Abstract
Ribonucleotide reductase (RNR) is an essential enzyme for all living organisms since is the responsible for the last step in the synthesis of the four deoxyribonucleotides (dNTPs) necessary for DNA replication and repair. In this work, we have investigated the expression of the three-RNR classes (Ia, Ib and III) during Escherichia coli biofilm formation. We show the temporal and spatial importance of class Ib and III RNRs during this process in two different E. coli wild-type strains, the commensal MG1655 and the enteropathogenic and virulent E2348/69, the prototype for the enteropathogenic E. coli (EPEC). We have established that class Ib RNR, so far considered cryptic, play and important role during biofilm formation. The implication of this RNR class under the specific growth conditions of biofilm formation is discussed.
Introduction
Escherichia coli biofilm development is a complex molecular process that involves a large number of genetic factors and genes. When the global gene expression profiles of biofilm and planktonic E. coli cells are compared, very significant differences are apparent [1]. Biofilm formation is underlying catheter-associated urinary tract infections (CAUTIs), urinary tract infections (UTIs) caused by uropathogenic Escherichia coli (UPEC), and the various types of diarrhea caused by enterohemorrhagic, enteroinvasive and enteroaggregative E. coli. The persistence of these biofilms may contribute to such infections becoming chronic conditions [2].
The complexity of biofilm formation makes it difficult to precisely identify the regulatory networks and the processes of alteration of gene expression which account for its development. In this context, it is important to understand how DNA synthesis is regulated and which factors participate in this process.
Although E. coli K-12 is not as proficient in making biofilms as other E. coli isolates, it has nonetheless been used in many studies, for instance to elucidate changes in transcriptomic profiles between planktonic cells and biofilm [1], [3], [4], [5]. Confocal analysis has evidenced a well-defined three-dimensional colonial structure with a mushroom form in K-12 strains and also the enteroaggregative E2348/69 E. coli strain [3], [6].
Ribonucleotide reductases (RNR) are essential enzymes in all living cells. These proteins catalyze the reduction of ribonucleotides (NTPs) to the corresponding deoxyribonucleotides (dNTPs), thus providing the buildings blocks for DNA synthesis and repair [7]. The three known RNR classes (I, II and III) use free radicals for catalysis but rely on different metallocofactors for the initiation of the radical reduction process, each exhibiting a different behavior towards oxygen [8]. Class I RNRs contain a stable tyrosyl radical and an oxygen-linked di-iron center required for the production of free radicals, and class I enzymes are only functional under aerobic conditions. This class of RNRs is further subdivided into classes Ia and Ib, because of significant differences in allosteric regulation and gene organization. Class Ia RNRs are encoded by the nrdA-nrdB genes, whereas class Ib RNRs are encoded by the nrdH-nrdI-nrdE-nrdF genes. It has been recently shown that class Ib small subunit (NrdF) stabilizes the tyrosyl radical by a dimanganese-oxo center [9], [10].Class II RNRs are coenzyme B12-dependent and can be active under both aerobic and anaerobic conditions. Lastly, class III RNRs, encoded by the nrdD-nrdG genes, carry a stable but oxygen-sensitive glycyl radical, and are only functional under anaerobic conditions. While almost all eukaryotic organisms encode exclusively class I RNRs, prokaryotes are known to encode more than one RNR class [8]. And additional protein, termed NrdR, was recently described as a novel transcriptional regulator capable of modulating the expression of all RNRs present in one organism [11].
E. coli and all enterobacteria encode three different RNR classes in their genome: Ia, Ib and III. While in E. coli class Ia RNR is active during aerobiosis and class III is active during anaerobiosis [12], [13], class Ib RNR has been considered a cryptic enzyme with no apparent function [7], [14]. However, a physiological role during growth of E. coli under iron starvation conditions has been assigned to class Ib. This enzyme requires manganese for tyrosyl radical generation and can replace the iron-dependent class Ia RNR [15]. A yet unanswered question is why E. coli encodes so many different RNRs which are apparently redundant. A hypothesis to be tested is that the different RNR classes present in E. coli are differentially expressed when cells deal with specific environmental signals, such as those found within the host or during biofilm formation. In this paper we evidence that when E. coli cells develop biofilms, expression of the different RNR enzymes is different to that observed in planktonic cells.
Results and Discussion
E. coli class Ib (nrdE) and class III (nrdD) RNR genes are key enzymes in biofilm formation
In this work we have used two different E. coli wild-type strains: the commensal MG1655 [16] and the enteropathogenic (EPEC) and virulent E2348/69 [17], this latter being the prototype for the EPEC E. coli strains involved in human disease and that still remains as the leading cause of infantile diarrhea in developing countries. EPEC colonize the proximal small intestine, where they adhere to epithelial cells forming microcolonies. Typically, EPEC strains form biofilms more complex in structure than MG1655 [6], [17]. Since class Ia RNR mutants are non-viable under normal laboratory growth conditions [14] we have studied only class Ib (ΔnrdE) and III (ΔnrdD) mutants. These mutants have been used to assay surface-associated biofilm formation on polyvinyl chloride (PVC) 96-well plates as described in the methods section.
Deletion of the nrdE gene in strains MG1655 and E2348/69 (ETS104 and ETS108) reduced by about 44% and 28% biofilm formation with respect to parental wild-type strains (Fig.1A). Quantification of viable cells in the ΔnrdE mutant revealed a 13.6% and 20.1% increase of planktonic cells when compared to parental wild-type strains, thus corroborating the noted decrease in biofilm formation (data not shown).
Figure 1. The Escherichia coli ΔnrdE and ΔnrdD mutants are defective in biofilm formation.
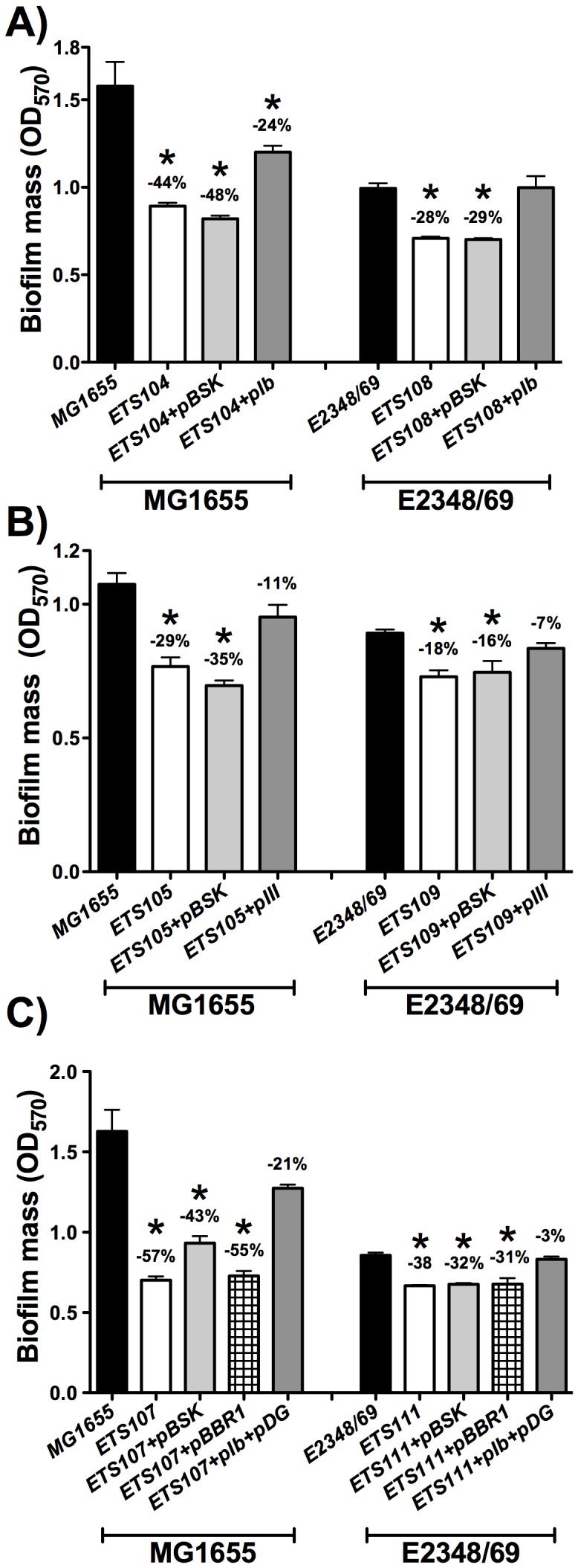
Biofilms of the wild-type E. coli strains (MG1655 and E2348/69) are compared to their isogenic ΔnrdE mutant in A), ΔnrdD mutant in B) and ΔnrdEΔnrdD double mutant in C). The values shown are the means of at least 4–5 independent experiments with six wells in each and the error bars represent standard deviations. Differences with respect to the wild-type strain were statistically significant for all the pairs of strains (*, P<0.05 by the Mann-Whitney test). Plots and statistics were generated using GraphPad Prism 5.0 software.
With respect to the nrdD gene, mutants lacking this enzyme (ETS105 and ETS109) produced in both strains 29% less biofilm formation than the corresponding parental wild-type strains (Fig.1B). To correlate loss of function of nrdD and nrdE genes and alterations in biofilm formation, these mutations were complemented with the corresponding wild-type alleles and biofilm formation was tested in these constructs. When plasmids containing the nrdHIEF and nrdDG operon (pIb and pIII) were transformed in the ETS104/ETS108 (Fig.1A) and ETS105/ETS109 strains (Fig.1B), the biofilm formation level was restored to levels comparable to those observed in the corresponding wild-type strains (MG1655 and E2348/69). We decided next to combine the two mutations. A double ΔnrdEΔnrdD mutation (ETS107 and ETS111) produced biofilm formation levels that were 57% and 38% lower than the wild-type strains (Fig.1C).
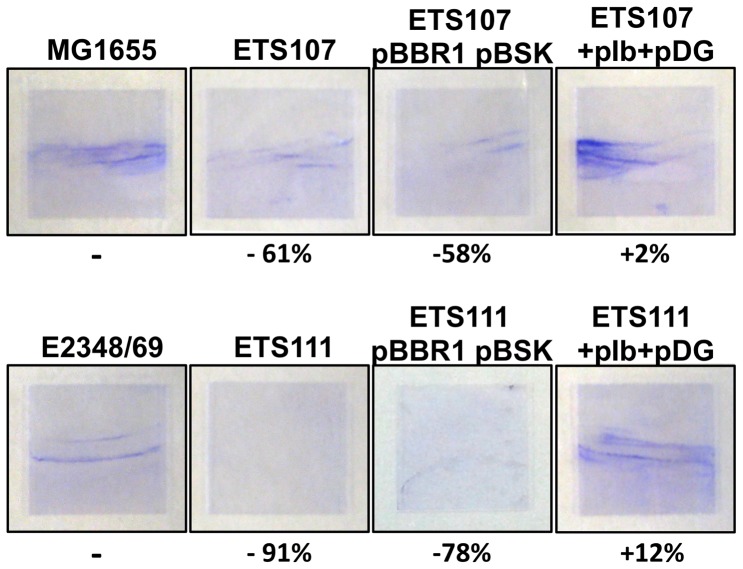
The absorbance levels of biofilm formation on a microtitter plate of strains MG1655 and E2348/69 are not high, but they proved to be highly reproducible. To corroborate the microtiter biofilm formation data, we performed additional assays in a glass tube, testing thus biofilm formation on a different surface. Fig.2 shows the biofilm formation of both wild-type E. coli strains and their corresponding double ΔnrdE and ΔnrdD mutant derivatives growing under aerobic conditions. Simultaneous deletion of the nrdE and nrdD genes in strains MG1655 or E2348/69 results either in a drastic reduction of biofilm (61% in strain ETS107) or almost completely abolishing it (91% in strain ETS111). When the double mutants ETS107 and ETS111 were complemented with plasmids pIb and pDG the biofilm formation level was restored to levels comparable to those of the corresponding parent strains (MG1655 and E2348/69). Glass tube biofilm formation was also checked by growing the different strains under microaerophlic conditions but significant biofilm growth could not be observed (data not shown).
Figure 2. Biofilm formation in glass surface also shows that double ΔnrdEΔnrdD mutants are defective in biofilm formation.
Biofilm formation was compared in wild-type E. coli MG1655 and E2348/69 strains to their isogenic double ΔnrdE and ΔnrdD mutants. Complementation experiment and controls are also shown.
All these results evidence that both class Ib and class III RNRs play a significant role during E. coli biofilm formation.
The transcriptional factor nrdR induces biofilm formation through the activation of class Ib and III genes
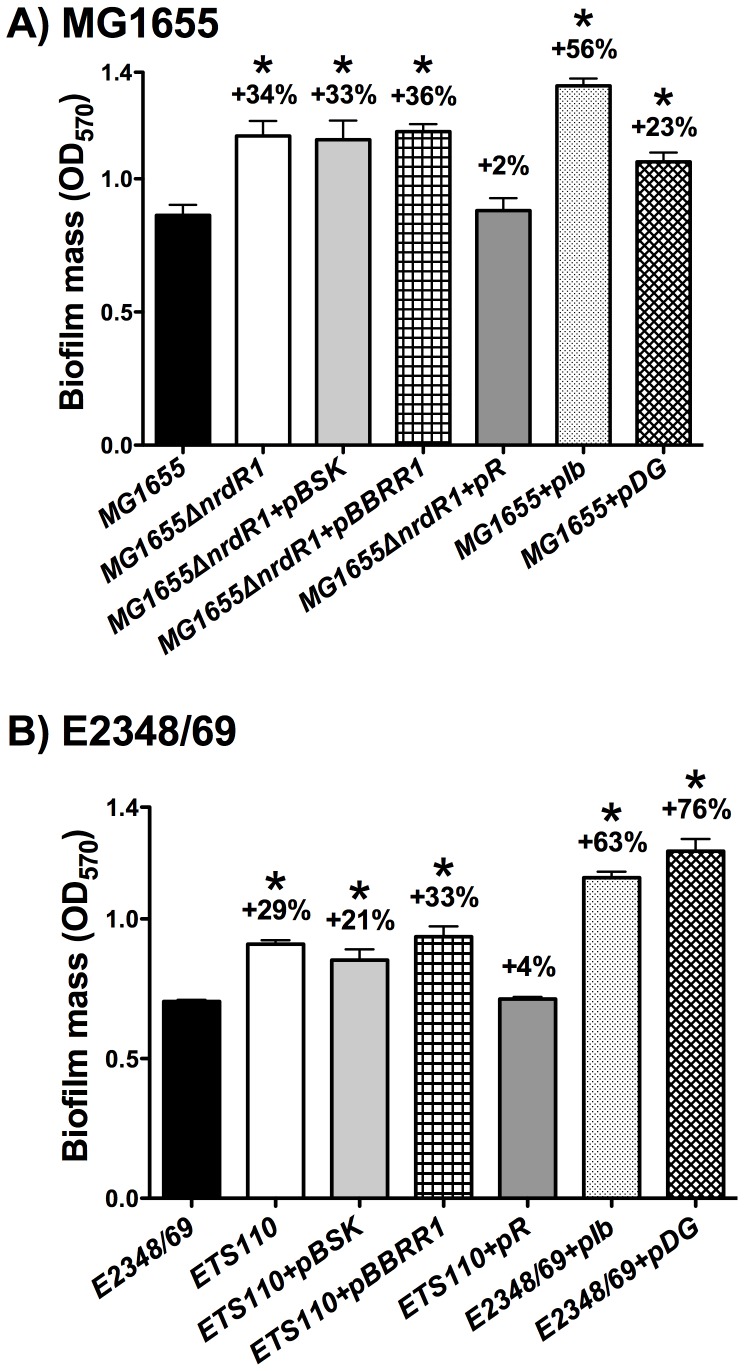
The transcriptional factor NrdR acts as a repressor of the three-nrd genes in E. coli [11]. We decided to test if this protein plays a role in biofilm formation. To this purpose, ΔnrdR mutant derivatives were constructed in both MG1655 and E2348/69 strains. When compared to the corresponding parent strains, biofilm formation was higher (by 34% and 29%) in the ΔnrdR mutant derivatives (MG1655ΔnrdR1 and ETS110) (Fig3a and 3b). This suggests nrdE and nrdD up-regulation in ΔnrdR cells. Furthermore, a complementation test using pR plasmid (Table1) restored the biofilm levels to those of the wild-type strains. With respect to the mechanism by which NrdR enhances E. coli biofilm formation, it should be commented here that it had previously been shown that NrdR deficiency strongly increases class Ib RNR transcription from 25 to 50 times and class III RNR from 6 to 10 times in strain MG1655 depending on the growth phase [11]. We decided to measure transcription of nrdE in E. coli E2348/69strain and verify the results in the MG1655 strain by RT-PCR in cells grown at OD550 = 0.2 and 0.8. The expression level of the nrdE (class Ib) gene resulted to be 15.24 and 23.84 fold higher (means of three different experiments) in the E2348/69ΔnrdR mutant compared to the wild-type E2348/69 cells. To test whether the increased biofilm formation in an nrdR mutant was due either to class Ib or III RNR, we independently overexpressed the entire class Ib operon (nrdHIEF) and class III operon (nrdDG) in wild-type strains MG1655 and E2348/69 by using plasmids pIb and pIII (see Table1). We found a 56% and 63% higher level of biofilm formation compared to that produced with the ΔnrdR mutant when class Ib RNR was overproduced and 23% and 76% when is overproduced class III RNR. Again, these results corroborate the importance of both RNR classes (Ib and III) during biofilm formation.
Figure 3. The E. coli ΔnrdR mutant enhances biofilm formation through the class Ib and III RNR classes.
Biofilms of the wild-type E. coli strains MG1655 in panel A and E2348/69 in panel B are compared to their isogenic ΔnrdR mutant. The values shown are the means of at least 7–8 independent experiments with six wells in each and the error bars represent standard deviations. Differences with respect to the wild-type strain were statistically significant for all the pairs of strains (*, P<0.05 by the Mann-Whitney test). Plots and statistics were generated using GraphPad Prism 5.0 software.
Table 1. Strains, plasmids and bacteriophages used in this study.
| Strain or plasmids | Description | Source |
| Plasmids | ||
| pGEM-T easy | A/T cloning vector (AmpR) | Promega |
| pBluescriptSK | High-copy number cloning vector (AmpR) | Stratagene |
| pBBR1MCS-5 | High-copy number cloning vector (GmR) | [36] |
| pETS130-GFP | Promoterless GFP (GmR) | [34] |
| pIb | nrdHIEF operon cloned into BamHI site of pBluescriptSK(+) | This work |
| pIII | nrdDG operon cloned into BamHI site of pBluescriptSK(+) | This work |
| pR | nrdR cloned into BamHI site of pBluescriptSK(+) | This work |
| pDG | nrdDG operon cloned into BamHI site pBBR1MCS-5 | This work |
| pETS150 | pETS130 derivative carrying nrdA promoter | This work |
| pETS151 | pETS130 derivative carrying nrdH promoter | This work |
| pETS152 | pETS130 derivative carrying nrdD promoter | This work |
| Strains | ||
| DH5α | recA1 endA1 hsdR17 supE44 thi-1 relA1 Δ(lacZYA-argF)U169 deoR Φ80dlacZM15 | Laboratory stock |
| JW2650 | E. coli K-12 strain BW25113 ΔnrdE::kan (KanR) | [30] |
| JW4197 | E. coli K-12 strain BW25113 ΔnrdD::kan (KanR) | [30] |
| JW5437 | E. coli K-12 strain BW25113 ΔrpoS::kan (KanR) | [30] |
| UA6068 | E. coli MC1061 ΔnrdD:: Cm (CmR) | [14] |
| MG1655 | E. coli MG1655 wild type | |
| MG1655ΔnrdR1 | E. coli MG1655 with a HpaI-Bsp119I nrdR deletion | [11] |
| ETS104 | E. coli MG1655 ΔnrdE::kan (KanR) | This work |
| ETS105 | E. coli MG1655 ΔnrdD::kan (KanR) | This work |
| ETS106 | E. coli MG1655 ΔnrdR::kan (KanR) | This work |
| ETS107 | E. coli MG1655 ΔnrdE::kan (KanR) ΔnrdD::Cm (CmR) | This work |
| ETS113 | E. coli MG1655 ΔrpoS::kan (KanR) | This work |
| E2348/69 | E. coli 0127:H6 E2348/69 wild type enteropathogenic | [17] |
| ETS108 | E. coli E2348/69 ΔnrdE::kan (KanR) | This work |
| ETS109 | E. coli E2348/69 ΔnrdD::kan (KanR) | This work |
| ETS110 | E. coli E2348/69 ΔnrdR::kan (KanR) | This work |
| ETS111 | E. coli E2348/69 ΔnrdE::kan (KanR) ΔnrdD::Cm (CmR) | This work |
| ETS112 | E. coli E2348/69 ΔrpoS::kan (KanR) | This work |
| Phagues | ||
| P1vir Phage | Laboratory stock | |
Differential expression of E. coli RNR genes during biofilm formation
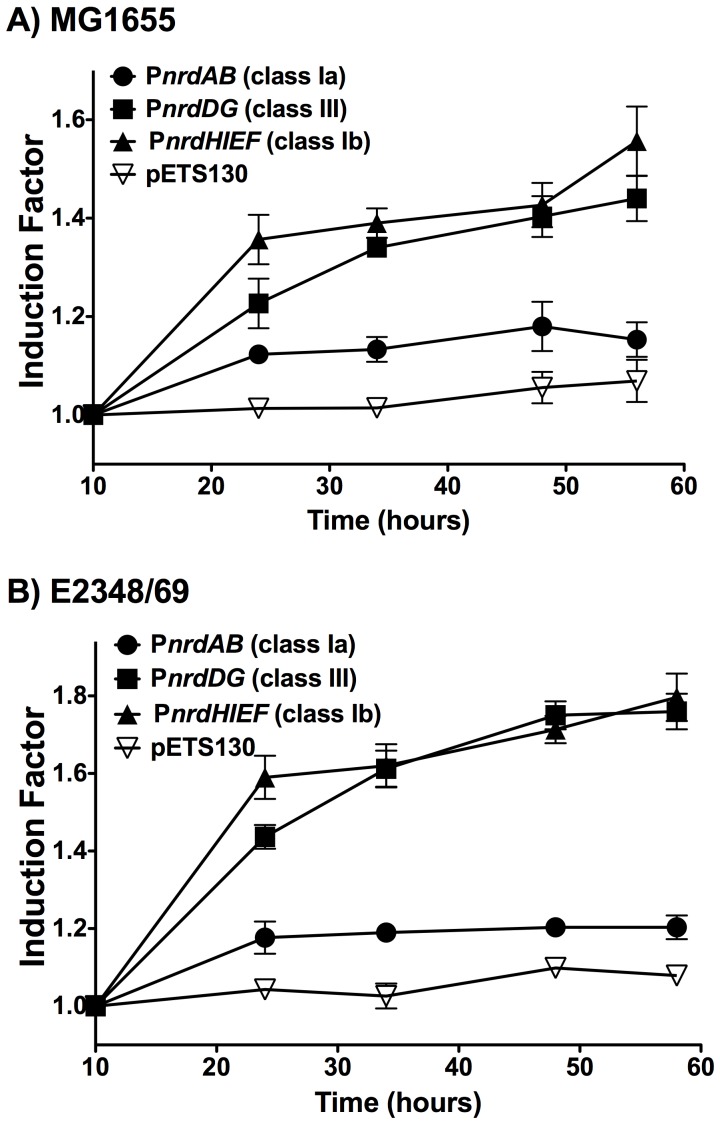
Previously published data (NCBI GEO dataset; GDS2768, GDS2753, GSE18362, GSE13418) have rendered contradictory results with respect to the differential expression of the nrd genes in E. coli biofilms vs. planktonic cells: some of the nrd genes have been found to be up-regulated in some studies and down-regulated in others. Due to these contradictory observations we decided to carry out gene expression analysis of each nrd promoter during the course of the biofilm formation. We used E. coli MG1655 and E2348/69 cells transformed with plasmids carrying the transcriptional fusion of each nrd promoter to the green fluorescent protein (pETS150, pETS151 and pETS152) and the control plasmid pETS130 as described in the material and methods section. Fig.4 shows induction of expression of different nrd genes indicated as a fold-change compared to the 10 h biofilm formation. In this experiments we clearly see the in vivo RNR class expression shift during the course of biofilm formation. We observed that E. coli MG1655 (Fig.4A) and E2348/69 (Fig.4B) exhibited the lowest expression of PnrdAB along the biofilm formation. However, the PnrdHIEF operon exhibited the highest increase of expression in the first hours of the biofilm formation. In a mature biofilm (≈55 h) both the PnrdHIEF and PnrdDG construction are significantly induced (1.6 and 1.8 fold induction in the E. coli MG1655 and E2348/69 respectively) and (1.4 and 1.8 fold induction in the E. coli MG1655 and E2348/69 respectively). In contrast the expression of the PnrdAB promoter remained the same along the time, thus demonstrating the scarce importance of this operon during the biofilm formation. Control plasmid did not show any expression variation along the experiment.
Figure 4. Expression of E. coli RNR classes during biofilm formation.
Curves show fold changes expression of the different RNR promoter classes (PnrdAB – class Ia; PnrdDG – class III; PnrdHIEF – class Ib; pETS130 as a empty vector) from different times points during the biofilm formation of MG1655 (A) and E2348/69 (B) E. coli strains. All values are normalized respect the expression detected in biofilm after 10 h. The values shown are the means of three independent experiments with six wells in each and the error bars represent standard deviations.
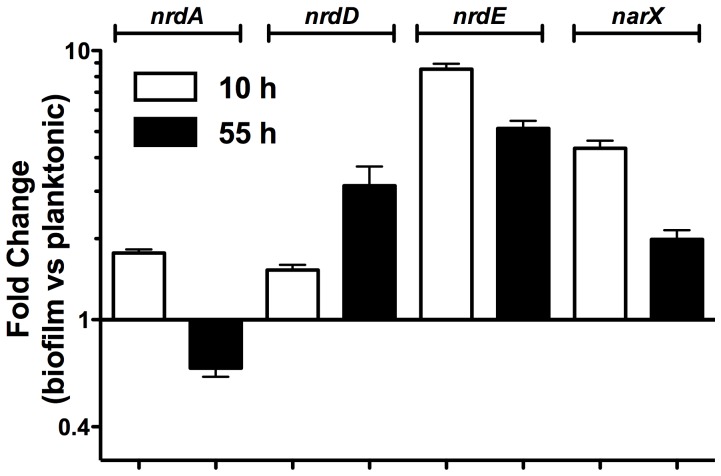
To corroborate these data we measured expression of nrdA, nrdD and nrdE by real-time PCR with probes from nrdA, nrdD and nrdE [11] in RNA samples extracted from biofilms and from planktonic MG1655 E. coli cells grown for 10 h and 55 h. Figure5 shows the differential expression pattern for each of the nrd genes. Biofilm formation led to nrdA (class Ia) gene repression along the biofilm formation (1.55 fold repression in cells collected at 55 h). In contrast, when compared to the level in planktonic cells, expression of nrdE (class Ib) and nrdD (class III) was higher in biofilm forming cells (from 1.5- to 9-fold). The highest expression level takes place in the nrdE gene at any time interval. It is important to note that the expression level of the nrdD gene is moderate (only 1.5 fold higher) at the beginning of biofilm formation (10 h) but increases considerably (to 3.2 fold) in a mature biofilm (55 h) in which there are areas where conditions are microaerophilic. To confirm microaerophilic conditions, expression of the narX gene, which is controlled by fnr a key transcription regulator in cells growing in anaerobic conditions and is up regulated under microaerophilic/anaerobic environments, was tested. narX expression was up regulated during biofilm growth, consistent with the development of microaerophilic conditions in the inner layers of E. coli biofilms (4.34 and 2 fold induction in 10 h and 55 h cultures).
Figure 5. Relative expression ratio of RNR genes in biofilm versus planktonic cells.
The expression level of the indicated genes was measured in cDNA samples derived from 10- and 55-h old E. coli MG1655 biofilms grown at 37°C in PVC plates. For each gene the relative expression was defined as the level in cells from biofilm compared to those from planktonic cells. The 16S rRNA sequence was used as an internal standard. Error bars represent the SD from n = 3 independent replicates.
Expression of class Ib genes depends on the Fur and RpoS proteins
Several studies have described an increased expression of various genes affected by oxidative stress or nutrient starvation during biofilm formation [18]. Previous studies have suggested that the expression of class Ib RNR is up-regulated under nutrient starvation and oxidative stress [19]. Nevertheless the molecular mechanisms triggering their expression remain to be elucidated. Considering that our results showed that class Ib RNR was required and highly expressed during biofilm formation and especially during the initial steps, we decided to explore the factor(s) inducing the expression of this particular RNR class. RpoS has been shown to be upregulated in biofilms [20]. Furthermore, it has been evidenced in different global transcriptomic analysis that expression of class Ib RNRs is RpoS dependent [21], [22]. We decided to further correlate RpoS with class Ib RNR expression during biofilm formation. We performed RT-PCR analysis of nrdE expression on E. coli MG1655 and E2348/69 wild-type cells and their corresponding isogenic rpoS mutant strain ETS112 and ETS113 at different points on the exponential growth curve (OD550 = 0.3 and 0.8). Expression levels of the nrdE (class Ib) gene were found to be 2.61 and 9.44 fold higher (means of three different experiments) in the wild-type MG1655 cells than in the rpoS mutant derivative and 3.56 and 10.11 in the wild-type E2348/69 cells than in its rpoS mutant derivative (data not shown). To further confirm these results we used western-blot analysis to measure the NrdF expression (class Ib) in cells grown exponential and stationary growth phases in E. coli MG1655 and E2348/69 wild-type and rpoS mutants. In Fig6A is shown the dependence of class Ib on the presence of the RpoS protein. Both E. coli strains showed a reduced class Ib RNR expression in the ΔrpoS mutant compared to the wild-type strain at exponential and stationary growth phases, thus demonstrating the potential role of RpoS as an activator of the class Ib RNR and especially in biofilm formation where rpoS gene is highly induced [1], [20]. The precise mechanisms by which induces RpoS induces class Ib transcription needs to be further analysed.
Figure 6. Class Ib RNR expression depend on the Fur and RpoS proteins.
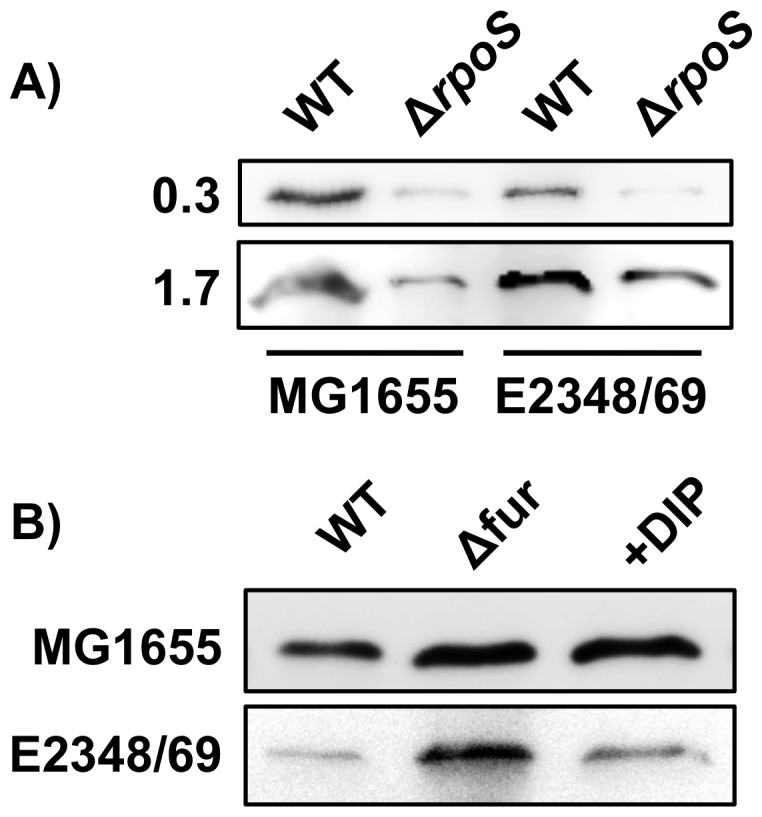
Western-blot analysis of the NrdF protein (class Ib RNR) in E. coli MG1655 and E2348/69 wild-type cells in A) rpoS mutant at stationary and exponential growth and B) fur mutant.
It has been recently shown that oxidative stress induces the expression of class Ib genes (nrdHIEF) by inactivating of the ferric uptake regulator protein (FUR) [15]. This has also been extensively studied in other stress environments [23], [24]. A fur box was previously described in class Ib RNR promoter region [25]. This effect of class Ib activation through FUR inactivation is further supports the role class Ib RNR expression in biofilm formation where a high level of oxidative stress occurs [26]. This result was previously described for the E. coli MG1655 strain but we also tested if the pathogenic E. coli E2348/69 strain showed the same induction in the nrdHIEF expression as the commensal MG1655 strain [15], [25] and response to the FUR transcriptional regulator. Bioinformatics analysis of the E. coli E2348/69 class Ib promoter region revealed an 8 mer FUR box (CGTAATCA) at the same position as previously described in the E. coli MG1655 [25]. In Fig6B we described the repressor behaviour of the FUR transcriptional regulator on the class Ib expression. Band quantification shows that the addition of an specific iron-chelator DIP (see Material and methods) into the medium created a condition of iron deficiency and led the expression of class Ib (1.6 and 1.7 times in E. coli MG1655 and E2348/69 respectively) as well as the one found in the fur mutant (1.7 and 2.5 times in E. coli MG1655 and E2348/69 respectively) demonstrating the dependence of the class Ib transcription on the FUR regulatory protein as was also previously demonstrated in strain MG1655 [25], [27].
All together, it seems clear that class Ib RNR genes are highly induced during biofilm formation, as the cells cope with nutrient starvation (iron deficiency) and oxidative stress, and we can hypothesize that the molecular mechanism that triggers its expression is mediated directly through the Fur transcriptional factor and by the rpoS sigma factor which is highly induced under these metabolic conditions. Any direct participation of the RpoS protein or an indirect effect through another transcriptional factor remains to be investigated.
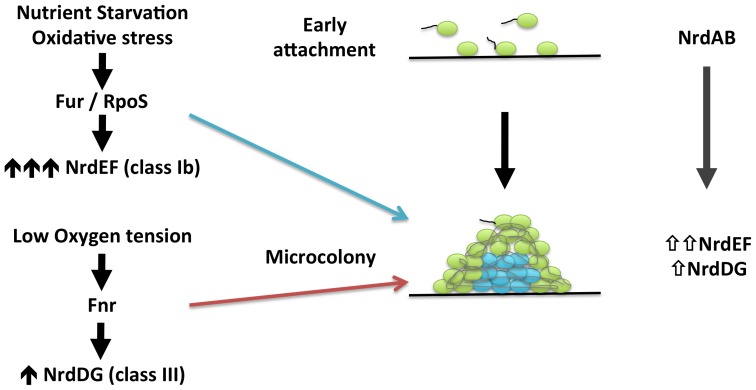
Model for roles of ribonucleotide reductase classes in Escherichia coli biofilm formation
In this paper, we evidence that the hitherto cryptic class Ib RNR is expressed when E. coli cells form biofilms (see Fig.1, 4 and 5). Class Ia is active during standard laboratory growth conditions and also in planktonic cells, and down-regulated when cells switch to a biofilm state (Fig.5). Our results evidence an induction of RNR classes Ib and III during biofilm conditions. Hence, a physiological role for class Ib RNR is shown in these conditions for the first time (Figs.1,2,3,4,5). The need for simultaneous expression of RNR classes Ib and III has also been addressed. It is likely that during the early stages of attachment, planktonic cells express RNR class Ia (Fig.7). After attachment, E. coli strains and especially enteropathogenic E. coli (EPEC) isolates form microcolonies that develop into three-dimensional structures within which some regions are microaerophilic [6], [18]. Under these circumstances, with conditions of nutrient starvation and oxidative stress [26] class Ib expression is triggered, probably directly by the inactivation of the FUR protein or indirectly by RpoS, and is enzymatically functional. In these conditions, parts of this structure can result in a nutrient and oxygen limitation creating parts of microaerophilic areas. Class III RNR is fully enzymatically active under microaerophilic conditions [14] and it is highly expressed anaerobically by the FNR protein, as already described [12], [13]. Note that in E. coli E2348/69 strain, which forms more complex biofilms where more anaerobic environments can be created and in which expresses more nrdD protein (Fig.4) compared to the E. coli MG1655 strain which forms less structured biofilms. Other types of cells located at the inner parts of the biofilm might express class Ib RNR which can be active under these conditions [28] and favour its expression under oxidative stress and nutrient starvation as specified before.
Figure 7. Proposed model for expression of ribonucleotide reductases in E. coli biofilm formation.
For the first time, we have established a significant physiological role for class Ib and III RNR in E. coli and particularly in context of the formation of biofilms. Accordingly, class Ib and III RNR should be considered as a target for new anti-proliferative agents. Such agents might be useful in combined therapies to reduce and eliminate biofilms, making the chronic colonization/infection by certain pathogenic E. coli cells less virulent.
Materials and Methods
Bacterial strains, plasmids and culture conditions
All strains and plasmids used in this study are listed in Table1. Escherichia coli cells were grown in Luria-Bertani (LB) at 37°C. Antibiotics and chromogenic substrates were used at the following final concentrations: 50 µg ampicillin ml−1, 50 µg kanamycin ml−1, 30 µg chloramphenicol ml−1, 30 µg X-Gal ml−1. Bacterial growth was measured by reading OD550. In order to create iron-limiting conditions, the iron chelator 2,2′-dipyridyl (DIP) (SIGMA) was added at 150 µM to LB liquid cultures.
Strain and plasmid construction
Plasmid DNAs were isolated using the QIAprep miniprep kit (Qiagen). PCRs were carried out using High-Fidelity PCR enzyme mix (Fermentas) according to the manufacturer's instruction. Other molecular biology techniques were carried out by standard procedures [29].
Different nrd mutant strains were constructed by introducing certain mutations (Table1) from the Keio collection E. coli strains [30] to the MG1655 and E2349/69 strains by P1 transduction [31] followed by selection of an appropriate drug resistance marker. All mutations were tested by PCR using a combination of gene-specific and transposon specific primers (Table2).
Table 2. Primers and probes used in this study.
| Name | Sequence (5′→3′) | Application |
| EcoliDG-BHI-up | AAGGATCCGCCGTGAATGGAAG | Clonning |
| EcoliDG-BHI-lw | AAGGATCCTCGACATTCTGGTCGGTCAG | Clonning |
| Ecoli nrdR up | AAGGATCCCAGTCTTGCCGGTGTTTTCG | Clonning |
| Ecoli nrdR lw | AAGGATCCGCAACGCGTGTACTTCGGC | Clonning |
| Operon-Ib-up (BHI) | AAAGGATCCGCATGGTCGTACTCGCGTCC | Clonning |
| Operon-Ib-lw (BHI) | AAAGGATCCATTATTTCCCTGCTGCGGGTAGTG | Clonning |
| pBBR1-up | CATCGCAGTCGGCCTATTGG | Check-Clonning |
| pBBR1-lw | CACTTTATGCTTCCGGCTCG | Check-Clonning |
| M13 dir | GTTTTCCCAGTCACGAC | Check-Clonning |
| M13 rev | CAGGAAACAGCTATGAC | Check-Clonning |
| k1 | CAGTCATAGCCGAATAGCCT | Check-Clonning |
| kt | CGGCCACAGTCGATGAATCC | Check-Clonning |
| k2 | CGGTGCCCTGAATGAACTGC | Check-Clonning |
| EcA-lw | TCAGATCTTACATGCGCCGC | Reverse transcription |
| EcD-lw | GGCTTCATCGCCTTTTGCTTCC | Reverse transcription |
| EcR-lw | GCGCGATCTCTTCGCCAAATTC | Reverse transcription |
| EcE-lw | AAAGTGCACAGGAGACGCAG | Reverse transcription |
| EcGAP-lw | GATGATGTTCTGGGAAGCGC | Reverse transcription |
| PnrdA BamHI up | AAAGGATCCATCATTTTCTATAAGACGG | Promoter-probe clonning |
| PnrdA ClaI lw | AAAATCGATCACCAGCAGATTCTGATTCATG | Promoter-probe clonning |
| PnrdD BamHI up | AAAGGATCCTTGAGGCTGTCTGGTGGTTAC | Promoter-probe clonning |
| PnrdD ClaI lw | AAAATCGATGCACTTTGCAGCCGTCTCG | Promoter-probe clonning |
| PnrdH BamHI up | AAAGGATCCAAAAATGATAATAAATACGCG | Promoter-probe clonning |
| PnrdH ClaI lw | AAAATCGATGTAAATAGTAATGCGCATG | Promoter-probe clonning |
To construct plasmids pETS154, pETS155, pETS156 and pETS157 the nrdHIEF (class Ib), nrdDG (class III) and nrdR operons were cloned into plasmids pBluescriptSK(+) and pBBR1MCS-5 under the control of their respective native promoters (Table1 and 2).
Comparison of the growth curves of the wild type, mutants and complemented strains did not show any significant differences (data not shown).
Biofilm formation
Biofilm formation was tested on polyvinyl chloride (PVC) 96-well plates using crystal violet staining as previously described [32]. Briefly, strains from an overnight culture were inoculated at an OD550 of 0.01 in LB with 0.2% glucose into wells of PVC microtiter plates (COSTAR) and incubated at 37°C, without agitation, for 48 h. The attached cells were stained with 0.1% crystal violet and biofilm formation was quantified by dissolving the CV in ethanol and measuring the absorbance at 570 nm (A570).
To study biofilm development on glass surface, we followed basically the method described previously by O′Toole [33]. 3 mL aliquot of diluted culture was added to a borosilicate glass tube containing a sterile 18×18 mm glass cover slip (Menzel-Gläser) and incubated at 37°C for 48 h. Excess of broth was removed and washed three times in PBS and attached cells were fixed in methanol and stained with 0.1% crystal violet. Biofilm was quantified by dissolving the attached cells in ethanol and absorbance was measured as described above.
RNA preparation and quantitative RT-PCR
Total RNA was extracted from E. coli cells using the RNeasy Mini kit (Qiagen) according the manufacturer's instruction. RNA quantity was measured using a NanoDrop spectrophotometer (ND-1000, NanoDrop). RNA samples were treated for DNA digestion with DNase I (Turbo DNA-free, Ambion) and 1 µg was reverse-transcribed with the SuperScript® III Reverse transcriptase kit (Invitrogen). Real-time PCR measurements were carried out using TaqMan primers and probes, and detection was performed using and ABI Prism 7700 Sequence Detection System from Applied Biosystems as described previously [11]. The 16S rRNA sequence was used as an internal standard.
Construction of GFP fusions
Transcriptional fusions in E. coli were constructed by PCR amplification of the promoter region of the nrdA (706 bp), nrdH (268 bp) and nrdD (462 bp) and cloning of these regions upstream of the promoterless gfp gene in pETS130-GFP [34]. Primers used for these cloning are described in Table2. Sequencing and PCR analysis confirmed the orientation and correctness of the inserts. GFP fluorescence was measured on an FLx800 Fluorescence microplate reader (BioTek). All assays were averages of at least three independent trials.
Detection of NrdE by Western blot analysis
SDS-page and immunobloting were performed as described previously [34]. Briefly, protein crude extracts were extracted with the BugBuster® extraction reagent (Novagen) from E. coli culture samples. 5 µg of protein from each condition were resolved on a 7.5% polyacrylamide gel and transferred to PVDF membranes (Immun-Blot™ PVDF membranes, Bio-Rad). The immunodetection of proteins was performed using rabbit polyclonal antibodies against NrdF protein [35] at dilution 1/1000 (Agrisera, Sweden). The detection of primary antibodies was performed using donkey anti-rabbit (Bio-Rad) horseradish-peroxidase-conjugated secondary antibodies at 1/50000 dilution, and visualized using the Amersham™ ECL™ Prime western blotting reagent (GE Healthcare) according the manufactured's protocol. The membrane was visualized and analyzed with an ImageQuant™ LAS4000 mini (GE Healthcare). The polyclonal antibodies cross-reacted with few proteins which was used as internal control for equal protein loading.
Acknowledgments
We are grateful to Maria del Carmen Jaramillo for her technical assistance.
Funding Statement
This work was supported by grants from the “ERA-NET Pathogenomics-BIO2008-04362-E” and from the Spanish Ministerio de Ciencia e Innovación (PI081062) and (BFU2011-24066) to ET and Consolider-Intermods (CDS2008-00013) from the Spanish MICINN to ET and AJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Schembri MA, Kjaergaard K, Klemm P (2003) Global gene expression in Escherichia coli biofilms. Mol Microbiol 48: 253–267. [DOI] [PubMed] [Google Scholar]
- 2. Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322. [DOI] [PubMed] [Google Scholar]
- 3. Domka J, Lee J, Bansal T, Wood TK (2007) Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol 9: 332–346. [DOI] [PubMed] [Google Scholar]
- 4. Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK (2004) Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol 64: 515–524. [DOI] [PubMed] [Google Scholar]
- 5. Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, et al. (2004) Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51: 659–674. [DOI] [PubMed] [Google Scholar]
- 6. Moreira CG, Palmer K, Whiteley M, Sircili MP, Trabulsi LR, et al. (2006) Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli . JBacteriol 188: 3952–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torrents E, Sahlin M, Sjöberg B-M (2008) The Ribonucleotide Reductase Family- Genetics and Genomics. In: Ribonucleotide Reductases (ed KK Andersson) Nova Science Publishers: pp. 17–77.
- 8. Lundin D, Torrents E, Poole AM, Sjöberg B-M (2009) RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cotruvo JA, Stubbe J (2011) Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry 50: 1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boal AK, Cotruvo JA Jr, Stubbe J, Rosenzweig AC (2010) Structural basis for activation of class Ib ribonucleotide reductase. Science 329: 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Torrents E, Grinberg I, Gorovitz-Harris B, Lundstrom H, Borovok I, et al. (2007) NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. JBacteriol 189: 5012–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boston T, Atlung T (2003) FNR-mediated oxygen-responsive regulation of the nrdDG operon of Escherichia coli . JBacteriol 185: 5310–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roca I, Ballana E, Panosa A, Torrents E, Gibert I (2008) Fumarate and nitrate reduction (FNR) dependent activation of the Escherichia coli anaerobic ribonucleotide reductase nrdDG promoter. Int Microbiol 11: 49–56. [PubMed] [Google Scholar]
- 14. Garriga X, Eliasson R, Torrents E, Jordan A, Barbe J, et al. (1996) nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli . Biochem Biophys Res Commun 229: 189–192. [DOI] [PubMed] [Google Scholar]
- 15. Martin JE, Imlay JA (2011) The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol 80: 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, et al (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462. [DOI] [PubMed] [Google Scholar]
- 17. Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, et al. (2009) Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. JBacteriol 191: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stewart PS, Franklin MJ (2008) Physiological heterogeneity in biofilms. Nat Rev Microbiol 6: 199–210. [DOI] [PubMed] [Google Scholar]
- 19. Monje-Casas F, Jurado J, Prieto-Alamo MJ, Holmgren A, Pueyo C (2001) Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. JBiol Chem 276: 18031–18037. [DOI] [PubMed] [Google Scholar]
- 20. Ito A, May T, Taniuchi A, Kawata K, Okabe S (2009) Localized expression profiles of rpoS in Escherichia coli biofilms. Biotechnol Bioeng 103: 975–983. [DOI] [PubMed] [Google Scholar]
- 21. Dong T, Kirchhof MG, Schellhorn HE (2008) RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol Genet Genomics 279: 267–277. [DOI] [PubMed] [Google Scholar]
- 22. Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R (2005) Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. JBacteriol 187: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varghese S, Wu A, Park S, Imlay KR, Imlay JA (2007) Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli . Mol Microbiol 64: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, et al. (2010) Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 11: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vassinova N, Kozyrev D (2000) A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli . Microbiology 146 Pt 12: 3171–3182. [DOI] [PubMed] [Google Scholar]
- 26. Boles BR, Singh PK (2008) Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad SciUSA 105: 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, et al. (2003) Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. JBiol Chem 278: 29478–29486. [DOI] [PubMed] [Google Scholar]
- 28. Torrents E, Jordan A, Karlsson M, Gibert I (2000) Occurrence of multiple ribonucleotide reductase classes in gamma-proteobacteria species. Curr Microbiol 41: 346–351. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual: Cold Spring Harbor Laboratory Press.
- 30.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006 0008. [DOI] [PMC free article] [PubMed]
- 31.Silhavy TJ, Berman ML, Enquist LW (1984) Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 32. Weiss-Muszkat M, Shakh D, Zhou Y, Pinto R, Belausov E, et al. (2010) Biofilm formation by and multicellular behavior of Escherichia coli O55:H7, an atypical enteropathogenic strain. Appl Environ Microbiol 76: 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O′Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, et al. (1999) Genetic approaches to study of biofilms. Methods Enzymol 310: 91–109. [DOI] [PubMed] [Google Scholar]
- 34. Sjöberg BM, Torrents E (2011) Shift in Ribonucleotide Reductase Gene Expression in Pseudomonas aeruginosa during Infection. Infect Immun 79: 2663–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Torrents E, Sjöberg BM (2010) Antibacterial activity of radical scavengers against class Ib ribonucleotide reductase from Bacillus anthracis . Biol Chem 391: 229–234. [DOI] [PubMed] [Google Scholar]
- 36. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]