Abstract
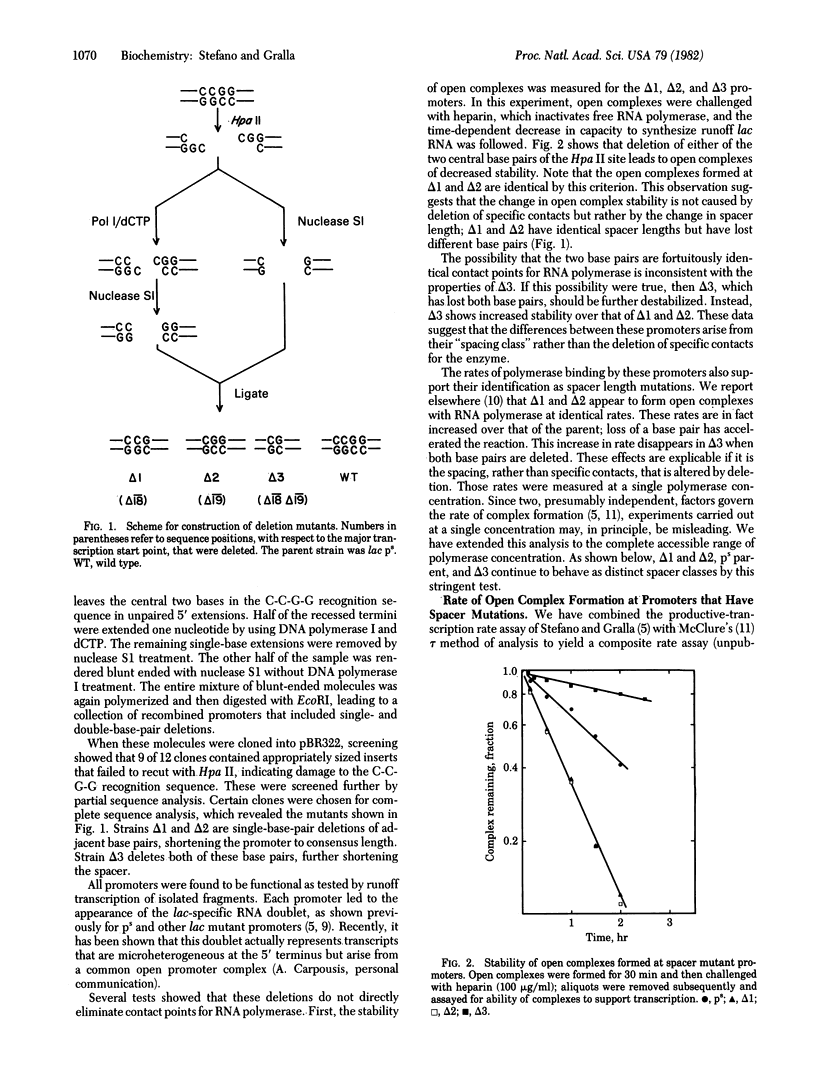
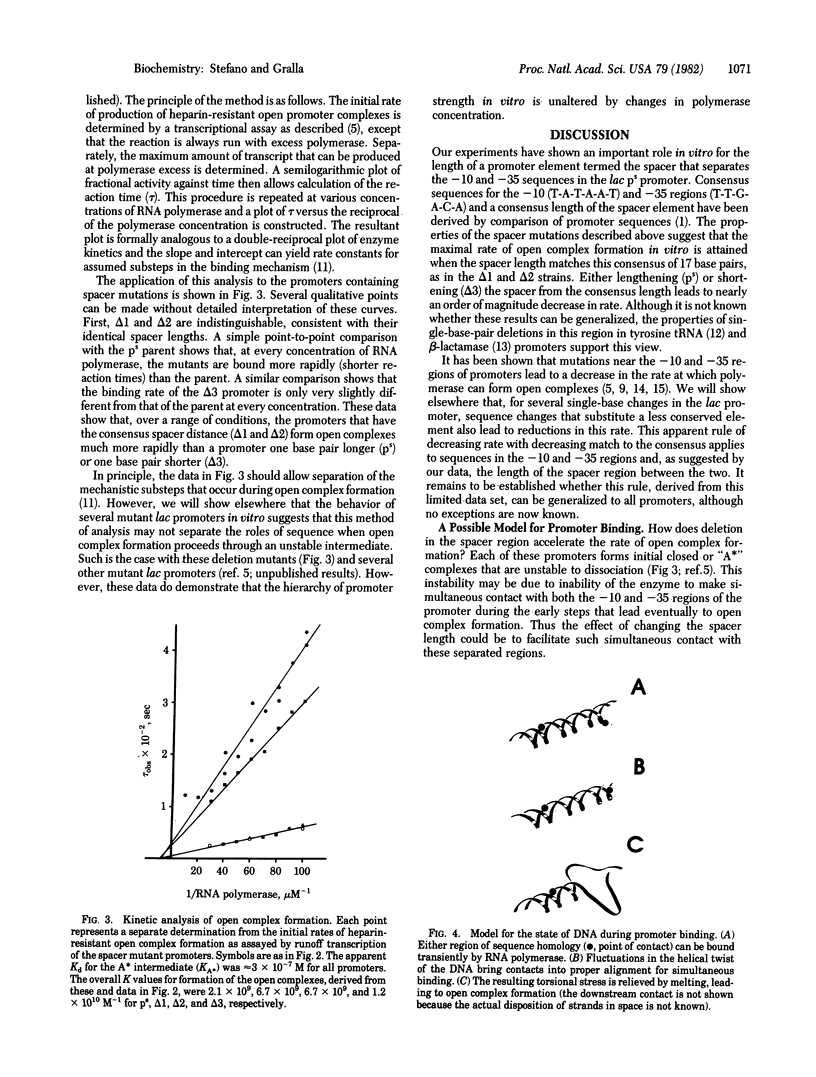
Mutations have been constructed that delete either one or two base pairs near position -19 in the lac ps promoter. Deletion of either of two adjacent base pairs increases the rates of open complex formation by nearly on order of magnitude. Two promoters that have different single-base deletions are indistinguishable by either their rates of open complex formation or stability of the open complexes once formed. However, simultaneous deletion of both base pairs produces a promoter that forms complexes at a rate similar to that of the unmodified DNA sequence. The maximal rate of open complex formation is achieved at a spacer length of 17 base pairs, the most frequently occurring spacer length among promoters. These results suggest that the spacing between the two strongly conserved regions of sequence homology is an important determinant of the rate of open complex formation. A model is suggested that proposes that three important promoter elements, the -10 region, the -35 region, and the spacer region, act simultaneously to facilitate open complex formation by RNA polymerase.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman M. L., Landy A. Promoter mutations in the transfer RNA gene tyrT of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4303–4307. doi: 10.1073/pnas.76.9.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. In vitro comparison of initiation properties of bacteriophage lambda wild-type PR and x3 mutant promoters. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6381–6385. doi: 10.1073/pnas.77.11.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Grundström T., Edlund T., Normark S. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981 Mar 19;290(5803):221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Reznikoff W. S. In vitro analysis of the Escherichia coli RNA polymerase interaction with wild-type and mutant lactose promoters. J Mol Biol. 1978 Nov 15;125(4):467–490. doi: 10.1016/0022-2836(78)90311-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Rate-limiting steps in RNA chain initiation. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Stefano J. E., Ackerson J. W., Gralla J. D. Alterations in two conserved regions of promoter sequence lead to altered rates of polymerase binding and levels of gene expression. Nucleic Acids Res. 1980 Jun 25;8(12):2709–2723. doi: 10.1093/nar/8.12.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano J. E., Gralla J. D. Kinetic investigation of the mechanism of RNA polymerase binding to mutant lac promoters. J Biol Chem. 1980 Nov 10;255(21):10423–10430. [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]



