Abstract
Objective
Current cerebrospinal fluid (CSF) tests for sporadic Creutzfeldt-Jakob disease (sCJD) are based on the detection of surrogate markers of neuronal damage such as CSF 14-3-3 which are not specific for sCJD. A number of prion protein conversion assays have been developed, including real-time quaking induced conversion (RT-QuIC). The objective of this study is to investigate whether CSF RT-QuIC analysis could be used as a diagnostic test in sCJD.
Methods
An exploratory study was undertaken which analysed 108 CSF samples from patients with neuropathologically confirmed sCJD or from control patients. Of the 108 CSF samples 56 were from sCJD patients (30 female, 26 male, aged 31–84 years; 62.3 ± 13.5 years) and 52 were from control patients (26 female, 26 male, aged 43–84 years; 67.8 ± 10.4 years). A confirmatory group of 118 patients were subsequently examined which consisted of 67 cases of neuropathologically confirmed sCJD (33 female, 34 male, aged 39–82 years; 67.5 ± 9.0 years) and 51 control cases (26 female, 25 male, aged 36–87 years; 63.5 ± 11.6 years).
Results
The exploratory study showed that RT-QuIC analysis had a sensitivity of 91% and a specificity of 98% for the diagnosis of sCJD. These results were confirmed in the confirmatory study which showed that CSF RT-QuIC analysis had a sensitivity and specificity of 87% and 100% respectively.
Interpretation
This study shows that CSF RT-QuIC analysis has the potential to be a more specific diagnostic test for sCJD than current CSF tests.
INTRODUCTION
Creutzfeldt-Jakob disease (CJD) belongs to a family of fatal neurodegenerative diseases known as transmissible spongiform encephalopathies (TSEs) or prion diseases. The most common form of CJD, known as sporadic CJD (sCJD), presents with a rapidly progressing dementia, ataxia and myoclonus, and death typically occurs within 6 months. Neuropathologically these diseases are characterised by the post-translational conformational change of a normal cellular protein, called prion protein (PrPC) into a disease-associated form, termed PrPSc. PrPSc is partially protease-resistant and can induce PrPC to undergo a conformational change and produce more PrPSc in a self-propagating manner by a seeded aggregation process. The PrPSc, thus produced, accumulates throughout the brain and this is accompanied by spongiform change of the neuropil, neuronal loss and degeneration.
Apart from brain biopsy, there is no disease-specific pre-mortem diagnostic test for sCJD. Current clinical diagnostic criteria rely on clinical features and the results of investigations such as EEG, brain MRI and the presence of 14-3-3 in the cerebrospinal fluid (CSF).1,2 Since its introduction into the diagnostic criteria for sCJD in 1998, the analysis of CSF for 14-3-3 has become a widely accepted investigation in patients with suspected sCJD.3–5 This protein is released into the CSF as a result of acute neuronal damage and as such is a surrogate marker for the degenerative changes associated with sCJD. Indeed, there have been a number of concerns raised about the specificity of CSF 14-3-3.6,7
This has prompted the search for a more specific and disease-related pre-mortem diagnostic test for sCJD. The ability of PrPSc to convert PrPC into aggregated protease-resistant isoforms has been exploited using a variety of techniques such as protein misfolding cyclic amplification (PMCA) and quaking induced conversion (QuIC) which mimic the PrPC to PrPSc conversion process in an accelerated in vitro format. Using such techniques PrPSc has been detected in the blood of pre-clinical scrapie-infected hamsters,8 in the CSF of hamsters experimentally infected with scrapie9 and in sheep with clinical scrapie.10
A recent adaptation of QuIC (real-time QuIC) has been described which incorporates thioflavin T (ThT) along with recombinant PrP in the reaction mixture. The ThT binds to the aggregated recombinant PrP causing a change in the ThT emission spectrum that can be monitored in real-time. Recent studies have shown that CSF samples from hamsters inoculated with experimental scrapie and from patients with sCJD can be correctly identified using real-time QuIC (RT-QuIC).9,11,12 We now describe the findings of a blinded retrospective investigation into the value of RT-QuIC in the diagnosis of sCJD.
MATERIALS AND METHODS
Patients
The study included an exploratory group of 108 patients consisting of 56 cases of neuropathologically confirmed sCJD14 (30 female, 26 male aged 31 – 84 years (mean 62.3 ± 13.5 years) at notification) and 52 control cases (26 female, 26 male aged 43 – 84 years (mean 67.8 ± 10.4 years) at notification). A confirmatory group of 118 patients was subsequently examined. This group consisted of 67 cases of neuropathologically confirmed sCJD14 (33 female, 34 male aged 39 – 82 years (mean 67.5 ± 9.0 years) at notification) and 51 control cases (26 female, 25 male aged 36 – 87 years (mean 63.5 ± 11.6 years) at notification). All the control cases consisted of patients who were initially suspected of having sCJD but subsequently had either a pathologically proven alternative diagnosis or an alternative clinical diagnosis provided by either the clinical team or by a member of the NCJDRSU (Supplementary material Table 1). Of all the sCJD cases investigated 59 were homozygous for methionine at codon 129 of the PRNP gene, 25 were heterozygous for methionine and valine and 20 were homozygous for valine.
CSF samples
Consecutive CSF samples which had greater than 0.5ml available for analysis were selected. CSF samples were sent to the laboratory on dry ice and stored at −80°C prior to analysis. Consent for research was obtained for each sample and ethical approval was given by the MREC Scotland A (05/MRE00/67). All samples were analysed blinded to the final diagnosis.
Sporadic CJD brain tissues
Brain tissue was provided by the MRC Brain Bank in the NCJDRSU from examples of each of the six clinico-pathological sCJD subtypes as defined by the combination of their PRNP-codon 129 genotype (MM, MV, VV) and their PrPres type (type 1 or 2) according to the nomenclature of Parchi and Gambetti 14,15 as MM1, MM2-cortical subtype, MV1, MV2, VV1 and VV2. All cases used had been thoroughly examined histologically and biochemically and the diagnosis reached using internationally accepted criteria. In each case grey matter enriched samples of ~100mg were taken from the frontal cortex for preparation as seed material for RT-QuIC.
The amounts of brain homogenate introduced into the RT-QuIC assays were normalized according to the titre of PrPres. Proteinase K (PK) digested (50μg/ml) samples of sCJD brain homogenate were compared alongside dilutions of purified human recombinant PrP (rHuPrP). The levels of PrPres per unit of brain extract were estimated by comparing the densities of the single Western blot band corresponding to purified rHuPrP and the combined densities of the three bands corresponding to the PrPres in the brain extracts. This was used for calculation of serial dilutions of the brain homogenates to 100fg PrPres/2μl.
Aliquots of 10% w/v brain homogenate (BH) were thawed at room temperature freshly for each RT-QuIC assay. The brain homogenates were diluted by serial 1:10 dilutions using N2 buffer (PBS containing 0.1% SDS and 1 × N2 supplement [Invitrogen]). A 10μl sample was added to 90μl of N2 buffer, with vortex mixing between each serial dilutions (4x). Brain was diluted so that a 2μl volume of the final dilution would contain 100fg PrPres. For sCJD frontal cortex tissue samples the dilution of 100% brain tissue required for this varied from 5 × 10−6 to 7.5 × 10−5.
Control brain tissues
Frontal cortex tissue was also taken from two Alzheimer’s disease (AD) cases. These cases were referred to the NCJDRSU as suspected cases of CJD, but were found to have neuropathological evidence of AD at post-mortem and as such were used as negative controls. In addition, two further negative controls from individuals without neurodegenerative diseases were obtained from the MRC Sudden Death Brain and Tissue Bank (Sudden Death (SD) controls). The AD and SD BHs were diluted 1×10−6 with N2 buffer. Ethical approval for the use of brain tissue from the MRC NCJDRSU Brain and Tissue Bank was covered by Tayside B ethics committee 11/ES/0022 Edinburgh Brain Bank/
Preparation of hamster recombinant PrP
Hamster recombinant PrPC (rPrPC), (residues 23–231; accession K02234) was prepared according to the method of Wilham et. al.11 with minor modifications. The concentration of rPrPC was determined by BCA reagent (Thermo Scientific) and adjusted to 0.2–0.5 mg/mL. Aliquots were stored at −80°C until use. Purity of rPrPC was estimated at 99%, by SDS-PAGE and immunoblotting (data not shown). Human rPrP was investigated as a possible substrate for RT-QuIC. However, despite having a shorter lag-phase, the RT-QuIC reactions showed an increased number of spontaneous aggregations in the unseeded reactions and non-CJD control brain homogenate seeded reactions. For this reason all subsequent investigations used hamster rPrP.
Real-time QuIC Analysis
RT-QuIC buffer composition was as follows: 10mM phosphate buffer (pH 7.4), 170 mM NaCl (total 300mM including phosphate buffer), 0.1 mg/mL rPrPC (Hamster 23–231), 10 μM Thioflavin-T (ThT), and 10 μM ethylenediaminetetraacetic acid tetrasodium salt (EDTA). Reactions were prepared in a black 96-well, optical bottomed plate (Nalgene Nunc International, #265301) in volumes of 98 μL for brain homogenate (BH) seeded reactions and 85 μL for CSF seeded reactions. Correspondingly, 2 μL of diluted BH or 15 μL neat CSF were added to the wells for a final reaction volume of 100 μL. Each sample was run in quadruplicate, allowing 4 control samples [unseeded, SD BH, AD BH, sCJD-MM1 BH] and 20 CSF samples to be tested on one plate. The plates were sealed and incubated in a BMG OPTIMA FLUOstar plate reader at 42°C for 90–120 h with intermittent shaking cycles: double orbital with 1 minute shake (600rpm), 1 minute rest. ThT fluorescence measurements (450nm excitation and 480nm emission) were taken every 15 minutes.
The plate reader measures ThT fluorescence in relative fluorescence units (rfu) with saturation occurring at 65,000. The average fluorescence for each quadruplicate sample was monitored against time. A baseline rfu of ~5,000 for CSF and unseeded reactions and ~8,000 for BH seeded reactions was to be expected for the initial readings. This difference in baseline readings is probably due to the presence of a small amount of SDS (0.002%) in the BH seeded reactions and its absence in the CSF and unseeded reactions. Following a lag phase of ~10hrs for sCJD BH and >30hrs for sCJD CSF, positive samples displayed a rise in rfu. By 90hrs the majority of the sCJD CSF seeded reactions had shown an increase in fluorescence. A positive response was considered to have occurred if the average of the two highest readings at 90 hours was 10,000 rfu or greater. A cut-off value of 10,000 rfu was chosen based on the mean ± 3SD (5696 ± 1962) of the first 20 non-CJD CSF samples analysed.
SDS-PAGE and immunoblotting
For SDS-PAGE analysis, BH and CSF seeded RT-QuIC products were incubated with 6ug/mL proteinase K (PK) in the presence of 0.27% N-lauroylsarcosine sodium salt for 1 hr at 37°C followed by the addition of 2x SDS-PAGE sample buffer (containing 4M urea). CSF seeded RT-QuIC products were diluted a further 6-fold with sample buffer. BH seeded RT-QuIC products were not additionally diluted. PK digested samples were immunoblotted using polyclonal anti-PrP antibody R20.
CSF 14-3-3 analysis
CSF 14-3-3 analysis was undertaken as previously described.16
PRNP codon 129 genotype and PrP isotyping
Prion protein isotyping was performed on all suspected cases of prion disease where frozen brain tissue was received by the NCJDRSU as previously described.17 DNA was extracted from brain using standard techniques and analysed using the Helsinki method.18
Statistical analysis
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and efficiency of each marker were obtained. Statistical significance between groups was assessed using two-tailed Mann-Whitney U-test using 3 replicates.
RESULTS
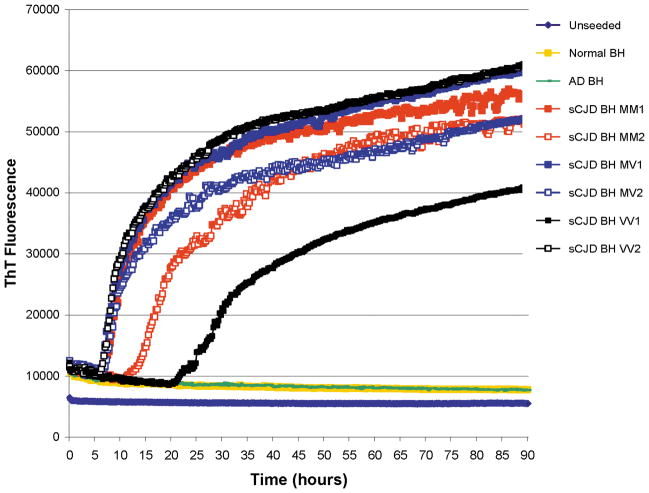
The kinetics of the RT-QuIC reactions seeded with sCJD, AD and SD BHs are shown in Figure 1. The BHs from all six subtypes of sCJD show positive responses which start after a lag phase of between 5 and 20 hours. This demonstrated that the hamster full-length rPrP could act as a universal substrate for all subtypes of sCJD. From Figure 1 it appears that the sCJD-MM2 (cortical subtype) and sCJD-VV1 BHs have longer lag phases than the other sCJD subtypes, however this was not a consistent finding. Flat responses were seen with the unseeded reactions and those reactions seeded with AD and SD BHs.
Figure 1.
The RT-QuIC responses from reactions seeded with the equivalent of 100fg PrPres from brain homogenates (BH) from six subtypes of sCJD, or an equivalent dilution (5 × 10−6) of an AD BH and a normal/sudden death (SD) BH. An unseeded reaction is also shown. Each point represents the mean of 4 replicate rfu readings.
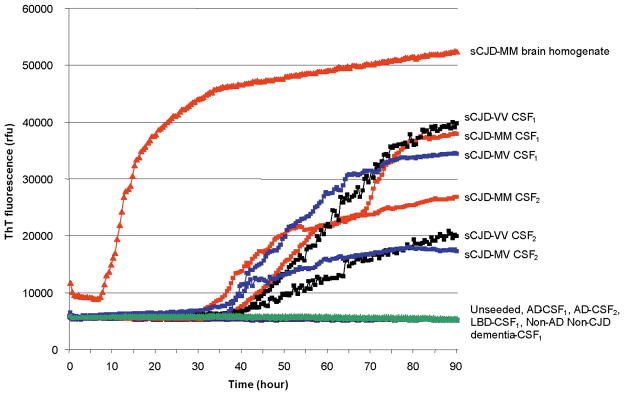
The RT-QuIC responses from reactions seeded with sCJD CSF samples show a similar pattern to those obtained using sCJD BHs, although there is a greater lag phase of approximately 30 hours and the maximal responses obtained are less (Figure 2).
Figure 2.
The RT-QuIC responses from reactions seeded with 100fg of sCJD-MM BH, 15μl of CSF from two sCJD-MM, two sCJD-MV and two sCJD-VV cases. The RT-QuIC responses from the unseeded reaction and from reactions seeded with two CSF samples from AD patients, one from a patient with Lewy body dementia and one from a patient with non-CJD, non-AD dementia are also shown. Each point represents the mean of 4 replicate rfu readings.
Positive RT-QuIC responses are seen in reactions seeded with sCJD CSF samples from all three genotypes of sCJD. There is marked variation in the rate of increase in the rfu signal between the sCJD CSF samples investigated, however by 90 hours the RT-QuIC response flattens out. The RT-QuIC responses at 90 hours for each CSF sample investigated in this study are shown in Figure 3. There is no relationship between the PRNP codon 129 status of the patient and the maximal rfu signal obtained by RT-QuIC analysis. In addition no correlation was found between the age at onset of disease, disease duration, timing of lumbar puncture and the final RT-QuIC response (r2 <0.05).
Figure 3.
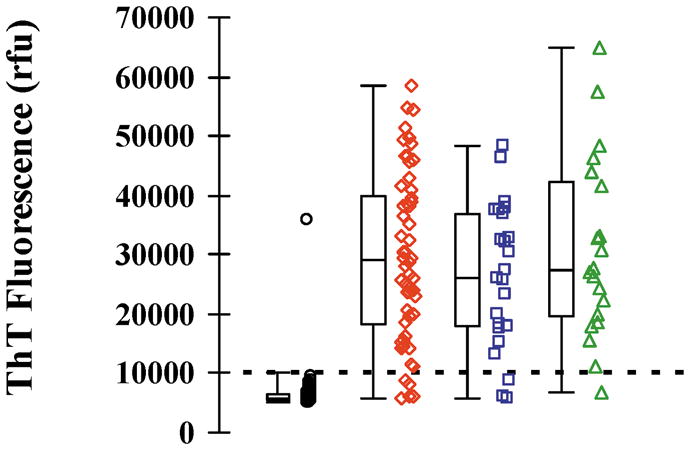
Combined scatter and box and whisker plots of the CSF RT-QuIC rfu in control patients (○), sCJD – MM genotype (◇), sCJD – MV genotype (□) and, sCJD – VV genotype (△). Each point represents the average of the two highest rfu readings for each patient investigated. Box and whisker plots show the 2nd, 25th, 75th and, 98th centiles.
Using the criteria of 10,000 rfu at 90 hours to investigate the exploratory group of patients we found that positive RT-QuIC responses were obtained in 51/56 CSF samples from sCJD patients and in one patient from the control group. This gave a sensitivity and specificity of 91% and 98% respectively (Table 1). These results were replicated in the confirmatory group of patients where positive RT-QuIC responses were found in 58/67 sCJD patients and in none of the 51 control patients investigated. This resulted in a sensitivity and specificity of 87% and 100% respectively.
Table 1.
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and efficiency for CSF RT-QuIC and 14-3-3 in neuropathologically confirmed sCJD for both the exploratory and confirmatory groups. Figures are given for each of the PRNP codon 129 genotypes of sCJD (MM: homozygous for methionine; MV heterozygous for methionine and valine and VV: homozygous for valine). The figures given are the number of positive or negative results over the total number of samples investigated. Efficiency was defined as:
| Exploratory Group | Confirmatory Group | Both Groups | ||||
|---|---|---|---|---|---|---|
| RT-QuIC | 14-3-3 | RT-QuIC | 14-3-3 | RT-QuIC | 14-3-3 | |
| sCJD | 51/56 | 52/56 | 58/67 | 64/67 | 109/123 | 116/123 |
| sCJD controls | 1/52 | 23/52 | 0/51 | 13/51 | 1/103 | 36/103 |
| Sensitivity (All) | 91% | 93% | 87% | 96% | 89% | 94% |
| Sensitivity (MM) | 90% | 97% | 90% | 93% | 90% | 95% |
| Sensitivity (MV) | 88% | 82% | 88% | 100% | 88% | 95% |
| Sensitivity (VV) | 100% | 100% | 92% | 100% | 95% | 100% |
| Specificity | 98% | 56% | 100% | 75% | 99% | 65% |
| PPV | 98% | 69% | 100% | 83% | 99% | 76% |
| NPV | 91% | 88% | 85% | 93% | 88% | 91% |
| Efficiency | 94% | 75% | 92% | 86% | 93% | 81% |
The sensitivity of RT-QuIC was comparable to CSF 14-3-3 a marker that is currently used for the investigation of patients with suspected sCJD (Table 1). Fourteen sCJD CSF samples were negative for RT-QuIC and there was no difference between these patients and those who where positive for RT-QuIC in terms of age at onset of disease, disease duration, timing of CSF sampling or PRNP codon 129 genotype. All but two of these 14 sCJD patients were positive for CSF 14-3-3 and one of the two remaining patients had triphasic waves on their EEG that were typical for sCJD. The remaining patient did not show any changes on the EEG or MRI that were typical for sCJD and a review of the neuropathology revealed changes that were consistent with a variably protease-sensitive prionopathy. This patient was homozygous for methionine at codon 129 of the PRNP gene; the brain showed spongiform change in the cerebral cortex and contained microplaques and plaque-like PrP deposits in the cerebellum, with plaque-like deposits in the cerebrum as recently described by Zou et al.19 The PrP deposits in this case showed more sensitivity to proteinase K digestion than those in sCJD (personal communication, James Ironside).
The specificity of RT-QuIC was substantially better than that of CSF 14-3-3 (Table 2). Only one control patient gave a positive RT-QuIC response. This patient presented with progressive memory loss, developed gait impairment and became mute. The patient had a positive CSF 14-3-3, the EEG showed non-specific changes and the MRI showed age-related vascular changes and the presence of a bilateral subdural haematoma. A clinical diagnosis of vascular dementia was made and after a disease duration of between 10–17 months the patient died. No post-mortem was performed. Further review of this patient’s notes by two independent neurologists concluded that a diagnosis of sCJD could not be excluded.
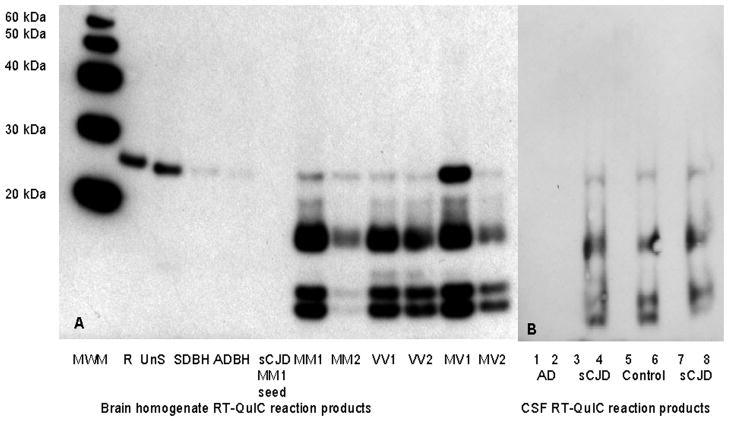
Because RT-QuIC uses an indirect measure (ThT binding) to assess rPrP aggregation state, the products formed by RT-QuIC reactions were subjected to mild PK digestion and Western blotting (Figure 4) to characterise the reaction product.
Figure 4.
SDS-PAGE and immunoblot of RT-QuIC products from reactions seeded with BHs from patients with sCJD, AD or SD. The unseeded RT-QuIC reaction products (UnS), the hamster rPrP (R) and the sCJD-MM1 BH seed are also shown. SDS-PAGE and immunoblot of RT-QuIC products from reactions seeded with CSF samples from AD (2), sCJD (4, 8) or from the control CSF sample that gave a positive reaction (6), the corresponding CSF samples used for seeding the reactions are shown in 1,3,5,7. The western blot was immunoblotted using R20 antibody. Protein molecular weight markers (MWM) are shown. All lanes, except R, have been treated with 6μg/ml PK for 1 hour at 37°C. A total of 4.5ng of untreated hamster rPrP was added to lane R. The amount of hamster rPrP in each of the RT-QuIC reaction mixtures loaded on the gel was 500ng, prior to PK digestion,
The gels were immunoblotted using R20, an antibody that binds to the C-terminus of the PrP20, which is included in the protease resistant core of PrPSc. Fig 4A shows that the products formed by RT-QuIC reactions seeded with sCJD BH contain a series of PK-resistant bands, the most prominent of which are less than 20kDa, in addition to a band at ~28kDa. The <20kDa bands are specific to RT-QuIC reactions seeded with sCJD BH whereas the ~28kDa band is found at variable intensity in all PK-treated RT-QuIC reaction products and represents residual full length rPrP that escaped proteolysis. A total of 4.5ng of untreated hamster rPrP was added to lane R, and, prior to PK digestion, the amount of hamster rPrP in each of the RT-QuIC reaction mixtures loaded on the gel was 500ng. The RT-QuIC reaction mixture seeded with MV1 sCJD BH had the largest amount of residual undigested hamster rPrP, which was estimated to be 9ng, based on the comparison of the band intensity to that of the untreated hamster rPrP (lane R). This means that <2% of the total rPrP present in each RT-QuIC reaction remained undigested with ~98% of the hamster rPrP having undergone at least partial PK digestion. Minor amounts of such residual full-length ~28 kDa rPrP bands are often seen in QuIC reaction products independent of the seeding status of the reactions10,21 and thus are inconsequential to the interpretation of the results. The molecular weights of the smaller PK-digested bands are similar to those previously reported for 263K scrapie seeded RT-QuIC and for 263K scrapie and sheep scrapie QuIC reactions.10,11 The banding pattern is similar in all of the subtypes of sCJD, although the intensity of the bands varies, with the MM2 and MV2 seeded reactions being less intense. RT-QuIC reactions which were unseeded or seeded with AD or SD BHs did not show any PK-digested bands in the <20kDa region. The products from RT-QuIC reactions seeded with sCJD CSF samples were found to be similar to those found in the sCJD BH (Fig 4B). No such PK-digested bands were found in the reaction seeded with an AD CSF sample. The RT-QuIC products from the reaction seeded with the control CSF sample which gave a positive RT-QuIC response, also showed the presence of PK-digested bands with a similar pattern and molecular weights to those found in sCJD BH and CSF samples.
DISCUSSION
The results from this small retrospective study show that RT-QuIC analysis of CSF samples from patients with suspected sCJD has a sensitivity and specificity of 89% and 99% respectively. This compares well with the results from Atarashi et al12 who reported a sensitivity of 80% and a specificity of 100%. There was found to be a single control patient who was unexpectedly positive by RT-QuIC. The electrophoretic pattern of the resultant RT-QuIC reaction products showed the presence of PK-digested bands with a similar pattern and molecular weights to those found in sCJD BH and CSF samples. Subsequent examination of the clinical records of this patient suggested that it was not possible to exclude a diagnosis of sCJD and as this patient died without a post-mortem a definitive diagnosis cannot be made.
The results from this study show that RT-QuIC has a comparable sensitivity to CSF 14-3-3 but is considerably more specific. There is no relationship between the RT-QuIC response obtained in sCJD cases and any clinical feature such as age at onset of disease, disease duration or timing of the lumbar puncture.
The products created from RT-QuIC reactions seeded with sCJD CSF samples contain hamster rPrP that has been converted to a partially PK resistant form, and is reminiscent of genuine disease associated PrP. It is partially protease resistant and present in N-terminally truncated forms. In contrast, as noted above, the ~28 kDa band most likely represents a small portion of the original full-length rPrP substrate that escaped exposure to PK, perhaps due to splashing up on the vessel wall or seed-independent, amorphous aggregation during the QuIC reaction (compare seeded and unseeded lanes in figure 4A). Variable amounts of such full-length rPrP residues have been observed previously in PK-digested QuIC reaction products without any correlation of the test sample to TSE.10,21 Although the nature of the material present in CSF of CJD patients that triggers the RT-QuIC reaction is not known it seems plausible that it is a disease-associated form of PrP and therefore specific to CJD. This is in contrast to CSF 14-3-3 which is a neuronal protein released into the CSF as a result of neuronal damage and is therefore a surrogate marker for CJD. We suggest that it is this difference that accounts for the higher specificity seen with RT-QuIC when compared to CSF 14-3-3.
The RT-QuIC products created from reactions seeded with sCJD BH or CSF samples show similar PK-resistance and PrP fragmentation pattern on Western blotting, suggesting that the ability to “seed” the RT-QuIC is similar in both BH and CSF samples from sCJD. It is not known whether the RT-QuIC products created from these seeding reactions are associated with infectivity. However, multiple intracerebral inoculations of hamster rPrP RT-QuIC products seeded with hamster scrapie BH have not induced scrapie in hamsters (J. Wilham and B. Caughey, unpublished data).
In conclusion, this study demonstrates that RT-QuIC analysis of CSF samples from patients with suspected sCJD has the potential to be a more accurate pre-mortem diagnostic test for sCJD than CSF 14-3-3, most probably because the analyte is a disease-specific rather than surrogate marker. However, further studies are required to assess the sensitivity of CSF RT-QuIC in each of the sCJD subgroups and to assess the complementary role that CSF 14-3-3 and RT-QuIC may have in the clinical diagnosis of sCJD. Further studies, such as eQuIC21 are being pursued to improve the sensitivity whilst maintaining the high degree of specificity of RT-QuIC. In addition, greater numbers of CSF samples need to be investigated from patients with the widest possible spectrum of TSEs to gain a better understanding of how this potential new test can be used in both clinical and surveillance settings.
Supplementary Material
Acknowledgments
This study has been funded by Alliance Biosecure, a Small Projects Grant from the University of Edinburgh Development Trust and the Scottish Government’s Chief Scientists Office, Government of Scotland (LMcG, AP). The National CJD Research & Surveillance Unit is funded by the Department of Health and the Scottish Home Office Department of Health (MA, MH, RW, RK, AG). This work was supported in part by the Intramural Research Program of the NIAID, NIH (CO, JW, BC) and by the Department of Health (England) (NA, GM). The Edinburgh Brain Bank is supported by the Medical Research Council (G1000681). We are indebted to Professor James Ironside (NCJDRSU) for providing brain tissue, neuropathological expertise and for his critical reading of this manuscript. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health.
Footnotes
Author Contributions
LMcG and AP performed the RT-QuIC analysis, CO and JW involved in training LMcG and AP in the RT-QuIC analysis, NA and GM prepared and supplied the hamster recombinant PrP, MA was responsible for the analysis of CSF 14-3-3, MH, BC, RW, RK and AG were responsible for the design and interpretation of the study.
Conflicts of Interest
There were no conflicts of interest
References
- 1.World Health Organisation. Report of a WHO consultation on global surveillance, diagnosis and therapy of human transmissible spongiform encephalopathies. Geneva, Switzerland: WHO; Feb, 1998. [Google Scholar]
- 2.Zerr I, Kallenberg K, Summers DM, et al. Updated clinical diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Brain. 2009;132:2659–2668. doi: 10.1093/brain/awp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Juan P, Green A, Ladogana A, et al. CSF tests in the differential diagnosis of Creutzfeldt-Jakob disease. Neurology. 2006;67(4):637–643. doi: 10.1212/01.wnl.0000230159.67128.00. [DOI] [PubMed] [Google Scholar]
- 4.Collins S, Boyd A, Fletcher A, et al. Creutzfeldt-Jakob disease: diagnostic utility of 14-3-3 protein immunodetection in cerebrospinal fluid. J Clin Neurosci. 2000;7(3):203–208. doi: 10.1054/jocn.1999.0193. [DOI] [PubMed] [Google Scholar]
- 5.Shi Q, Gao C, Zhou W, et al. Surveillance for Creutzfeldt-Jakob disease in China from 2006–2007. BMC Public Health. 2008;8:360. doi: 10.1186/1471-2458-8-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh K, Shirabe S, Eguchi H, et al. 14-3-3 protein, total tau and phosphorylated tau in cerebrospinal fluid of patients with Creutzfeldt-Jakob disease and neurodegenerative disease in Japan. Cell Mol Neurobiol. 2006;26:45–52. doi: 10.1007/s10571-006-9370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geschwind MD, Martindale J, Miller D, et al. Challenging the clinical utility of the 14-3-3 protein for the diagnosis of sporadic Creutzfeldt-Jakob disease. Arch Neurol. 2003;60(6):813–6. doi: 10.1001/archneur.60.6.813. [DOI] [PubMed] [Google Scholar]
- 8.Saa P, Castilla J, Soto C. Presymptomatic detection of prions in blood. Science. 2006;313:92–94. doi: 10.1126/science.1129051. [DOI] [PubMed] [Google Scholar]
- 9.Atarashi R, Wilham JM, Christensen L, et al. Simplified ultrasensitive detection of recombinant PrP conversion with shaking. Nat Methods. 2008;5:211–212. doi: 10.1038/nmeth0308-211. [DOI] [PubMed] [Google Scholar]
- 10.Orru CD, Wilham JM, Hughson AG, et al. Human variant CJD and sheep scrapie PrPres detection using seeded conversion of recombinant prion protein. Protein Engineering, Design and Selection. 2009;22(8):515–21. doi: 10.1093/protein/gzp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilham JM, Orru CD, Bessen RA, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathogens. 2010;6(12):e1001217. doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atarashi R, Satoh K, Sano K, et al. Ultrasensitive human prion detection in cerebrospinal fluid by real-time quaking induced conversion. Nature Medicine. 2011;17(2):175–8. doi: 10.1038/nm.2294. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie D, Ironside J. Clinical and Neuropathological Investigations in Creutzfeldt-Jakob disease. Adv Clin Neurosci Rehabilitation. 2006;5:620–22. [Google Scholar]
- 14.Gambetti P, Kong Q, Zou W, et al. Sporadic and familial CJD:classification and characterisation. Br Med Bull. 2003;66:213–39. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 15.Parchi P, Giese A, Capellari S, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;42(2):224–33. [PubMed] [Google Scholar]
- 16.Chohan G, Pennington C, MacKenzie JM, et al. The role of cerebrospinal fluid 14-3-3 and other proteins in the diagnosis of sporadic Creutzfeldt-Jakob disease in the United Kingdom: a 10 year review. J Neurol Neuropath Psych. 2010;81(11):1243–8. doi: 10.1136/jnnp.2009.197962. [DOI] [PubMed] [Google Scholar]
- 17.Hill A, Joiner S, Wadsworth JDF, et al. Molecular classification of sporadic Creutzfeldt-Jakob disease. Brain. 2003;126:1333–1346. doi: 10.1093/brain/awg125. [DOI] [PubMed] [Google Scholar]
- 18.Nurmi MH, Bishop M, Strain L, et al. The normal population distribution of PRNP codon 129 polymorphism. Acta Neurol Scand. 2003;108:374–378. doi: 10.1034/j.1600-0404.2003.00199.x. [DOI] [PubMed] [Google Scholar]
- 19.Zou W-Q, Puoti G, Xiao X, et al. Variably protease-sensitive prionopathy: a new sporadic disease of the prion protein. Ann Neurol. 2010;68:162–172. doi: 10.1002/ana.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caughey B, Raymond GJ, Ernst D, Race RE. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orrú CD, Wilham JM, Raymond LD, et al. Prion disease blood test using immunoprecipitation and improved quaking-induced conversion. MBio. 2011;2(3):e00078-11. doi: 10.1128/mBio.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.