom position, comprising the family of converging structures discussed in the manuscript.
Protein purification
hApe1 expression and purification followed protocols described previously (17). BL21(DE3) pLysS bacteria, transformed with an hApe1-containing pET-28A plasmid, was grown in minimal medium to a culture density of A600 = 0.9–1, at 37°C. At this point, addition of 1 mM IPTG and additional 6 hr incubation at the same temperature induced hApe1 expression. Bacteria were then harvested by centrifugation, suspended in 25 mM HEPES, pH 7.9, buffer containing 5 mM EDTA, 0.5 mM DTT, and 100 mM NaCl, and disrupted by sonication. Subsequent, fifteen minutes centrifugation at 12,000 rpm (Sorvall, SS-34 rotor) at 4°C clarified the protein extract, which was then injected in a two-column system composed of a fast flow Q-Sepharose connected to a fast flow S-Sepharose. hApe1 protein was eluted from the S-Sepharose column using a linear gradient of 0.1 to 1.5 M of NaCl over 200 ml. Fractions containing hApe1 were concentrated and loaded into a Superdex-75 column (1.6 × 60 cm, Pharmacia), pre-equilibrated with 25 mM HEPES buffer, pH 7.9, 0.5 mM EDTA, 0.5 mM DTT and eluted using the same buffer. Purified hApe1 run as a single band in a 12% SDS-PAGE. The Bradford protein assay (Bio-Rad, Inc.) was used to determine hApe1 concentration using bovine serum albumin as standard.
hApe1 Digestion Experiments
Duplex incision reactions were performed by duplicate in 20µl reaction volumes, as described previously (17). Reaction mixtures contained 100 nM of duplex substrate and 0.03 nM of purified hApe1 dissolved in 25 mM HEPES buffer, pH 7.6, containing 50 mM NaCl, 50 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 6% glycerol and 0.5 mM EDTA. Reaction mixtures stood on ice for 15–20 minutes prior to incubation at 37°C. Duplex samples were 5’end-labeled with γ-[31P] APT (Amersham) using T4 polynucleotide kinase (Roche Diagnostics), as previously described (17). Cleavage reactions products were resolved by 6% SDS-PAGE, visualized and quantified using a PhosphorImager (Molecular Dynamics Inc.).
RESULTS
Non-exchangeable Proton Spectra of the (AP)2−1 Duplex
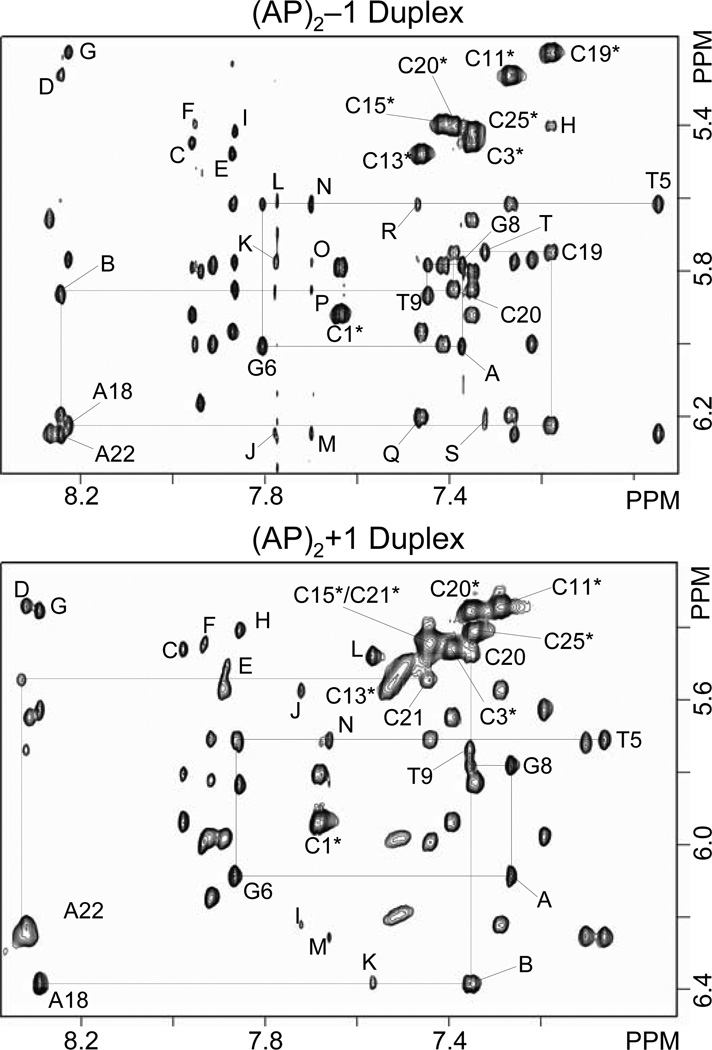
NOESY spectra collected for the (AP)2−1 duplex in D2O buffer are characterized by the presence of sharp signals with few overlapped resonances. Sequence specific assignment of the non-exchangeable protons follows the analysis of two-dimensional NOESY, COSY and TOCSY spectra collected at room temperature, using standard procedures (44–46). The top panel of Figure 2 shows an expanded contour map of a NOESY spectrum (300ms mixing time) depicting interactions between the base (7.0–8.5 ppm) and the sugar H1’ (5.2–6.4 ppm) protons of the (AP)2−1 duplex. Identification of NOE cross peaks between the pyrimidine H6 (or purine H8) protons and their own and 5’-flanking sugar H1’ protons indicates that the (AP)2−1 duplex is right-handed helix. Similarly, cross peaks between cytosine H5 and 5’-flanking base (purine-H8 or pyrimidine-H6) protons (Figure 2, top panel, peaks C-I) and between adenine H2 and sugar H1’ protons on the same and complementary strands (Figure 2, top panel, peaks J-T) are further evidence of the right-handedness of the helix and proper stacking throughout the (AP)2−1 duplex. We observe two sequential NOE cross peaks, G8(H8)-G6(H1’) and A22(H8)-C20(H1’, between the residues that flank both AP site lesions of the duplex (Figure 2, top panel, peaks A and B), which are consistent with an extra-helical arrangement of both abasic residues in the (AP)2−1 duplex. The complete assignment of the base-sugar H1’ proton region (NOESY walk) is illustrated in Figure 1S (Supplementary Material).
Figure 2.
Expanded regions of phase sensitive NOESY spectra (300ms mixing time) recorded at 25°C with the samples dissolved in D2O buffer (25 mM Na2PO4, 50 mM NaCl), pH 6.8, showing interactions between the base (7.0–8.4 ppm) and sugar H1’ (5.2–6.5 ppm) protons. Solid lines connect sequential interactions for the (top) (AP)2−1 (bottom) and (AP)2+1 duplexes on the central (…T5-G6-F7-G8-T9…) segment, and broken lines do the same on the complementary strand. Cross peaks labeled with residue numbers indicate intra-residue base (purine-H8/pyrimidine-H6) to H1’ sugar interactions and the asterisks denote cytosine H5–H6 cross peaks. In the (AP)2−1 duplex (top panel), other labels are assigned as follows: A, G8(H8)-G6(H1’); B, A22(H8)-C20(H1’); C, G2(H8)-C3(H5); D, A10(H8)-C11(H5); E, G12(H8)-C13(H5); F, G14(H8)-C15(H5); G, A18(H8)-C19(H5); H, C19(H6)-C20(H5); I, G24(H8)-C25(H5); J, A22(H2)-A22(H1’); K, A22(H2)-T23(H1’); L, A22(H2)-T5(H1’); M, A4(H2)-A4(H1’); N, A4(H2)-T5(H1’); O, A4(H2)-T23(H1’); P, A4(H2)-G24(H1’); Q, A10(H2)-A10(H1’) and C13(H6)-C13(H1’); R, A10(H2)-C11(H1’); S, A18(H2)-A18(H1’) and A18(H2)-A10(H1’); T, A18(H2)-C19(H1’). In the (AP)2+1 duplex (bottom panel) labeled cross peaks are assigned as follows: A, G8(H8)-G6(H1’); B, C20(H6)-A18(H1’); C, G2(H8)-C3(H5); D, A10(H8)-C11(H5); E G12(H8)-C13(H5); F, G14(H8)-C15(H5); G, A18(H8)-C20(H5); H, G24(H8)-C25(H5); I, A10(H2)-A10(H1’); J, A10(H2)-C11(H1’); K, A18(H2)-A18(H1’); L, A18(H2)-C20(H1’); M, A22(H2)-A22(H1’); N, A22(H2)-T5(H1’).
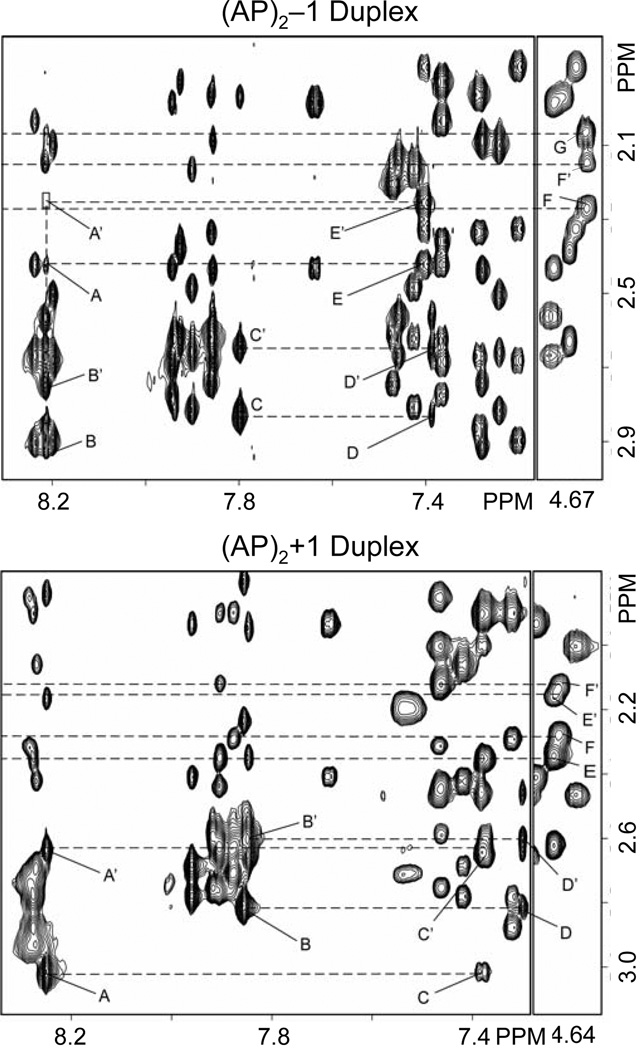
NOE cross peaks between the base (prymidine-H6 or purine-H8) and sugar H2’/2” protons provide further evidence of the spatial proximity of lesion flanking residues in the (AP)2−1 duplex (Figure 3, top panel). Specifically, a pair of weak cross peaks between G8(H8)-G6(H2’) and G8(H8)-G6(H2”) indicate proximity of G8 and G6 residues, which are separated in the duplex sequence by F7 (Figure 3, top panel, peaks D and D’). On the complimentary strand, the weak NOE peak between A22(H8)-C20(H2”) indicates proximity of the F21-flanking residues but no interaction is observed between A22(H8)-C20(H2’) (Figure 3, top panel, peak A and box A’). This is an indication that the abasic residues in the (AP)2−1 duplex are not extruded identically from the helix. The observation of the G6(H8)-G8(H8) and G8(H8)-G6(H3’) NOE peaks (data not shown) between F7 neighboring residues, but not of the corresponding A22(H8)-C20(H6) and A22(H8)-C20(H3’) interactions on the complementary strand, further suggests that the AP residues are differently extruded from the helix in the (AP)2−1 duplex.
Figure 3.
Expanded regions of phase sensitive NOESY spectra (300ms mixing time) recorded at 25°C with the samples dissolved in D2O buffer (25 mM Na2PO4, 50 mM NaCl), pH 6.8, showing interactions between the base (7.0–8.4 ppm) and sugar H2’/H2” (1.8–3.0 ppm) protons in the (AP)2−1 (top) and (AP)2+1 (bottom) duplexes. Labeled peaks are assigned as follows: top panel, A, A22(H8)-C20(H2”); A’, A22(H8)-C20(H2’); B, A22(H8)-A22(H2”); B’, A22(H8)-A22(H2’); C, G6(H8)-G6(H2’); C’, G6(H8)-G6(H2”); D, G8(H8)-G6(H2’); D’, G8(H8)-G6(H2”); E, C20(H6)-C20(H2”); E’, C20(H6)-C20(H2’); F, F21(H3’)-F21(H2”) or F21(H2’); F’, F21(H3’)-F21(H2”) or F21(H2’); and G, F7(H3’)-F7(H2”) and F7(H2’). Bottom panel: A, A18(H8)-A18(H2’); A’, A18(H8)-A18(H2”); B, G6(H8)-G6(H2’); B’, G6(H8)-GG6(H2”); C, C20(H6)-A18(H2”); C’, C20(H6)-A18(H2’); D, G8(H8)-G6(H2’); D’, G8(H8)-G6(H2”); E, F7(H3’)-F7(H2” or H2’); E’, F7(H3’)-F7(H2” or H2’); F, F19(H3’)-F19(H2” or H2’) and F’, F19(H3’)-F19(H2” or /F19H2’).
The sugar H1’/1” protons of the AP residues F7 and F21 are readily identified with the aid of COSY spectra by their unique location at 3.9–4.1 ppm, but are not stereo-specifically assigned (Figure 3S). These protons show strong COSY interactions with neighboring H2’/2” protons (Figure 3S, peaks A and B), which in turn exhibit NOE cross peaks with their vicinal H3’ protons (Figure 3, top panel, peaks F, F’ and G). Only a few sequential interactions are present between the AP and adjacent residues in the (AP)2−1 duplex suggesting mobility of the AP residues in solution. Weak sequential NOE peaks are observed between G8(H8) and F7(H1’/1”) and F7(H4’) protons, as well as between A22(H8)-F21(H1’/1”) in the complementary strand (data not shown). However, no inter-residue cross peak is seen between H2’/2” protons of the AP residues and flanking base protons (Figure 3A). Table 1S (Supplementary Material) lists the proton chemical shifts in the (AP)2−1 duplex.
(AP)2+1 Duplex
NOESY spectra collected for the (AP)2+1 duplex in D2O buffer are also characterized by the presence of sharp signals with few overlapped resonances. As before, the sequence specific assignment of the non-exchangeable protons follows the analysis of two-dimensional NOESY, COSY and TOCSY spectra, recorded at room temperature, using standard procedures (44–46). The bottom panel on Figure 2 shows a contour plot of an expanded region of a NOESY spectrum (300ms mixing time) displaying interactions between the base (7.0–8.5 ppm) and the sugar H1’ (5.2–6.4 ppm) protons of the (AP)2+1 duplex. Cross peak connectivity between the pyrimidine H6 (or purine H8) protons and their own, and 5’-flanking, sugar H1’ protons indicates that the (AP)2+1 duplex is right-handed helix in solution. Furthermore, NOE peaks between cytosine H5 and 5’-flanking purine H8 protons (Figure 2, bottom, peaks C to H) and between adenine H2 and sugar H1’ protons (Figure 2, bottom, peaks I to N) are additional evidence of the right-handedness of the helix. As seen in the (AP)2−1 duplex case, strong inter-residue cross peaks between G8(H8)-G6(H1’), C20(H8)-A18(H1’), and A18(H2)-C20(H1’) (Figure 2, bottom panel, peaks A, B and L) indicate proximity between the AP flanking residues. These cross peaks are consistent with an extra helical arrangement of both AP residues in the (AP)2+1 duplex too. The complete NOESY walk is illustrated in Figure 2S (Supplementary Material).
NOE peaks between the base (purine H8 and pyrimidine H6) and the sugar H2’/2” protons offer further evidence of the proximity of lesion flanking residues in the (AP)2+1 duplex (Figure 3, bottom panel). NOE cross peaks seen between G8(H8)-G6(H2’) and G8(H8)-G6(H2”) and between C20(H6)-A18(H2”) and C20(H6)-A18(H2’) (Figure 3, bottom, peaks D/D’ and C/C’, respectively) indicate proximity of these residues in the structure and support the extra helical arrangement of F7 and F19 on the (AP)2+1 duplex. Similarly, G6(H8)-G8(H8) and G8(H8)-G6(H3’) NOEs, between the F7 flanking residues, and C20(H6)-A18(H8) and C20(H6)-A18(H3’) interactions, between residues flanking F19 (data not shown), suggest that both AP residues adopt a similar extra helical conformation in the (AP)2+1 duplex. It is interesting to notice that these observations are in contrast to the case of the (AP)2−1 duplex, where only the residues flanking F7 produced those cross peaks.
Assignment of the F7 and F19 protons follows the identification of the strong H1’/H1”-H2’/H2” COSY cross peaks (Figure 4S, peaks A and B), in a region of the spectrum that lacks other proton interactions. On the NOESY (300 ms mixing time) spectrum of the (AP)2+1 duplex, we observe a pair of cross peaks en between G8(H8) and F7(H1’/1”) protons and, within the tetrahydrofuran rings, between H3’ and H2’/2” protons of AP residues, F7 and F21, (Figure 3, bottom, peaks, E, E’, F, and F’). However, as in the case for the (AP)2−1 duplex, there is no inter-residue cross peak between the H2’/2” protons of AP residues and any of the flanking base protons (Figure 3). Table 2S (Supplementary Material) lists the proton chemical shifts in the (AP)2+1 duplex.
Exchangeable Proton Spectra of the (AP)2−1 Duplex
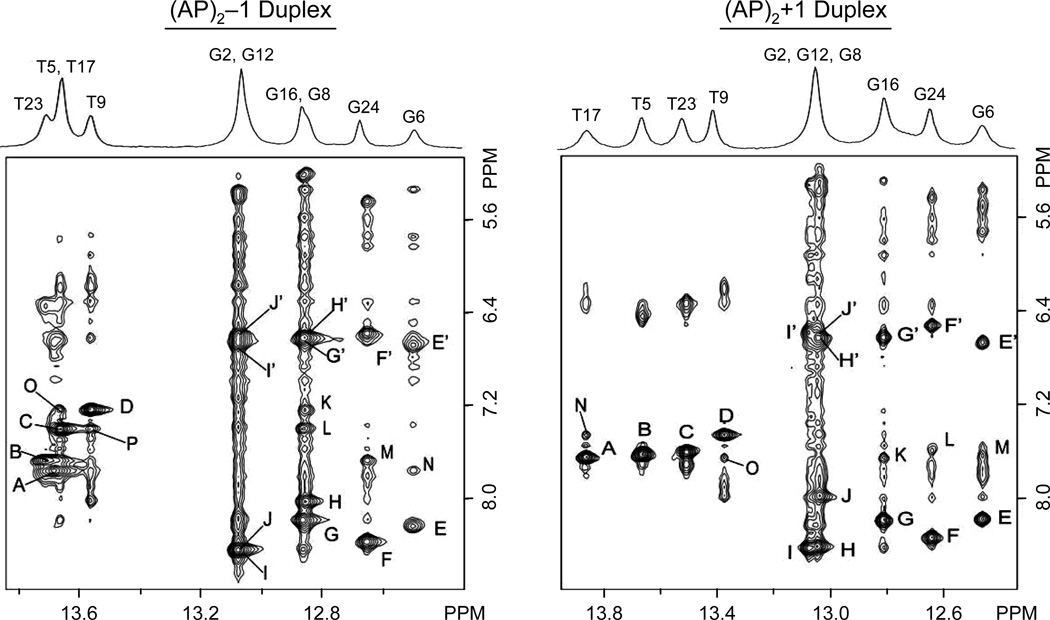
Figure 4 (left panel) shows the one-dimensional proton spectrum of the (AP)2−1 duplex, recorded in H2O buffer, pH 6.8, at 5°C along with an expanded region of a NOESY (220ms mixing time) spectrum depicting interactions between these and the base/amino protons. At this temperature, seven partially resolved signals account for all imino protons of the duplex, with T(NH3) signals resonating between 13.4–14.0 ppm and G(N1H) peaks appearing between 12.4–13.2 ppm. Sequence specific assignment of the exchangeable protons follows the analysis of the NOESY spectrum recorded under identical conditions. Strong NOE cross peaks between T(NH3) and A(H2) protons distinguish all four WC A•T pairs of the (AP)2−1 duplex (Figure 4, left panel, peaks A-D). The previous identification of A(H2) protons on the NOESY data collected in D2O buffer permits the assignment of all T(NH3) signals. Similarly, strong NOE peaks correlate G(N1H) with the hydrogen-bonded and exposed cytosine amino protons across G•C base pairs of the duplex (Figure 4, left panel, peaks E/E’-J/J’). Subsequent identification of intra-residue NOE peaks between the cytosine H5 and NH2 protons (data not shown) allows the sequence specific assignment of non-terminal C(N4H2) signals and, in turn, that of the G(N1H) protons. Notably, the orphan G6 and C20 residues display the NOE interactions that characterize WC alignments (Figure 4, left panel, peaks E-E’), indicating the formation of a canonical G6•C20 base pair at the lesion site of the (AP)2−1 duplex. Furthermore, the observation of sequential NOE interactions between imino and A(H2) protons (Figure 4, left panel, peaks K-P) and among imino protons of sequential base pairs (Figure 5S, Supplementary Material, peaks A-H) suggests proper base pair stacking throughout the (AP)2−1 duplex. Due to their fast solvent exchange, the imino protons of terminal residues fail to show NOE interactions with other DNA protons and only the observation of exchange cross peaks with solvent permits their identification on the spectrum. Table 1S (Supplementary Material) lists the chemical shifts of the exchangeable proton in the (AP)2−1 duplex.
Figure 4.
Expanded regions of NOESY (220ms mixing time) spectra recorded at 5°C in 25 mM phosphate H2O buffer, pH 6.8, containing 50 mM NaCl and 1.0 mM EDTA. Labeled cross peaks are assigned as follows: (AP)2−1 duplex, A, T5(N3H)-A22(H2); B, T23(N3H)-A4(H2); C, T17(N3H)-A10(H2); D, T9(N3H)-A18(H2); E/E’, G6(N1H)-C20(N4Hhb)/C20(N4Hex); F/F’, G24(N1H)-C3(N4Hhb)/C3(N4Hex); G/G’, G16(N1H)-C11(N4Hhb)/C11(N4Hex); H/H’, G8(N1H)-C19(N4Hhb)/C19(N4Hex); I/I’, G2(N1H)-C25(N4Hhb)/C25(N4Hex); J/J’ G12(N1H)-C15(N4Hhb)/C15(N4Hex); K, G8(N1H)-A18(H2); L, G16(N1H)-A10(H2); M, G24(N1H)-A4(H2); N, G6(N1H)-A22(H2); O, T17(H3H)-A18(H2) P, T9(N3H)-A10(H2). (AP)2+1 duplex, A, T17(N3H)-A10(H2); B, T5(N3H)-A22(H2); C, T23(N3H)-A4(H2); D, T9(N3H)-A18(H2); E/E’, G6(N1H)-C21(N4Hhb)/C21(N4Hex); F/F’, G24(N1H)-C3(N4Hhb)/C3(N4Hex); G/G’, G16(N1H)-C11(N4Hhb)/C11(N4Hex); H/H’, G2(N1H)-C25(N4Hhb)/C25(N4Hex); I/I’, G12(N1H)-C15(N4Hhb)/C15(N4Hex); J/J’, G8(N1H)-C20(N4Hhb)/ C20(N4Hex); K, G16(N1H)-A10(H2); L, G24(N1H)-A4(H2); M, G6(N1H)-A22(H2); N, T17(N3H)-A18(H2); O, T9(N3H)-A10(H2).
(AP)2+1 Duplex
The one-dimensional proton spectrum recorded in H2O buffer, pH 6.8, at 5°C, shows that the imino proton signals of the (AP)2+1 duplex are better resolved than those of the (AP)2−1 cluster (Figure 4, right panel). Sequence specific assignment of these protons follows the analysis of NOESY (140 and 220 ms mixing time) spectra recorded under identical conditions. Strong interactions between T(N3H) and A(H2) protons identify all WC A•T base pairs of the (AP)2+1 duplex (Figure 4, right panel, peaks A-D). Similarly, NOE peaks between G(N1H) and C(N4H2) protons (Figure 4, right panel, peaks E/E’-J/J’) permits the assignment of all internal G•C base pairs of the duplex. As it was the case of the (AP)2−1 duplex, we observe NOE interactions between orphans G8 and C20 residues (Figure 4, right panel, peaks J/J’) indicating the formation of a canonical G•C base pair at the lesion site of the (AP)2+1 duplex. Sequential NOE interactions between imino and A(H2) protons (Figure 4, left panel, peaks K-O) and among imino protons of sequential base pairs (Figure 5S, Supplementary Material, peaks A-I) indicate proper base pair stacking throughout the (AP)2+1 duplex. As before, the imino protons of terminal residues fail to show NOE interactions with other DNA protons and only the observation of exchange cross peaks with solvent permits their identification. Table 2S, supplementary material, lists the chemical shift of the exchangeable protons in the (AP)2+1 duplex.
Thermal Stability
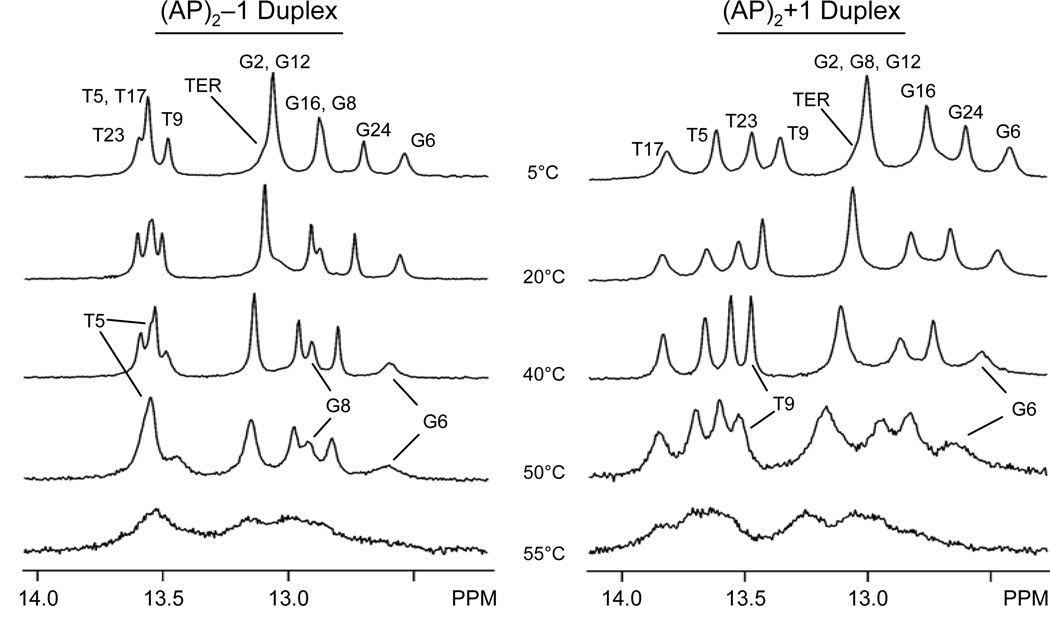
Duplex ‘melting’ provides a direct assessment of the impact that lesions have on the stability of duplex formation. Analysis of UV260 melting curves revealed that both (AP)2 duplexes have similar melting temperature values, which are about 17% lower than that of the unmodified control duplex (data not shown). Figure 5 shows the imino proton region of one-dimensional spectra of (AP)2 duplexes recorded in H2O buffer, pH 6.8, in the 5°C to 55°C range. Due to fast water solvent, the imino proton signals of terminal base pairs in both duplexes are almost undetected, even at low temperature, appearing as a broad shoulder at the base of the overlapping G2(N1H)-G12(N1H) peak (Figure 5). During the transition to random coil, both clusters behave similarly, melting inwards from the ends of the duplexes. As the temperature increases, the T5(N3H), G6(N1H), and G8(N1H) signals of the (AP)2−1 duplex are seen on the 40°C and 50°C spectra, indicating the persistence of lesion site T5•A22, G6•C20 and G8•C19 base pairs at high temperature (Figure 5, left panel). In the (AP)2+1 duplex, the G8(N1H) signal overlaps with the G2(N1H)-G12(N1H) peak and, thus, cannot be monitored independently. However, G6(N1H) and T9(N3H) signals are still seen in the 50°C spectrum, establishing the presence of G6•C21 and T9•A18 base pairs at that temperature and suggesting that G8•C20 may be present, as well (Figure 5, right panel). Further temperature increase causes general broadening of the remaining imino proton signals, indicating extensive ‘melting’ of both (AP)2 duplexes at 55°C.
Figure 5.
Temperature dependence of imino proton signals on the (left) (AP)2−1 and (right) (AP)2+1 duplexes, recorded in 25 mM phosphate buffer, pH 6.8, containing 50 mM NaCl and 1.0 mM EDTA.
Three-Dimensional Structures
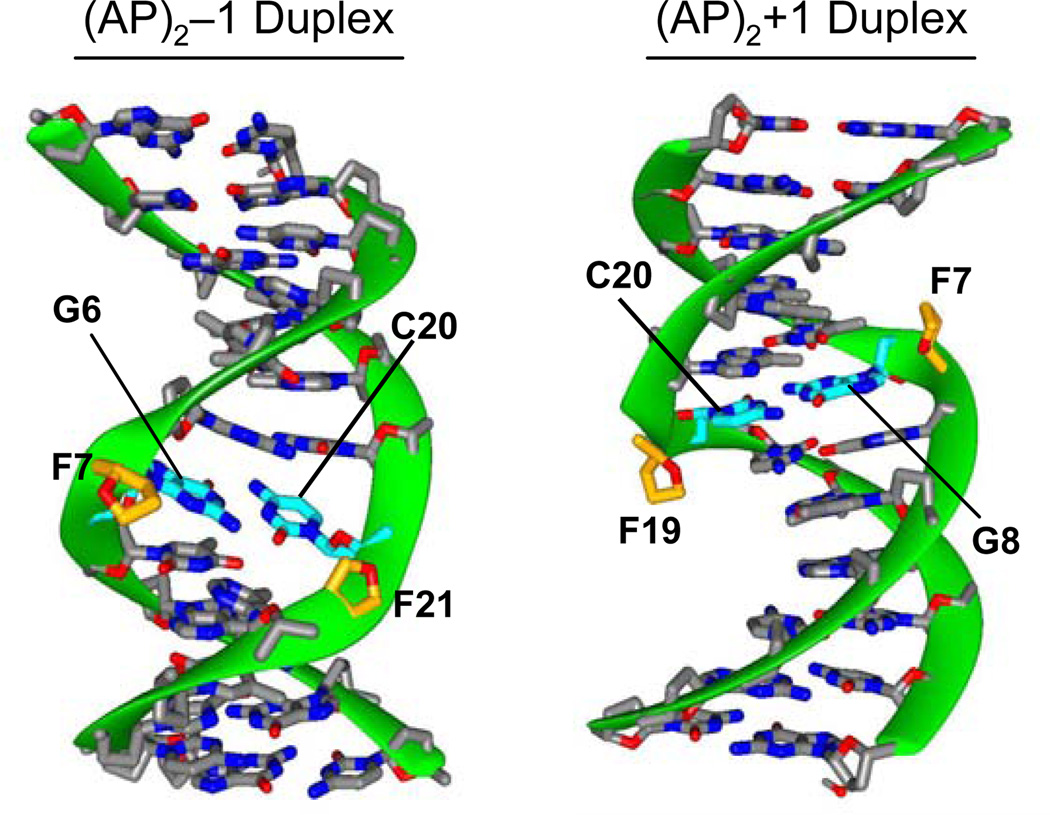
Analysis of the RMSD matrix identified a family of distance-refined structures for each duplex that converged with pair wise values <1.5 Å. The refined structures are in good agreement with of the experimental NMR data and have no violation of the NOESY-spectra-derived inter proton distances > 0.1 Å. In addition, back calculation of the NOE intensities using the refined (AP)2−1 and (AP)2+1 duplex structures show the residual factors (NMR R-factors) dropping to about 13.2% and 12.8%, respectively, indicating that the three dimensional models quantitatively agree with the experimental data. Figures 6S and 7S (Supplementary Material) show stereo views of the refined models and Table 1 lists statistics of the refinement. As shown in Figure 6, the (AP)2−1 and (AP)2+1 duplexes are regular right-handed helices with WC base pair alignments throughout. There is compression at the lesion site of the duplexes, which produces bulged AP sites and shortening of the end-to-end distances by 7% and 5% on the (AP)2−1 and (AP)2+1 duplexes, respectively. The helical axis of both duplexes is bend in the direction of the major groove, 30° in the (AP)2−1 cluster and 15° in the (AP)2+1 cluster, partially contributing to the helix shortening. Table 2 summarizes structural parameters of the (AP)2 duplexes.
Table 1.
Molecular dynamics statisticsa
| Covalent Geometry Violations (RMSD) | (AP)2−1 Duplex | (AP)2+1 Duplex |
|---|---|---|
| Bond lengths (Å) | 0.02 | 0.01 |
| Bond Angles (°) | 5 | 4 |
| Improper Angles (°) | 0.7 | 0.5 |
| Van der Waals Energy (kcal/mol) | −286 | −262 |
| Experimental Restraints Violations (RMSD) | ||
| NOE distances (Å) (number of distances) | 0.04 (436) | 0.02 (441) |
| Constrained Dihedrals (°) (number of constrains) | 0.3 (216) | 0.2 (216) |
| NMR R-factors (%) (values on initial structures) | 13.2 (16.5) | 12.8 (16.3) |
Averaged values computed from the five converging structures.
Figure 6.
Ribbon representations (49, 50) of the averaged distance-refined structure shown with the minor groove prominent in the (AP)2−l duplex and the major groove prominent in the (AP)2+l duplex. Unmodified nucleotides are colored by atom type and AP residues appear with the O4’ atom in red. In the (AP)2−l duplex, F7, to a greater extent, and F21 are extruded towards the minor groove. In the (AP)2+1 duplex (B), both AP residues are similarly extruded into the major groove.
Table 2.
Structural Properties of (AP)2 Duplexesa
| Duplex Curvature and Lesion Site Base Parameters | (AP)2−1 duplex | (AP)2+1 duplex |
|---|---|---|
| Helix Shortening (%)b | 5.8–8.6 | 4.6–6.2 |
| Helix Bend (°) | 22–39 | 10–21 |
| Bend Direction | Major Groove | Major Groove |
| G6 | G8: X-Dips (Å), Y-Disp (Å), Incl (°) | −0.25, +0.22, 15 | −3.17, +0.39, 7 |
| C20: X-Dips (Å), Y-Disp (Å), Incl (°) | −0.07, −0.38, −3 | −2.51, −0.93, 4 |
| Central G•C pair: X-Disp (Å), Y-Disp (Å), Incl (°) | −0.16, 0.30, 9 | −2.84, 0.66, 2 |
| Sugar Pucker | ||
| G6 | G8 | C3’-exoc | C2’-endo |
| C20 | C1’-exoc | C2’-endo |
| F7 | C2’-exo | C2’-endod |
| F21| F19 | C2’-endo | C2’-endo |
Percentage relative to the length of the 13-mer B-form duplex control.
One structure in the C2’-endo range.
One structure in the C2’-exo range. Normal values in B-form DNA are: X-Displacement = −0.71 Å; Y-Displacement = 0 Å; Inclination = 6°.
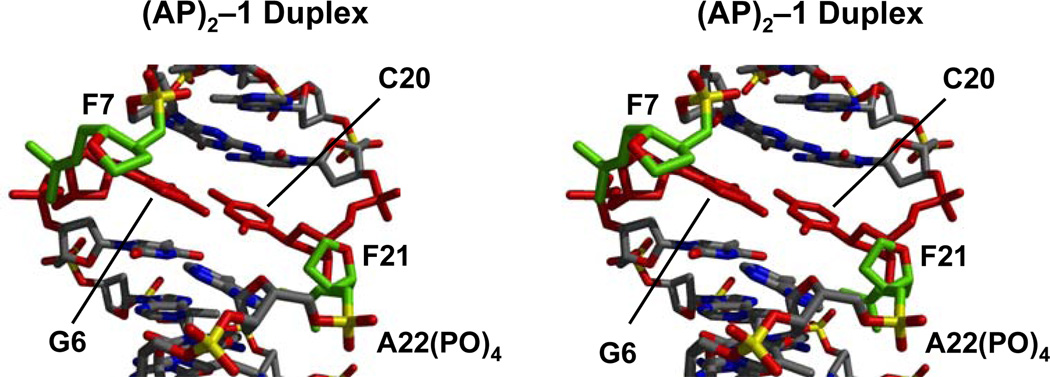
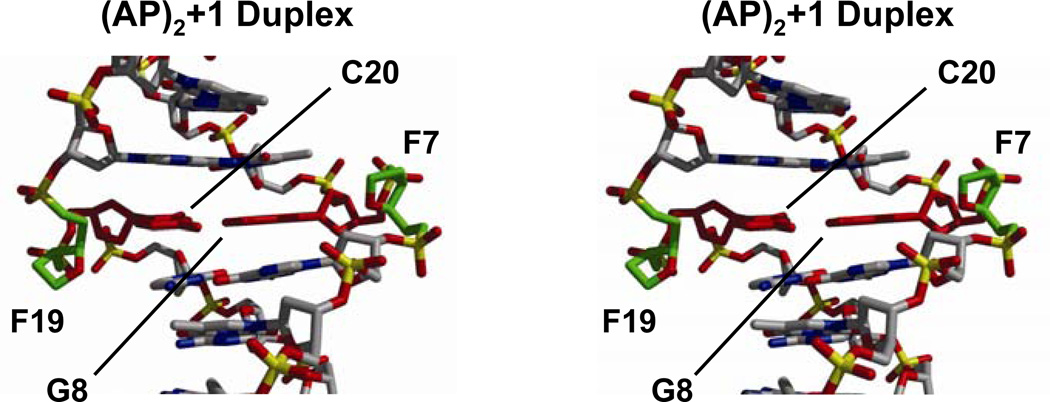
Figures 7 and 8 show cross-eye stereo views of the (AP)2 duplexes depicting the lesion site conformation. In the (AP)2−1 cluster, AP residues are unequally extruded from the helix. F7 forms an abasic bulge, inducing a kink in the sugar-phosphate backbone, while F21 is mostly in line with the backbone, slightly pushing the A22 phosphate towards the solvent (Figure 7). By contrast, F7 and F19 are completely extra helical in the (AP)2+1 duplex, forming similar AP bulges on both strands (Figure 8). A remarkable structural difference between the duplexes is the position of the abasic residues, which appear in the minor grove and are closer to each other in the (AP)2−1 duplex (Figure 7) while they locate in the major groove and are farther apart in the (AP)2+1 duplex (Figure 8). However, since the AP residues are small, groove width and depth dimensions in both duplexes remain, however, unaltered by their presence.
Figure 7.
Stereo views (58, 59) of the lesion site of the (AP)2−l duplex seen with the minor groove prominent. AP site atoms are colored in green, the central G•C base pair in red and other atoms by type.
Figure 8.
Stereo views (58, 59) of the lesion site of the (AP)2+l duplex seen with the major groove prominent. AP site atoms are colored in green, the central G•C base pair in red and other atoms by type.
Local structural perturbations are more significant in the (AP)2−1 duplex. Both, G6 and C20 move toward the major groove of the duplex, showing a base pair X-Displacement (X-Disp) of −0.16 Å as opposed to −0.71 Å in B-form DNA. Mostly G6 and C20, to a lesser extent, are inclined bringing them to a same plane and allowing the formation of a WC base pair at the lesion site (Figure 7). In addition, they undergo compensatory Y-Displacement (Y-Disp) facilitating formation of the G6•C20 base pair. Sugar conformations are somewhat flexible in the (AP)2−1 cluster. G6 and C20 sugars appear in the C3’-exo and C1’-exo conformations, respectively, in four out of five refined structures, and F7 adopts C2’-exo and F21 C2’-endo puckers in all the refined models.
In contrast to the (AP)2−1 cluster, structural perturbations at the lesion site of the (AP)2+1 duplex are relatively minor. The lesion site base pair, G8•C20, appears displaced to the minor groove and in the direction of the F7 strand backbone, without changing its inclination value. Furthermore, all lesion site sugars are in the C2’-endo range, with F7 in a C2’-exo conformation in only one of the refined models. Structural parameters at the lesion site of (AP)2 duplexes are summarized in Table 2.
hApe1 Incision of (AP)2 Duplexes
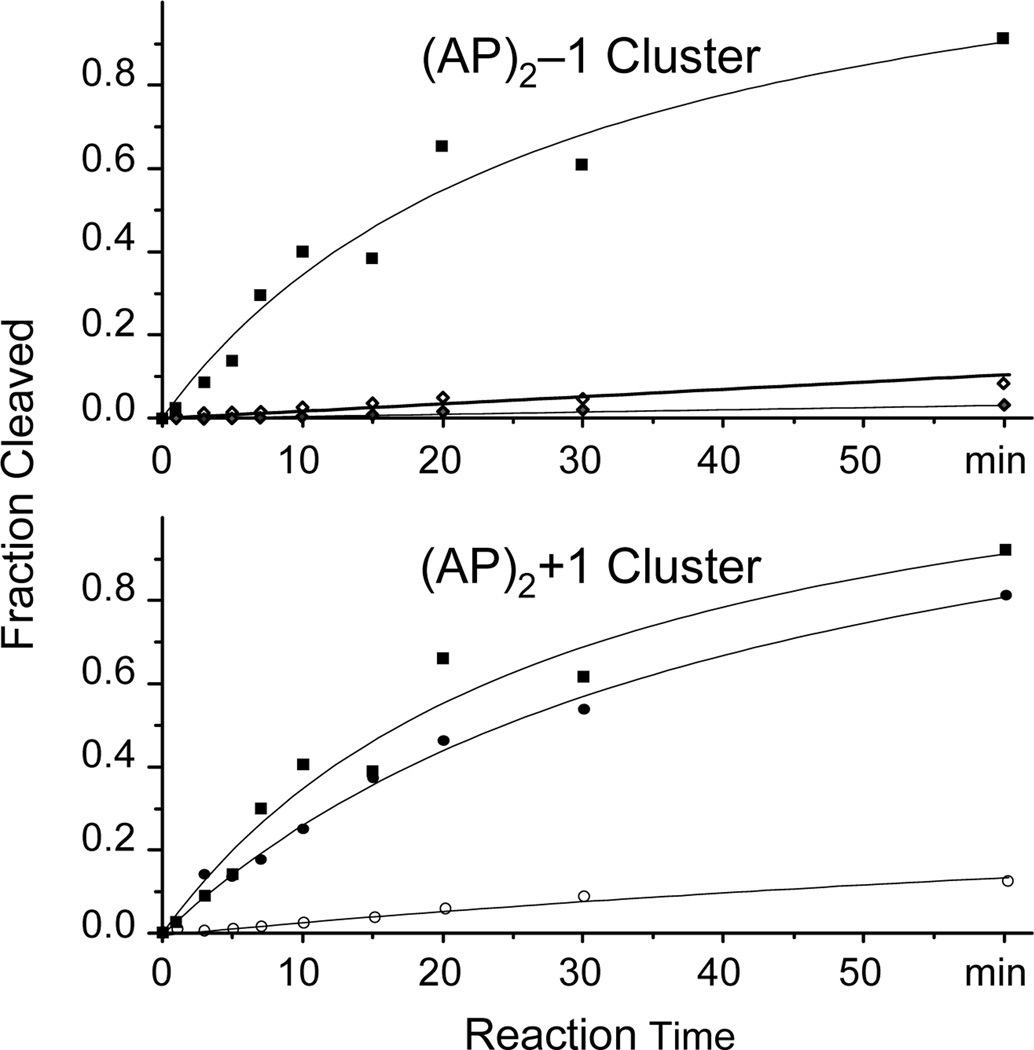
As shown in Figure 9, recombinant Ape1 processes (AP)2 clusters with different efficiency that depended on the lesion orientation within the cluster and their sequence context. The enzyme shows weak incision activity on the (AP)2-1 cluster, cleaving less than 10% of either abasic site under conditions that cleave close to 90% of a control duplex having a single AP site in the F7 sequence context (Figure 9, top panel). Further addition of hApe1 increased the amount of (AP)2-1 the incised fractions only slightly, reaching about 35% of the substrate after overnight incubation (data not shown). In contrast, hApe1 exhibits strong activity on the (AP)2+1 cluster, cutting the F7 AP site with efficiency similar to that recorded for the control duplex, while the other AP site (F19) shows reduced level of cleavage (Figure 9, bottom panel). In addition, after supplementing the reaction with an additional amount of hApe1, overnight incubation shows over 70% incision at F19 (data not shown).
Figure 9.
Incision of bi-stranded (AP)2±l duplexes by human Ape1 endonuclease. Solid and empty symbols indicate incision at F7 and F19 (or F21), respectively. Solid squares show cleavage of a control duplex sample having a single AP site in the context of F7 lesion.
DISCUSSION
NMR Spectra and Solution Structure of (AP)2 Duplexes
The directionality of NOE peaks between base and sugar H1’ protons (Figures 1S and 2S) and the observation of hydrogen bonded base imino protons (Figure 4) establish that (AP)2 duplexes are right-handed helices stabilized by WC base pairs (44–46). There is compression at the damage site of the duplexes pushing the abasic residues out of the helix and bringing the lesion flanking nucleotides close in space (Figure 2, peaks A and B). However, disparities on NOE interactions between base and sugar H2’, H2” and H3’ protons reveal that the extra helical character of the abasic residues in these duplexes is different. Sequential NOE peaks observed between G6 and G8 in one strand and A18 and C20 in the other strand of the (AP)2+1 duplex are equivalent (Figure 3), indicating that F7 and F19 are extruded from the helix to a similar extent. By contrast, the corresponding interactions between C20 and A22 residues are missing in the (AP)2−1 duplex (Figure 3), suggesting a different extra helical conformation for F21 in the (AP)2−1 cluster. Notwithstanding this difference, lesion counter residues can form a WC G•C base pair in both duplexes (Figure 4), preserving the loss of double stranded character at their lesion site.
Restrained MD reduced the NMR R-factors of the initial (AP)2−1 and (AP)2+1 models to about 20 and 22%, respectively (Table 1), confirming that the refined structures are accurate representations of the experimental NMR data. On both duplexes the AP residues bulge out appearing in the major groove of the (AP)2+1 cluster (Figures 6 and 8) or in the minor groove of the (AP)2−1 duplex (Figures 6 and 7) where F7 is partially extra helical. The extra helical location of the AP residues in the (AP)2+1 duplex bears striking similarity to the conformation of abasic frame-shift intermediates, where the single AP residue forms a bulge in the major groove of the duplex without perturbing the global DNA conformation (32, 33). The structure of bi-stranded (AP)2±1 clusters is also quite different from that of duplexes having a single abasic residue, where both the apyrimidinic site and counter nucleotide are always intra helical (19–21, 25–31) or the apurinic site and sometimes its partner base position outside the helix (21, 23, 26–27). Structurally, then, bi-stranded (AP)2±1 clusters are unique lesions that do not have the characteristics of two single AP sites put together.
In this study, we designed the (AP)2±1 duplexes to have one AP residue in each cluster opposite dC and the other opposite dG and establish whether the relative orientation of the lesions affected the putative formation of a central G•C base pair. Our results show that while lesion directionality within the cluster determines the groove location of the AP residues, formation of a proper G•C base pair occurs in both cases. Previously we determined the structures of bi-stranded (AP)2±1 duplexes, having dG and dA as AP site partners. Similar to the structures reported here, there was helix compression in both clusters bringing the orphan purine residues to register with the formation of G•A mismatches. In the +1 orientation, the AP residues bulged out toward the major groove and the resulting G•A alignment was highly propeller twisted and non-coplanar. In the −1 orientation, however, AP residues remained mostly aligned with the sugar-phosphate backbone, facilitating the formation of a coplanar G•A mismatch. Hydrogen bond interactions were absent in both mismatchs (34). While many more combinations of orphan bases are possible, it is apparent that helix compression, duplex collapse and subsequent formation of a base pair at the lesion site is a general structural feature of bi-stranded (AP)2±1 duplexes. The identity of the AP site partners will determine the quality of the lesion site base pair, matched or mismatched, affecting in this way the stability of the damaged duplex. The observation that, following the formation of the central G•C base pair, the duplexes have similar melting temperatures (Figure 5) fully supports the concept that orphan residues are main stability determinants on bi-stranded (AP)2±1 clusters. On the other hand, cluster orientation will determine the position of the abasic residues, which will be fully bulged out and in the major groove of the duplex for the +1 orientation (Figure 8) or aligned with the backbone, or partially extruded in the minor groove, for the alternative orientation (Figure 7).
Ape1 processing of (AP)2 duplexes
Several groups have shown that cleavage of bi-stranded (AP)2 lesions by recombinant hApe1, or their processing by nuclear cell extracts, is orientation specific such that (AP)2+1 clusters are incised more efficiently than (AP)2−1 lesions (10–17). In agreement with these reports, we observe here that purified hApe1 preferentially incises AP sites with the +1 orientation, preference that is remarkable in the case of F7 that has identical sequence context in both duplexes (Figure 9). The reasons for this differential processing, however, remain largely unknown. It has been proposed that bi-stranded abasic site lesions disrupt in general the footprint of hApe1 or that the presence of the second AP lesion hinders specific enzyme-DNA contacts. Alternatively, it was suggested that clustered lesions strongly destabilize the helix, increasing the partial single stranded character of the duplex at the lesion site and, thus, reducing the affinity for hApe1. Without answering all the questions, the results reported here can rule out some of the possibilities.
The lesion site G•C pair and the lesion flanking G•C and A•T base pairs are present in both duplexes (Figure 4), indicating preservation of the double helical character at their lesion site. In addition, the duplex melting temperatures are almost identical (Figure 5), establishing minimal stability differences between these clusters. Therefore, any putative increase of single stranded character at the lesion site of the duplex or thermal stability variations between bi-stranded (AP)2±1 clusters are not the reasons for their differential recognition by hApe1.
The X-ray structure of a damaged DNA/hApe1 complex shows that, upon binding, the enzyme kinks the DNA helix by 35° and extrudes the abasic residue into its preformed active site pocket. Protein residues contact the duplex at both the major and minor grooves and the insertion of protein loop at the lesion site stabilizes the AP site counter residue that remains intra helical (47, 48). Molecular modeling shows that, irrespective of its orientation, a second AP site can fit in the structure of the DNA/hApe1 complex without causing relevant atomic clashes (data not shown). Therefore, it is very unlikely that hindrance of specific enzyme-DNA contacts by bi-stranded (AP)2±1 substrates would account for their differential processing by hApe1.
The different groove location of AP residues in these clusters may partially explain the variations on the hApe1 efficiency. The X-ray structure of the hApe1/DNA complex showed several protein-DNA interactions that specifically defined the orientation of the scissile AP residue in the enzyme catalytic pocket. Our structure of the (AP)2+1 duplex shows the AP residues extruded on the major groove in a conformation that is close to that of the hApe1/DNA complex, suggesting that structural changes following encounter with the enzyme will be small and have low energy barriers. In contrast, minor groove location of abasic residues in the (AP)2−1 duplex is quite far from that on the productive hApe1/DNA complex and will require major structural adjustments to reach the active site of the enzyme and, thus, have higher energy costs. In addition, the abasic residues are more exposed and enzyme accessible in the (AP)2+1 than the (AP)2−1 duplex, favoring the formation of a protein/DNA complex in the former case.
Conclusions
The use of ionizing radiation is a vital strategy in the treatment for various cancers where cell killing is largely the result of DSB induced apoptosis. Abortive repair of clustered lesions can be the source of additional DSB increasing the lethal action of radiation. Unrepaired clustered damages will induce DNA mutations, increasing the toxicity of ionizing radiation. We solved the structures of bi-stranded (AP)2±1 clusters and confirmed previous measurement of differential cleavage by hApe1. Our results showed that the different groove location of the AP residues most likely explain for their differential hApe1 processing, ruling out thermal stability considerations or disruption of specific enzyme/DNA interactions as possible explanations. Ionizing radiation also forms bi-stranded clusters having non-adjacent lesion and BER enzymes process them with different efficiency. However, it cannot be assumed that these lesions will follow the same structural principles than (AP)2±1 duplex. Thus, additional studies are needed to establish their conformation and understand and the mechanisms responsible for their differential processing.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported NIH grants CA77094 and CA47995. Thanks to Mr. Erich Bremer for providing computer support services and for information that proved helpful in the creation of figures, and to Ms. Cecilia Torres for synthesis and purification of modified oligodeoxynucleotides. Molecular graphics images were produced using the MidasPlus program from the Computer Graphics Laboratory, University of California, San Francisco (supported by NIH RR-01081).
Footnotes
Data bank accession numbers
Atomic coordinates were deposited in the RCSB Protein Data Bank: ID codes 1YCW and 1YCT for the (AP)2-1 duplex and the (AP)2+1 duplex, respectively.
REFERENCES
- 1.Ward JF. DNA Damage Produced by Ionizing Radiation in Mammalian Cells: Identities, Mechanisms of Formation and Reparability. Prog. Nucleic Acids Res. Mol. Bio. 1998;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 2.Halliwell B, Aruoma OI. DNA damage by oxygen derived species: Its mechanism and measurement in mammalian systems. FEBS Let. 1991;281:9–19. doi: 10.1016/0014-5793(91)80347-6. [DOI] [PubMed] [Google Scholar]
- 3.Newcomb TG, Loeb LA. Oxidative Damage and Mutagenesis. In: Nickoloff JA, Hoekstra MF, editors. DNA Damage and Repair, Vol. 1: DNA Repair in Higher Eukaryotes. Totowa, NJ: Humana Press, Inc; 2001. pp. 65–84. [Google Scholar]
- 4.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered Damages and Total Lesions Induced in DNA by Ionizing Radiation: Oxidized Bases and Strand Breaks. Biochemistry. 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered Damages induced in isolated DNA and human cells by low doses of ionizing radiation. P.N.A.S. USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem. Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 7.Wallace SS. In: Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Scandalios JG, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 49–90. [Google Scholar]
- 8.Wilson DM, III, Engelward BP, Samson L. In: Prokaryotic Base Excision Repair, in DNA Damage and Repair Vol 1: DNA Repair in Prokaryotes and Lower Eukaryotes. Nickoloff JA, Hoekstra MF, editors. New Jersey: Humana Press; 1988. pp. 29–64. [Google Scholar]
- 9.Blaisdell JO, Wallace SS. Abortive base excision repair of radiation-induced clustered DNA lesions in Escherichia coli. P.N.A.S. USA. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgakilas AG, Bennet PV, Wilson DM, III, Sutherland BM. Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoetic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhury MA, Weinfeld M. Reactivity of Human Apurinic/Apyrimidinic Endonuclease and Escherichia coli Exonuclease III with Bistranded Abasic Sites in DNA. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhury MA, Weinfeld M. The Action of Escherichia coli Endonuclease III on Multiply Damaged Sites in DNA. J. Mol. Biol. 1995;249:914–922. doi: 10.1006/jmbi.1995.0348. [DOI] [PubMed] [Google Scholar]
- 13.Harrison L, Hatahet Z, Wallace SS. In vitro Repair of Synthetic Ionizing Radiation-induced Multiply Damaged DNA Sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 14.Weinfeld M, Rasouli-Nia A, Chaudhury MA, Britten RA. Response of Base Excision Repair Enzymes to Complex DNA Lesions. Radiat. Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.David-Cordonnier M-H, Cunniffe SMT, Hickson ID, O’Neill P. Efficiency of Incision of an AP Site within Clustered DNA Damage by the Major Human AP Endonucleases. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 16.Lomax ME, Cunniffe S, O’Neill P. Efficiency of Repair of an Abasic Site within DNA Clustered Damage Sites by Mammalian Cell Nuclear Extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 17.Tian K, McTigue M, de los Santos C. Sorting the consequences of ionizing radiation: processing of 8-oxoguanine/abasic site lesions. DNA Repair. 2002;1:1039–1049. doi: 10.1016/s1568-7864(02)00163-5. [DOI] [PubMed] [Google Scholar]
- 18.Wilson DM, III, Takeshita M, Grollman AP, Demple B. Incision Activity of Human Apurinic Endonuclease (Ape) at Abasic Site Analogs in DNA. J. Biol. Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 19.Kalnik MW, Chang C-N, Grollman AP, Patel DJ. NMR Studies of Abasic Sites in DNA Duplexes: Deoxyadenosine Stacks into the Helix Opposite Cyclic Analogue of 2’-Deoxyribose. Biochemistry. 1988;27:924–931. doi: 10.1021/bi00403a013. [DOI] [PubMed] [Google Scholar]
- 20.Kalnik MW, Chang C-N, Johnson F, Grollman AP, Patel DJ. NMR Studies of Abasic Sites in DNA Duplexes: Deoxyadenosine Stacks into the Helix Opposite Acyclic Lesions. Biochemistry. 1989;28:3373–3383. doi: 10.1021/bi00434a037. [DOI] [PubMed] [Google Scholar]
- 21.Cuniasse Ph, Fazakerley GV, Guschlbauer W, Kaplan B, Sowers LC. The Abasic Site as a Challenge to DNA Polymerase. A Nuclear Magnetic Resonance Study of G, C, and T Opposite a Model Abasic Site. J. Mol. Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- 22.Withka JM, Wilde JA, Bolton PH. Characterization of Conformational Features of DNA Heteroduplexes Containing Aldehidic Abasic Sites. Biochemistry. 1991;30:9931–9940. doi: 10.1021/bi00105a017. [DOI] [PubMed] [Google Scholar]
- 23.Singh MP, Hill GC, Péoc’h D, Rayner B, Imbach J-L, Lown JW. High-Field NMR and Restrained Molecular Modeling Studies on a DNA Heteroduplex Containing a Modefied Apurinic Abasic Site in the Form of Covalently Linked 9-Aminoellipticine. Biochemistry. 1994;33:10271–10285. doi: 10.1021/bi00200a007. [DOI] [PubMed] [Google Scholar]
- 24.Goljer I, Kumar S, Bolton PH. Refined Solution Structure of a DNA Heteroduplex Containing an Aldehydic Abasic Site. J. Biol. Chem. 1995;270:22980–22987. doi: 10.1074/jbc.270.39.22980. [DOI] [PubMed] [Google Scholar]
- 25.Wang KY, Parker SA, Goljer I, Bolton PH. Solution Structure of a Duplex DNA with an Abasic Site in a dA Tract. Biochemistry. 1997;36:11629–11639. doi: 10.1021/bi971464l. [DOI] [PubMed] [Google Scholar]
- 26.Coppel Y, Berthet N, Coulombeau C, Coulombeau C, Garcia J, Lhomme J. Solution Conformation of an Abasic DNA Undecamer Duplex d(CGCACXCACGC).d(GCGTGTGCG): The Unpaired Thymine Stacks Inside the Helix. Biochemistry. 1997;36:4817–4830. doi: 10.1021/bi962677y. [DOI] [PubMed] [Google Scholar]
- 27.Beger RD, Bolton PH. Structures of Apurinic and Apyrimidinic Sites in Duplex DNAs. J. Biol. Chem. 1998;273:1565–1573. doi: 10.1074/jbc.273.25.15565. [DOI] [PubMed] [Google Scholar]
- 28.Hoehn ST, Turner CJ, Stubbe J. Solution structure of an oligonucleotide containing an abasic site: evidence for an unusual deoxyribose conformation. Nucleic Acids Res. 2001;29:3413–3423. doi: 10.1093/nar/29.16.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de los Santos C, El-khateeb M, Rege P, Tian K, Johnson F. Structural Impact of C1’(OH) Conformation on Duplex DNA Containing Abasic Sites. Biochemistry. 2004;43:15349–15357. doi: 10.1021/bi048400c. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Dupradeau FY, Case DA, Turner CJ, Stubbe J. Nuclear Magnetic Resonance Structural Studies and Molecular Modeling of Duplex DNA Containing Normal and 4'-Oxidized Abasic Sites. Biochemistry. 2007;46:3096–3107. doi: 10.1021/bi6024269. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Dupradeau FY, Case DA, Turner CJ, Stubbe J. DNA Oligonucleotides with A, T, G or C Opposite an Abasic Site: Structure and Dynamics. Nucleic Acids Res. 2008;36:253–162. doi: 10.1093/nar/gkm622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin Z, Hung K-N, Grollman AP, de los Santos C. Solution Structure of Duplex DNA Containing an Extrahelical Abasic Site Analog Determined by NMR Spectroscopy and Molecular Dynamics. Nucleic Acid Res. 1998;26:2385–2391. doi: 10.1093/nar/26.10.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuniasse Ph, Sowers LC, Eritja R, Kaplan B, Goodman MF, Cognet JAH, Le Bert M, Guschlbauer W, Fazakerley GV. Abasic Frameshift in DNA. Solution Conformation Determined by Proton NMR and Molecular Mechanics Calculations. Biochemistry. 1989;28:2018–2026. doi: 10.1021/bi00431a009. [DOI] [PubMed] [Google Scholar]
- 34.Lin Z, de los Santos C. NMR Characterization of Clustered Bistrand Abasic Site Lesions: Effect of Orientation on their Solution Structure. J. Mol. Biol. 2001;308:341–352. doi: 10.1006/jmbi.2001.4587. [DOI] [PubMed] [Google Scholar]
- 35.States DJ, Haberkorn RA, Ruben DJ. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Res. 1982;48:286–292. [Google Scholar]
- 36.Plateau P, Gueron M. Exchangeable proton NMR without baseline distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 37.Brunger A. X-PLOR Version 3.1 A System for X-ray Crystallography and NMR. New Haven, CT: Yale University Press; 1993. [Google Scholar]
- 38.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and axis of curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 39.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 40.Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- 41.Majumdar A, Hosur RV. Simulation of 2D NMR spectra for determination of solution conformations of nucleic acids. Prog. NMR Spectrosc. 1992;24:109–158. [Google Scholar]
- 42.Rinkel LJ, Altona C. Conformational analysis of deoxyrybofuranos ring in DNA by means of sums of proton-proton coupling constants: a graphical method. J. Biomol. Struct. Dyn. 1987;4:621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- 43.Ryckert J-P, Ciccoti G, Berendsen HJC. Numerical integration of Cartesian equations of motion of a system with constraints-molecular-dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 44.van de Ven FJ, Hilbers CW. Nucleic acids and nuclear magnetic resonance. Eur. J. Biochem. 1988;178:1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- 45.de los Santos C. Probing DNA structure by NMR Spectroscopy. In: Kool E, Barton D, Nakanishi K, Meth-Cohn O, editors. Comprehensive Natural Products Chemistry, Vol. 7: DNA and Aspects of Molecular Biology. Oxford, UK: Elsevier Science Ltd; 1999. pp. 55–80. [Google Scholar]
- 46.Hare DR, Wemmer DE, Chou SH, Drobny G, Reid B. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J. Mol. Biol. 1983;171:319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- 47.Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS. The crystal structure of the human DNA repair endonuclease HAPE1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mol CD, Izuml T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 49.Ferrin TE, Huang CC, Jarvis LE, Lngridge R. The MIDAS display system. J. Mol. Graphics. 1988;6:13–27. [Google Scholar]
- 50.Huang CC, Pettersen TE, Ferrin TE, Lngridge R. Conic: A faster render for space-filling molecules with shadows. J. Mol. Graphics. 1991;9:230–236. doi: 10.1016/0263-7855(91)80016-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.