Abstract
Background
In recent years, Staphylococcus epidermidis ( Se) has become a major nosocomial pathogen and the most common cause of infections of implanted prostheses and other indwelling devices. This is due in part to avid biofilm formation by Se on device surfaces. However, it still remains unknown that how the process of Se biofilm development is associated with relapsed infection in such patients.
Results
We have identified clinical Se isolates displaying enhanced biofilm dispersal and self-renewal relative to reference strain. These isolates also exhibit enhanced initial cell attachment, extracellular DNA release, cell autolysis and thicker microcolonies during biofilm development relative to reference strain. Our genetic analyses suggest that these clinical isolates exhibit significant downregulation of RNAIII, the effector molecule of the agr quorum sensing system, and upregulation of the autolysin gene atlE. Isogenic deletion of the agr system in Se 1457 confirmed that agr negatively regulating atlE resulted in enhanced initial cell attachment, extracellular DNA release, cell autolysis and biofilm formation abilities. In contrast, double deletion of agr and atlE significantly abolished these features.
Conclusions
Collectively, these data reveal the role of agr system in long-term biofilm development and pathogenesis during Se caused indwelling devices-related relapsed infection.
Keywords: Staphylococcus epidermidis, Biofilm, Autolysis, Extracellular DNA
Background
In recent years, coagulase-negative Staphylococcus epidermidis ( Se) has become the leading cause of infections related to indwelling medical devices such as vascular catheters, prosthetic joints and artificial heart valves [1,2]. Pathogenicity of Se is attributed to its formation of biofilm on the surface of medical devices, thereby enhancing Se resistance to antibiotics and host defenses in this setting [3,4]. In general, Se biofilm formation is a two-step process, in which bacteria first adhere to the surface (initial attachment phase) and subsequently form cell–cell aggregates and a multilayered architecture (accumulative phase) [5,6]. One autolysin protein, AtlE, facilitates bacterial attachment to the surface of medical devices and dictates pathogenesis for Se biofilm-associated infections in vivo [7,8]. In the accumulative phase, the polysaccharide intercellular adhesin (PIA), a linear poly-Nacetyl-1,6-β-glucosamine (PNAG) encoded by the icaADBC locus, is the major pathogenic determinant for intercellular adhesion [9,10]. In addition, we have previously demonstrated that extracellular DNA is generated during Se growth through AtlE-mediated lysis of a subpopulation of the bacteria; moreover, this process is required for initial bacterial attachment to surfaces and biofilm development [11]. An important negative regulator of biofilm formation by Se and Staphylococcus aureus is the accessory gene regulator ( agr) quorum sensing system, and agr mutation promotes biofilm formation by increasing the capacity of Se for initial cell attachment [12-14]. The agr system of Se and S. aureus consists of 4 genes ( agrAagrCagrD, and agrB) that are cotranscribed (RNAII) and the gene for the effector molecule of the agr system, RNAIII, which also encodes the gene for δ-toxin ( hld) [12,15].
Medical device-associated biofilms facilitate recalcitrant or recurrent infections despite use of appropriate antibiotics. However, there are only limited data about the long-term Se biofilm development, especially clinical isolates recovered from indwelling medical devices infection. It still remains unknown that how the process of Se biofilm development is associated with relapsed infection in such patients. Moreover, the molecular mechanisms causing such repeated infection also needs to be investigated. In the current study, we compared the long-term (~7 days) biofilm development and dispersal between Se clinical isolates causing indwelling medical devices infection and reference strain in the flow-chamber systems. We also compared the biofilm-related events (initial attachment, PIA synthesis, extracellular DNA release etc.) and biofilm-associated gene profiles in these clinical isolates and reference strain.
Methods
Bacterial strains, growth media and reagents
4 Se clinical isolates, referred to as Se-1, Se-2, Se-3 and Se-4, were recovered from 4 different patients at the Zhongshan Hospital (Shanghai, China) with indwelling catheter-associated infections as defined by the presence of fever, bacterial growth from peripheral blood samples collected from catheter sites. Se biofilm-positive strain 1457 wild type and agr mutants were kindly provided by Dr. Min Li (Huashan Hospital, Shanghai, China), as described previously [13]. The agr/ atlE double mutant was constructed as described previously [11]. The mutation was confirmed by Southern blotting and direct sequencing (data not shown), and we also independently confirmed that the 1457 agr mutant or agr/ atlE double mutant does not affect bacterial growth (see Additional file 1: Figure S1). Se biofilm-positive ATCC 35984 (also referred as RP62A) and biofilm-negative ATCC 12228 reference strains were purchased from American Type Culture Collection (ATCC). Tryptic soy broth (TSB; Oxoid) medium containing 0.25% glucose was used to support biofilm formation in the microtitre plates. AB medium [16] supplemented with 0.3 mM glucose and 3% TSB was used for biofilm cultivation in the flow-chamber system. SYTO 9 and propidium iodide (PI) (Live_Dead reagents, Molecular Probes) were used at a concentration of 1 μM for staining live or dead bacteria in biofilms, respectively. DDAO [7-hydroxy-9 H-(1,3-dichloro-9,9-dimethylacridin-2-one)] (Molecular Probes) was used at a concentration of 1 μM to stain extracellular DNA in biofilms [11]. TRITC (tetramethyl rhodamine isothiocyanate)-labeled wheat germ agglutinin (Molecular Probes, Eugene, OR) was used at a concentration of 0.1 mg/mL to stain the PIA in biofilms [17]. Hemoglobin was purchased from Sigma and used as indicated concentrations. The Ethics Committee of the Zhongshan Hospital of Fudan University and the East Hospital of Tongji University both exempted this study from review because the current study only focused on bacteria.
Cultivation of bacterial biofilms
Biofilm cultivation in polystyrene microtitre plates was carried out as described previously [11]. Briefly, overnight cultures of Se strains grown in TSB (0.25% glucose) medium were diluted 1:200. The diluted cultures were transferred to wells of polystyrene microtitre plates (200 μL per well) and incubated at 37 °C for 24 h. After washing, the wells were stained with 2% crystal violet for 5 min. Then, the plate was rinsed, air-dried, redissolved in ethanol and the absorbance was determined at 590 nm. For cultivation of Se biofilms in the flow-chamber system, the flow-chamber system was first assembled and prepared as described previously [18]. Briefly, the flow chambers were inoculated by injecting 350 μL overnight culture diluted to OD600 = 0.001 into each flow channel with a small syringe. After inoculation, flow channels were left without flow for 1 h, after which medium flow (0.2 mm/s) was started using a Watson-Marlow 205 S peristaltic pump.
Microscopy
All microscopic observations and image acquisition were performed using a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Jena) equipped with detectors and filter sets for monitoring SYTO 9, PI, DDAO and TRITC fluorescence. Images were obtained using an x63/1.4i objective or an x40/1.3i objective. Simulated 3D images and sections were generated using the IMARIS software package (Bitplane).
Bacterial attachment assays
Initial cell attachment was tested as described previously [11]. Briefly, cell suspensions from the mid-exponential phase of bacterial growth were diluted to OD600 = 0.1 in PBS, and then incubated in wells (1 mL per well) of cover-glass cell culture chambers (Nunc) for 30 min at 37°C, after which attached cells were calculated by microscopy.
Quantification of extracellular DNA
Extracellular DNA was quantified as described previously [11]. Overnight cultures were diluted to OD600 = 0.001 in AB medium supplemented with 0.5% glucose, 0.05 mM PI and 10% TSB. The diluted cultures were transferred to wells of polystyrene microtitre plates (150 μL per well) and incubated for 24 h at 37°C, upon which PI absorbance was measured at 480 nm and cell density was measured by OD600 using a Wallac microtitre plate reader. Relative amounts of extracellular DNA per OD600 unit were calculated.
qRT-PCR
After cultured for 1 d or 6 d, biofilm cells were resuspended in 500 mL buffer containing 25% sucrose, 10 mM Tris/HCl (pH 7.5) and 0.2 mg lysostaphin (Sigma-Aldrich). After incubation at 37°C for 10 min, total RNA was isolated using the RNeasy Mini kit according to the manufacturer’s instructions (QIAGEN). cDNA was synthesized from equivalent concentrations of total RNA using the SuperScript III First-Strand Synthesis SuperMix Kit (Invitrogen) according to the manufacturer’s instructions. Coding sequences for bacterial genes (and gyrB for internal controls) were amplified using iQ SYBR Green Supermix (Bio-rad). Custom primer sequences used for amplification experiments are included in Additional file 2: Table S1. Amplification was carried out using an iCycler IQ Real-Time PCR Detection System, and cycle threshold (Ct) values determined in duplicate for target gene transcripts and gyrB for each experiment. “No template” (water) and “no-RT” controls were used to ensure minimal background DNA contamination. Fold changes for experimental groups relative to assigned controls were calculated using automated iQ5 2.0 software (Bio-rad).
PCR and sequencing
Genomic DNA was extracted by using Wizard Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. The primers included in Additional file 2: Table S1 were designed from conserved sequences of agr, which are common to agr groups I, II and III, to amplify a 1022 bp fragment [19]. The PCR production was purified by using QIAquick PCR Purification Kit (Qiagen) then sequenced (Operon), and alignment analysis was performed by using Vector NTI Advance 9 software (Invitrogen).
Cell autolysis assays
Autolysis assays for Se strains were performed as described previously [11]. Briefly, cell samples (50 mL) were collected from exponential-phase cultures growing in TSB medium (OD600 = 0.6 ~ 0.8) containing 1 M NaCl, and cells were pelleted by centrifugation. The cells were washed twice with 50 mL ice-cold water and resuspended in 50 mL 0.05 M Tris/HCl (pH 7.2) containing 0.05% (v/v) Triton X-100. The cells were then incubated at 30°C with shaking, and OD600 was measured at 30 min intervals. The lysis rate induced by Triton X-100 was calculated as: OD0-ODt/OD0.
Results
Se isolates associated with catheter infection exhibit more avid self-renewal in long-term cultured biofilm assays
We first observed long-term (~7 days) cultured biofilm formation for Se-1-4 in the flow-chamber systems, together with one biofilm-positive Se reference strain (ATCC 35984). All strains displayed similar biofilm development during long-term cultivation, although they displayed heterogeneity for biofilm architecture (Figure 1). After one day in culture, the chamber surface was almost completely covered by bacterial biofilms, and many dead cells were present in the center of microcolonies. After 2 days, most of the dead cells were detached from the microcolonies, forming vacuoles. After 3-4 days, the residual cells in the old biofilms proliferated and occupied the vacuolated areas, forming a “renewed” biofilm in which only a few dead cells were observed. After 6-7 days, a large number of dead cells reappeared in the center of microcolonies. Notably, Se-1, Se-2, Se-3 and Se-4 displayed much bigger microcolonies, more dead cells, and more significant cell dispersal with much more vacuole formation relative to the reference strain ATCC 35984 (Figure 1).
Figure 1.
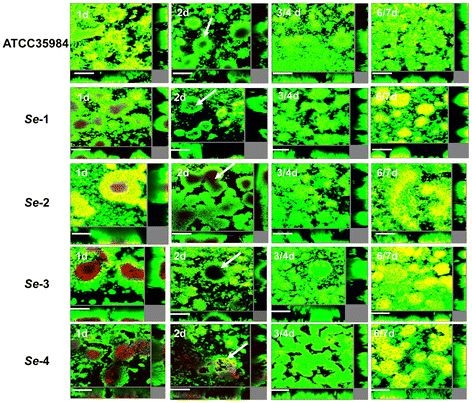
S. epidermidisisolates associated with catheter infection exhibit greater biofilm self-renewal. Laboratory strain ATCC 35984 and clinical isolates Se-1, Se-2, Se-3 and Se-4 were grown for ~7 days in flow chambers irrigated with minimal medium, and stained with SYTO 9 and PI at indicated time points to identify live and dead cells, respectively. Microscopic investigation was performed using confocal laser scanning microscopy (CLSM). The central pictures show horizontal optical sections, and the flanking pictures show side views. Live cells appear green and dead cells appear yellow/red. Bars, 50 μm.
Se isolates associated with catheter infection exhibit greater extracellular DNA content and capacity for cell attachment
We next compared biofilm formation capacity for these clinical isolates and the reference strain using the microtitre plates. These results first confirmed that all 4 Se clinical isolates displayed stronger biofilm biomass than ATCC 35984 by crystal violet staining (Figure 2A). Interestingly, we also found significantly more extracellular DNA release from these clinical isolates relative to the reference strain during biofilm formation (Figure 2B). Our previous study demonstrated that extracellular DNA is a major component required for initial bacterial attachment to surfaces, as well as subsequent early phases of biofilm development by Se[11]. In agreement with these results, we found that our clinical isolates exhibited a greater capacity for cell attachment relative to the reference strain (Figure 2C). PIA plays an important role in cell-cell adhesion during phase II of Se biofilm formation [10], and Jager et al. have previously reported detection of PIA synthesis in mature biofilms using TRITC-labeled wheat germ agglutinin staining [17]. However, we did not observe obvious differences in PIA synthesis between our Se clinical isolates and the reference strain (data not shown).
Figure 2.
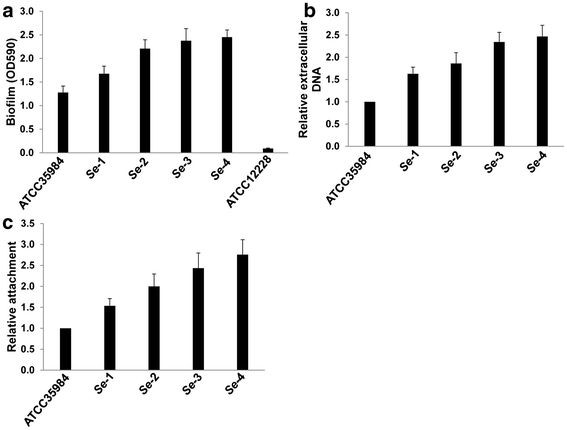
S. epidermidisisolates associated with catheter infection display more biofilm formation, extracellular DNA release and initial attachment than laboratory strain.(a) Cultures were grown in microtitre plates for 24 h at 37°C, and biofilm biomass was quantified using a crystal violet assay. (b) Cultures were grown for 24 h in minimal medium supplemented with 0.05 mM PI, whereupon PI absorbance (OD480) and cell density (OD600) were measured and relative amounts of extracellular DNA per OD600 unit were calculated. (c) Initial attachment of S. epidermidis strains in static chambers was measured as described in Methods. Error bars represent the S.E.M. for three independent experiments.
The agr system regulates initial cell attachment and cell autolysis during biofilm formation for se
Next, we compared the biofilm-associated gene profiles for our Se clinical isolates and the reference strain by qRT-PCR. Our results indicated that expression of atlE, the major autolysin gene of Se required for initial cell attachment, extracellular DNA release and Triton X-100 induced autolysis [7,11,13], was significantly increased in all the 4 clinical isolates (~2-7 fold) relative to the reference strain for 1 d- or 6 d-biofilm cells (Figure 3, Additional file 3: Figure S2). In contrast, there were no appreciable differences for expression of icaA, the gene encoding N-acetylglucosaminyltransferase and required for PIA synthesis and cell-cell aggregation among them. Notably, expression of RNAIII, a gene encoding an effector molecule of the agr quorum sensing system, was significantly reduced for all the Se clinical isolates relative to the reference strain (Figures 3, Additional file 3: Figure S2). Further experiments revealed that all the 4 clinical isolates displayed stronger cell autolysis abilities than ATCC35984 induced by Triton X-100 (Figure 4).
Figure 3.
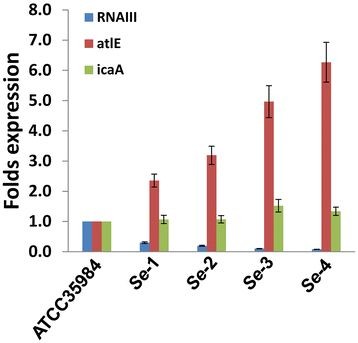
S. epidermidisisolates associated with catheter infection exhibit differential expression of genes associated with biofilm formation. The expression profiles of RNAIII, atlE and icaA were compared for 24-h biofilm cells of laboratory strain and clinical isolates using qRT-PCR as described in Methods. Error bars represent the S.E.M. for three independent experiments.
Figure 4.
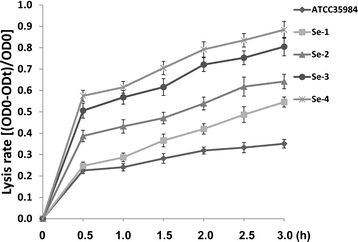
S. epidermidisisolates associated with catheter infection exhibit higher cell autolysis abilities. Triton X-100 induced cell autolysis assays were performed as described in Methods, and error bars represent the S.E.M. for three independent experiments.
Agr mutant increases initial cell attachment and cell death during biofilm formation through upregulation of atlE
To further clarify the roles of agr in cell attachment, cell death and biofilm formation, we assessed these endpoints for Se 1457 wild type (wt), agr mutant (△agr) and agr/ atlE double mutant (△agr/atlE) strains using our flow-chamber systems. We found more dead cells in the center of microcolony structures for 1457 △agr mature biofilms than 1457 wt (Figure 5A, B), while only few dead cells were seen in 1457 △agr/atlE (Figure 5C). Also, 1457 △agr displayed thicker microcolony structure during biofilm formation than 1457 wt (Figure 5D, E), in contrast, the biofilm formation ability of 1457 △agr/atlE was seriously impaired because it only formed very thin and loose biofilm structure (Figure 5F). Of note, cell dispersal, vacuole formation, and self-renewal biofilms were also observed after long-term culture in flow-chamber systems (data not shown). Crystal violet staining further confirmed that 1457 △agr formed stronger biomass than 1457 wt in the microtitre plate assays, while 1457 △agr/atlE only formed poor biomass (Figure 5G). Finally, 1457 △agr exhibited significantly increased initial cell attachment relative to 1457 wt, whereas which was impaired in 1457 △agr/atlE (Figure 5H).
Figure 5.
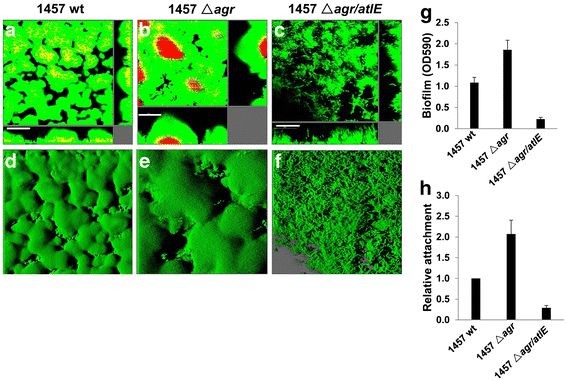
S. epidermidis agrsystem regulates biofilm formation and initial cell attachment throughatlE. ( a-d) S. epidermidis 1457 wild type (wt, a and d), agr mutant (△ agr, b and e) and agr/atlE double mutant (△ agr/atlE, c and f) were grown for 24 h in flow chambers irrigated with minimal medium, and were then stained with SYTO 9 and PI, upon which microscopic investigation was performed by CLSM. The 3-D images (d-f) were generated using the IMARIS, bars, 50 μm. (g) Biofilm biomass in microtitre plates was quantified using a crystal violet assay. (h) Initial attachment of S. epidermidis strains in static chambers was quantified as described in Methods. Error bars represent the S.E.M. for three independent experiments.
Agr regulates se release of extracellular DNA and autolysis through suppression of atlE
Our previous study revealed that mutation of atlE in Se 1457 significantly reduced extracellular DNA release and impairs biofilm formation [11]. Consistent with those results, qRT-PCR revealed that expression of atlE was significantly increased for 1457 △agr, but almost no atlE transcripts were detected in 1457 △agr/atlE (Figure 6A). Our qRT-PCR also confirmed that no RNAIII transcripts were detected in Se 1457 △agr, when compared with its wt strain (Figure 6A). Furthermore, 1457 △agr exhibited increased extracellular DNA relative to 1457 wt using both microtitre plate assays and DDAO staining in the flow-chamber systems (Figure 6C-F), while 1457 △agr/atlE abolished most extracellular DNA (Figure 6B6G-H). In addition, 1457 △agr displayed higher cell autolysis abilities than its wt strain, when induced by Triton X-100, whereas poor cell autolysis was seen in 1457 △agr/atlE (Additional file 4: Figure S3). Notably, expression of icaA transcripts was almost unchanged for 1457 △agr relative to its wt strain, however, icaA transcripts were partially reduced in 1457 △agr/atlE (Figure 6A).
Figure 6.
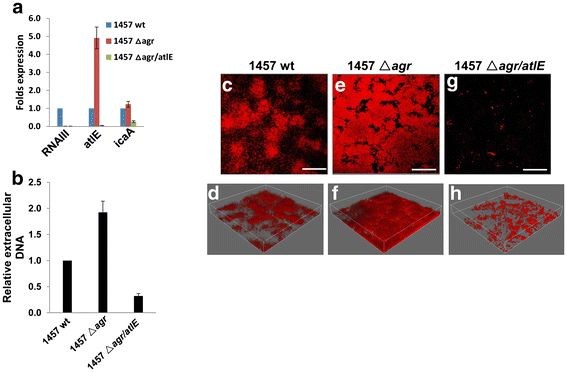
S. epidermidis agrsystem controls extracellular DNA release throughatlE.(a) Biofilm-associated gene transcripts were compared between 1457 wt, △ agr and △ agr/atlE by using qRT-PCR. (b) Extracellular DNA release from cultures in microtitre plates was quantified as described above. Error bars represent the S.E.M. for three independent experiments. (c-h)S. epidermidis 1457 wild type (wt, c-d) agr mutant (△ agr, e and f) and agr/atlE double mutant (△ agr/atlE, g and h) were grown for 24 h in flow chambers irrigated with minimal medium, and were then stained with DDAO for extracellular DNA in biofilms, upon which microscopic investigation was performed by CLSM. The 3-D images ( d/ f/ h) were generated using the IMARIS, bars, 50 μm.
Chemical inhibition of agr increases biofilm formation, initial attachment and cell autolysis through upregulation of atlE
A recent study has revealed that inhibition of S. aureus agr system by human hemoglobin promotes surface colonization [20]. To confirm the roles of agr in biofilm-associated events we found in Se 1457 genetic mutants above, here we treated Se 1457 wt strain with or without human hemoglobin (40 or 200 μg/mL). The results indicated that hemoglobin significantly reduced RNAIII transcripts (~40%-70% of inhibition) while increased atlE (~2.5-5.5 folds) but almost not affecting icaA (Figure 7). Functional assays further confirmed that hemoglobin increased biofilm formation, initial attachment, extracellular DNA release and cell autolysis in a dose-dependent manner (Figure 7), while which does not affect bacterial growth (data not shown).
Figure 7.
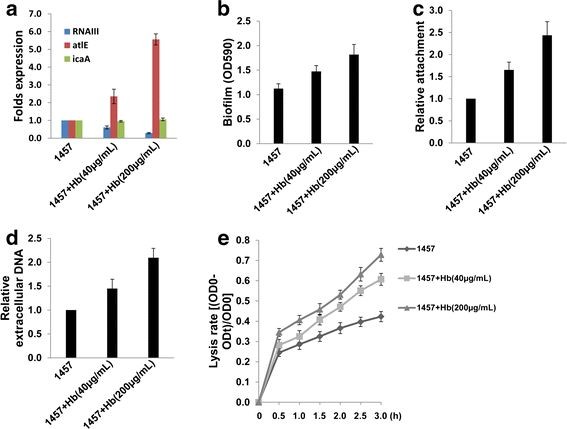
Chemical inhibition ofagrexhibit increased biofilm formation, extracellular DNA release and cell autolysis through upregulation ofatlE.S. epidermidis 1457 was treated with or without hemoglobin (40 or 200 μg/mL), then (a) Biofilm-associated gene transcripts were measured by using qRT-PCR; (b) Biofilm biomass was quantified using a crystal violet assay; (c-e) Initial attachment, extracellular DNA release and cell autolysis were determined as described above, respectively. Error bars represent the S.E.M. for three independent experiments.
Discussion
Se biofilm formation on implanted medical devices may result in recurrent or refractory infection unless the devices are removed, and removal and replacement of these devices incurs significant cost and risk for the patient. Flow-chamber systems simulate blood or other body-fluid flow in the vasculature of patients [18]. Using this and other complimentary approaches, we found that clinical Se isolates from patients with implanted catheter infections display greater microcolony densities, spontaneous cell death, and self-renewal capacity during biofilm development relative to reference strains. Bacteria in biofilms are 100 ~ 1000 times more resistant to antibiotics than planktonic cells [21-23], although our study does not directly address antibiotic sensitivity for our clinical isolates. Staphylococcal biofilm dispersal is associated with severe infection, including endocarditis, pneumonia and sepsis [24-26]. In addition, dispersal cells help bacteria establish new biofilms in more suitable niches, resulting in infection within multiple tissues [27]. Of interest, we collected the detached and “flow-out” cells in the flow-chamber systems for our clinical isolates and found living cells capable of forming new biofilms as quickly as their parent cells (Qin et al., unpublished data).
Interestingly, expression of RNAIII, a gene for the effector molecule of the agr system, was significantly reduced in all 4 Se clinical isolates, suggesting that the functions of agr quorum-sensing system were impaired in these isolates. Besides its regulatory function, RNAIII also encodes a δ-toxin, which effectively reduces cell attachment and subsequent biofilm formation of a Se agr mutant [13]. Our work does not address how RNAIII transcripts might be downregulated in our clinical isolates. Ongoing studies have found some potential spontaneous mutations present in the agr conserved region from Se-1, Se-2, Se-3 when compared with ATCC35984 strain (see Additional file 5: Figure S4), while Se-4 displayed some larger fragment variations for unknown reasons (Qin et al., unpublished data). However, it still requires further investigations to identify these potential spontaneous mutations responsible for RNAIII transcripts downregulation in these clinical isolates. Interestingly, about ~25% of S. aureus and ~17% of Se clinical isolates are naturally occurring agr mutants [19,28]. One recent study indicated that Se agr mutant showed increased biofilm development and colonization in a rabbit model [29]. In addition, nonfunctional agr occurred more frequently among strains isolated from infections of joint prostheses, which includes some mutations caused by insertion of an IS256 element [29]. Moreover, polymorphisms within the agr locus for staphylococci are associated with its pathogenicity [19,29,30]. We have also observed that agr-positive (with normal RNAIII transcription) Se clinical isolates retain capacity for self-renewal in long-term culture (Qin et al., unpublished data), suggesting that other mechanisms are responsible for self-renewal for these isolates. Another recent study reported that addition of a cyclic autoinducing peptide (AIP) to activate agr in S. aureus agr–positive strains mediated dramatic detachment of S. aureus biofilms through an increase in expression of Aur metalloprotease and the SplABCDEF serine proteases [31]. However, it is unclear whether these proteases may have similar functions in biofilms formed by agr–positive Se strains.
Expression of the gene encoding autolysin, atlE, was significantly increased in all 4 our clinical isolates. Previous data indicate that atlE expression is essential for initial cell attachment and biofilm formation by Se[7,11,13]. We previously reported that isogenic deletion of atlE in Se 1457 significantly reduced cell attachment, extracellular DNA release, cell autolysis and final biofilm formation [11]. We and others found that atlE transcripts were significantly increased in Se 1457 agr mutants, which exhibited enhanced cell attachment, extracellular DNA release, cell death ( atlE-mediated autolysis) and subsequent biofilm formation [13]. In contrast, we found that Se 1457 agr/atlE double mutant seriously impaired these features mentioned above in the current study. In fact, we think that increased densities of microcolonies in Se mutant mature biofilms will cause more cell death and detachment due to nutrition deficiency, oxygen stress or other reasons required further investigation. In addition, other mechanisms have also been recently reported to be related with staphylococcal extracellular DNA release and biofilm dissemination, including the cidA murein hydrolase regulator [32] and the β subclass of phenol-soluble modulins (PSMs) [26].
Conclusion
Taken together, our studies illuminate a novel mechanism for enhancing Se biofilm self-renewal: repression of agr and induction of atlE expression, which results in increased cell autolysis/death, biofilm dispersal and final promoting new biofilm formation. Better understanding the process and mechanisms of Se biofilm self-renewal in patients will help us develop more effective strategies against Se biofilm-related infection.
Competing interests
All authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: LY, ZQ and SM. Performed the experiments: LD, LY, VJF and ZQ. Analyzed the data: LD and ZQ. Contributed reagents/materials/analysis tools: VJF, CP and SM. Wrote the manuscript: CP, SM and ZQ. All authors read and approved the final manuscript.
Supplementary Material
Figure S1.S. epidermidis 1457 agr mutation does not affect bacterial growth. Growth curves for S. epidermidis 1457 wild type and agr mutant and agr/atlE double mutant cultivated in TSB batch cultures are shown. Data shown represent one of 3 independent experiments.
Figure S2.S. epidermidis isolates associated with catheter infection exhibit differential expression of genes associated with biofilm formation. The expression profiles of RNAIII, atlE and icaA were compared for 6-d biofilm cells of laboratory strain and clinical isolates using qRT-PCR as described in Methods. Error bars represent the S.E.M. for three independent experiments.
Figure S3.S. epidermidis agr system regulates cell autolysis through atlE. Triton X-100 induced cell autolysis assays were performed as described in Methods, and error bars represent the S.E.M. for three independent experiments.
Figure S4. Sequence alignment analysis of agr conserved regions from ATCC 35984, Se-1, Se-2 and Se-3. The agr conserved regions were amplified and sequenced as described in Methods, then alignment analysis was performed by using Vector NTI Advance 9 software (Invitrogen).
Table S1. Primer sequences for qRT-PCR in this study.
Contributor Information
Lu Dai, Email: dailu@musc.edu.
Liang Yang, Email: yangliang8418@hotmail.com.
Chris Parsons, Email: parsonch@musc.edu.
Victoria J Findlay, Email: findlay@musc.edu.
Søren Molin, Email: sm@bio.dtu.dk.
Zhiqiang Qin, Email: zqin@lsuhsc.edu.
Acknowledgement
This work was supported by grants from the National Natural Science Foundation for Young Scientist of China (81101791 to Z.Q.). Z.Q. was also supported by the DANIDA fellowship during his visit at DTU. L.Y. was supported by a grant from the Danish Research Council for Independent Research (09-073917).
References
- Raad II, Bodey GP. Infectious complications of indwelling vascular catheters. Clin Infect Dis. 1992;15(2):197–208. doi: 10.1093/clinids/15.2.197. [DOI] [PubMed] [Google Scholar]
- Rupp ME, Archer GL. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin Infect Dis. 1994;19(2):231–243. doi: 10.1093/clinids/19.2.231. quiz 244-235. [DOI] [PubMed] [Google Scholar]
- von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis. 2002;2(11):677–685. doi: 10.1016/S1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- Vadyvaloo V, Otto M. Molecular genetics of Staphylococcus epidermidis biofilms on indwelling medical devices. Int J Artif Organs. 2005;28(11):1069–1078. doi: 10.1177/039139880502801104. [DOI] [PubMed] [Google Scholar]
- Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43(6):1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Mack D, Becker P, Chatterjee I, Dobinsky S, Knobloch JK, Peters G, Rohde H, Herrmann M. Mechanisms of biofilm formation in Staphylococcus epidermidis and Staphylococcus aureus: functional molecules, regulatory circuits, and adaptive responses. Int J Med Microbiol. 2004;294(2–3):203–212. doi: 10.1016/j.ijmm.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24(5):1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183(7):1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178(1):175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Riedewald J, Rohde H, Magnus T, Feucht HH, Elsner HA, Laufs R, Rupp ME. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect Immun. 1999;67(2):1004–1008. doi: 10.1128/iai.67.2.1004-1008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153(Pt 7):2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182(6):1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(6):1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170(9):4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JD, Maaloe O. DNA replication and the cell cycle in Escherichia coli cells. J Mol Biology. 1967;23(2):99–112. [Google Scholar]
- Jager S, Mack D, Rohde H, Horstkotte MA, Knobloch JK. Disintegration of Staphylococcus epidermidis biofilms under glucose-limiting conditions depends on the activity of the alternative sigma factor sigmaB. Appl Environ Microbiol. 2005;71(9):5577–5581. doi: 10.1128/AEM.71.9.5577-5581.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller S, Sternberg C, Andersen JB, Christensen BB, Ramos JL, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64(2):721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Guan M, Jiang XF, Yuan FY, Xu M, Zhang WZ, Lu Y. Genetic polymorphism of the accessory gene regulator (agr) locus in Staphylococcus epidermidis and its association with pathogenicity. J Med Microbiol. 2004;53(Pt 6):545–549. doi: 10.1099/jmm.0.05406-0. [DOI] [PubMed] [Google Scholar]
- Pynnonen M, Stephenson RE, Schwartz K, Hernandez M, Boles BR. Hemoglobin promotes Staphylococcus aureus nasal colonization. PLoS Pathog. 2011;7(7):e1002104. doi: 10.1371/journal.ppat.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC, Holmes CJ. Effect of vancomycin hydrochloride on Staphylococcus epidermidis biofilm associated with silicone elastomer. Antimicrob Agents Chemother (Bethesda) 1987;31(6):889–894. doi: 10.1128/aac.31.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser BL, Taylor D, Dix BA, Cleeland R. Method of evaluating effects of antibiotics on bacterial biofilm. Antimicrob Agents Chemother (Bethesda) 1987;31(10):1502–1506. doi: 10.1128/aac.31.10.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37(6):1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitz AM, Yu F, Hermsen ED, Rupp ME, Fey PD, Olsen KM. Vancomycin susceptibility trends and prevalence of heterogeneous vancomycin-intermediate Staphylococcus aureus in clinical methicillin-resistant S. aureus isolates. J Clin Microbiol. 2011;49(1):269–274. doi: 10.1128/JCM.00914-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair CG, Gorman SP, Feron BM, Byers LM, Jones DS, Goldsmith CE, Moore JE, Kerr JR, Curran MD, Hogg G. et al. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 1999;25(10):1072–1076. doi: 10.1007/s001340051014. [DOI] [PubMed] [Google Scholar]
- Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19(9):449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides. 2001;22(10):1603–1608. doi: 10.1016/S0196-9781(01)00495-8. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190(8):1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- Moore PC, Lindsay JA. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J Clin Microbiol. 2001;39(8):2760–2767. doi: 10.1128/JCM.39.8.2760-2767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(19):8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.S. epidermidis 1457 agr mutation does not affect bacterial growth. Growth curves for S. epidermidis 1457 wild type and agr mutant and agr/atlE double mutant cultivated in TSB batch cultures are shown. Data shown represent one of 3 independent experiments.
Figure S2.S. epidermidis isolates associated with catheter infection exhibit differential expression of genes associated with biofilm formation. The expression profiles of RNAIII, atlE and icaA were compared for 6-d biofilm cells of laboratory strain and clinical isolates using qRT-PCR as described in Methods. Error bars represent the S.E.M. for three independent experiments.
Figure S3.S. epidermidis agr system regulates cell autolysis through atlE. Triton X-100 induced cell autolysis assays were performed as described in Methods, and error bars represent the S.E.M. for three independent experiments.
Figure S4. Sequence alignment analysis of agr conserved regions from ATCC 35984, Se-1, Se-2 and Se-3. The agr conserved regions were amplified and sequenced as described in Methods, then alignment analysis was performed by using Vector NTI Advance 9 software (Invitrogen).
Table S1. Primer sequences for qRT-PCR in this study.


