Abstract
Drosophilists have identified many, or perhaps most, of the key regulatory genes determining sex using classical genetics, however, regulatory genes must ultimately result in the deployment of the genome in a quantitative manner, replete with complex interactions with other regulatory pathways. In the last decade, genomics has provided a rich picture of the transcriptional profile of the sexes that underlies sexual dimorphism. The current challenge is linking transcriptional profiles with the regulatory genes. This will be a complex synthesis, but the prospects for progress are outstanding.
Keywords: drosophila, transcriptome, sex determination, effector, selector
ACTIVATORS, SELECTORS AND REALIZATORS
In the heady days when the idea that genes controlled development in a hierarchical manner was gaining ground, Garcia-Bellido [1] formalized the classification of genes responsible for patterning the anterior–posterior axes of Drosophila. In this model, activators gather positional information to turn on (or off) selector genes for particular fates, which in turn deploy a battery of genes that do the work of producing the phenotype. A tree is a good metaphor for this concept, where the activators are the roots, the selectors are trunks and branches and the realizators are leaves.
Molecular genetics has been incredibly adept at identifying and understanding the regulatory selectors in the trunk. Loss of these genes results in clean and overt phenotypes. Loss of activators often results in broad phenotypes, and molecular genetics is quite useful for finding these through a combination of expression analysis in mutants and genetic interactions. Molecular genetics does not do a very good job with the leaves. This is almost certainly due to the fact that many realizator genes, by definition, make smaller contributions to the phenotype. Losses of a few leaves here and there are easy to overlook, but these small additive or synergistic effects are exactly the type of phenotypes that we will need to understand when thinking about oligogenic genotypes associated with disease risk in humans, or underlying the small differences in fitness that drive evolution. Identifying and understanding the role of the genes in the canopy permits a full understanding of a developmental pathway.
Transcript profiling first by microarrays and now by RNA-Seq is an appealing tool for surveying the number and types of leaves on the tree. More importantly, gene expression can be used to cluster sets of co-expressed genes [2]. It was hoped that these batteries of co-regulated genes could be easily linked to transcription factors by profiling mutants and then discovering the enriched motifs in the cis-regulatory sequences proximal to the members of those gene batteries. These studies have seen mixed success, as it became clear that genes that are co-expressed are not usually co-regulated by a clean set of conserved selectors [3]. Even though working backwards from the leaves is too complex due to multiple solutions to a given transcriptional output, there is progress being made working outwards from known genes using combinations of expression profiling in selector gene mutants, determining where transcription factors bind by chromatin immunoprecipitation (ChIP), and of course genetics [3–6].
Early in the age of gene discovery Morgan, Sturtevant and others identified mutations in transformer (tra) [7] and intersex (ix) [8] that transformed females into males and revealed that sex determination in flies is genetic in nature and is broadly regulated by a few key genes. Bridges clearly understood that the contribution of many genes located on the X chromosome was important for determining sex [9]. The core roots, trunk and major branches of the sex determination pathway were established by 1980 [10] and have stood the test of time [11]. The following decades of genetics and biochemistry gave form to our molecular understanding of somatic sex determination that terminates with the transcription factors doublesex (DSX) and fruitless (FRU) for sexual dimorphisms, and the chromatin-modifying dosage compensation complex (DCC) for sex-specific viability [12] (Figure 1). Thanks to the capabilities of genome-wide transcription profiling, we can now see the leaves. Still unresolved are the branches between the leaves and trunk, which are key to understanding how sex-specific phenotypes and transcriptomes are linked to the trunk. Here we describe as much of the tree as is currently visible with a concentration on the structure and diversity of the canopy.
Figure 1:
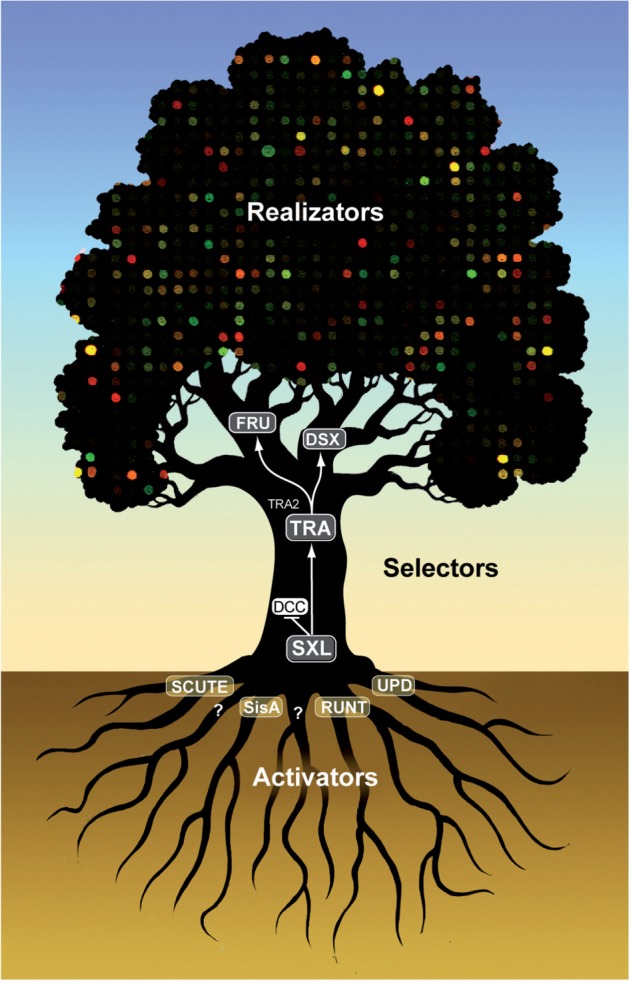
The somatic sex determination pathway of Drosophila melanogaster. SisA, sisterless-A; UPD, unpaired; SXL, sex lethal; DCC, dosage compensation complex; TRA, transformer; TRA2, transformer2; DSX, doublesex; FRU, fruitless.
THE ROOTS AND TRUNK
The activators of somatic sex determination that lie at the roots of the somatic sex determination pathway are a collection of genes located on the X chromosome (Scute, Sisterless-A, Runt and unpaired) whose protein products induce expression at the ‘early’ promoter (Pe) of Sex lethal (Sxl), the first selector gene of the pathway [13]. It is thought that the Pe responds directly to the concentrations of these X-linked gene products and that the threshold concentration for Pe activation is reached in females but not in males [13]. SXL protein produced from the Pe-produced mRNA acts to ensure continuous SXL protein production only in females by blocking splicing of a Sxl mRNA isoform from the maintenance promoter (Pm) that produces an untranslatable Sxl isoform containing stop codons in the open reading frame [14]. Without SXL protein produced from the Pe in males, Sxl mRNA produced from the Pm is spliced into the untranslatable form and thus Sxl is kept ‘off’ in males and ‘on’ in females [13, 14]. SXL protein functions in sex determination in females by directing the splicing of tra mRNA into a translatable form while in males, where Sxl is absent, tra mRNA is spliced into an isoform containing an exon with stop codons in all reading frames located near the start of the open reading frame that prohibits translation of the full-length mRNA [15, 16]. The presence of TRA protein in females enables splicing of dsx mRNA into a female-specific isoform (dsxF), whereas in males, where TRA protein is not made, dsx mRNA is spliced into a male-specific isoform (dsxM) [17–19]. Male and female DSX isoforms both encode zinc finger proteins that are identical at the amino terminus where the DNA binding domain is found but differ at their carboxy termini [20–22]. TRA is also responsible for splicing a subset of fru mRNAs into female- and male-specific isoforms. In females, fru mRNA from the P1 promoter is spliced under direction of TRA into an mRNA that upon translation, produces a truncated nonfunctional protein, whereas in males, fru mRNA from the P1 promoter encodes an intact male-specific Fru protein isoform (FruM) characterized as a BTB-POZ transcription factor [23–26].
DSX is responsible for almost all somatic sex determination throughout the Drosophila body including functions in the CNS while FRU functions exclusively in the CNS [24, 25, 27]. Biochemical analysis of DSXF and DSXM proteins revealed that they both bind directly to specific DNA sequences in the enhancer of the sex-biased expressed yolk protein (Yp) genes in the adult fatbody, and it was concluded that DSX functions as a terminal transcription factor in this pathway [21, 22]. Other genes that have roles in somatic sex determination include intersex (ix) and hermaphrodite (her). ix functions in parallel or downstream of dsx and has been identified as a protein partner for the female-specific DSXF that is required for DSXF function [10, 28–30]. HER encodes a zinc finger protein whose function is dependent on DSX for some phenotypes (female-specific foreleg bristles and pigmentation of tergites 5 and 6) and independent of DSX for a role in Yp gene expression activation in females and development of vaginal teeth. With the core members of the somatic sex determination [31, 32] pathway identified and the observation that genes in the terminal positions of the sex determination pathway are transcription factors, the pursuit of traditional genetic screens to identify other genes with global roles in sex determination in Drosophila would unlikely be fruitful. The study of sex determination in Drosophila was ripe for the genomics era.
THE LEAVES
The development of high-throughput techniques for genome-wide transcriptome analysis has provided insight into the impact of sex on gene expression. Some of the earliest microarray studies performed in Drosophila included analysis of male and female adult transcriptomes and revealed thousands of genes with sex-biased gene expression [33–37]. The early reports of widespread sex-biased gene expression with arrays that assayed portions of the Drosophila genome have been corroborated with arrays that complement the ever updating annotations of the fully sequenced Drosophila genome [38–46] and now RNA-sequencing (RNA-seq) [47]. Studies using a variety of microarray platforms and analysis methods or RNA-sequencing report that 17–73% of the genome is expressed in a sex-biased manner in whole adult flies of various Drosophila melanogaster strains [35, 36, 40–42, 46, 47]. The exact number of sex-biased genes depends on thresholds used for significance. Genes with male-biased or male-specific expression are observed in greater numbers and with overall higher expression levels compared to genes with female-biased or female-specific expression, a characteristic that is observed in multiple Drosophila species [34, 42, 43, 46–48]. Expression studies of whole flies, ovaries and testes, gonadectomized male and female adults or tudor mutant adult males or females that lack a germline revealed that the majority of sex-biased gene expression in adults occurs in the gonads [33, 34, 42, 49]. Transcript-specific microarrays and RNA-seq have enabled the study of sex-biased gene expression at the transcript level in whole flies and dissected tissues where hundreds of transcript isoforms with sex-biased transcription have been detected [41, 47, 50–52]. McIntyre et al. [41] estimate that up to 22% of genes represented by multiple transcripts in the Drosophila genome express isoforms in a sex-biased manner, revealing that sex-biased use of transcript isoforms is a widely used mechanism to diversify a sex-biased transcriptome. As described above, sex-biased transcript isoform production is certainly important for establishment and maintenance of the somatic sex determination pathway as all selector genes use alternative splicing to restrict protein products to a particular sex [14, 16–19].
The interest in the extent of sex-differential gene expression and sexual differentiation of diverse tissues has led researchers to perform microarray, RNA-seq, serial analysis of gene expression and some medium-throughput transcriptome experiments on many sexed tissues and developmental stages of Drosophila, including intact head and dissected CNS [50, 53–55], genital disc [56], leg [57, 58], developmentally staged pupae [59], male accessory gland [60], embryonic gonads [61], ovaries and testes [34, 42, 43, 51] and bam mutant ovaries and testes that arrest at early stages of germ cell development in both female and males and are therefore enriched for germ stem cells and early stage germ cell cysts [51, 62]. A list of publications using medium- to high-throughput techniques to identify sex-differentially expressed genes is presented in Table 1. Publicly available databases for genes with sex-biased expression in Drosophila melanogaster, Drosophila simulans and Anopheles gambiae include Sebida, FlyAtlas and MozAtlas [63–65]. Comparing genes identified with sex-biased expression in various dissected tissues reveals that tissues typically express unique sets of genes in a sex-biased manner (Table 1). Furthermore, when sex-differential expression of whole flies was analyzed over a time course of development during metamorphosis, different stages were associated with distinct sets of genes with few genes with sex-biased gene expression shared between stages [59]. These observations suggest that many genes participate in sexual development and differentiation and the connection of branches to leaves will depend on spatial and temporal context.
Table 1:
Publications reporting medium- to high-throughput sex-biased gene expression in Drosophila melanogaster: whole flies or dissected tissues
| References | Tissue | Journal |
|---|---|---|
| Andrews et al. [34] | Gonad | Genome Research 10:2030–43 |
| Swanson et al. [60] | Male accessory gland | PNAS 98:7375–9 |
| Jin et al. [35] | Whole adult | Nature Genetics 29:389–95 |
| Arbeitman et al. [33] | Whole adult | Science 297:2270–75 |
| Dauwalder et al. [54] | Head | Genes and Development 16:2879–92 |
| Fujii and Amrein [55] | Head | EMBO J 21:5353–63 |
| Xu et al. [58] | Leg | Cell Tissue Research 307:381–92 |
| Parisi et al. [43] | Whole adult, Gonad | Science 299:697–700 |
| Ranz et al. [36] | Whole adult | Science 300:1742–5 |
| Meiklejohn et al. [37] | Whole adult | PNAS 100:9894–9 |
| Stolc et al. [44] | Whole adult | Science 306:655–60 |
| Gibson et al. [40] | Whole adult | Genetics 167:1791–99 |
| Arbeitman et al. [49] | Whole adult | Development 131:2007–21 |
| Parisi et al. [42] | Gonad, whole adult and gonadectomized adult | Genome Biology 5:R40 |
| Barmina et al. [57] | Leg | Developmental Biology 288:528–44 |
| McIntyre et al. [41] | Whole adult | Genome Biology 7:R79 |
| Baker et al. [39] | Whole adult | BMC Genomics 8:454 |
| Chintapalli et al. [64] | Multiple tissues, Whole adult | Nature Genetics 39:715–20 |
| Goldman et al. [53] | Head | PLoS Genetics 3:e216 |
| Zhang et al. [46] | Whole adult | Nature 450:233–7 |
| Ayroles et al. [38] | Whole adult | Nature Genetics 41:299–307 |
| Lebo et al. [59] | Whole pupae | BMC Genomics 10:80 |
| Casper and Van Doren [61] | Embryonic gonad | Development 136: 3821–30 |
| Telonis-Scott et al. [45] | Whole adult | Genetics 181: 421–34 |
| Parisi et al. [66] | Adult carcass | BMC Genomics 11:346 |
| Gan et al. [51] | Gonad | Cell Research 20:763–83 |
| Chang et al. [50] | Head | BMC Genomics 12:364 |
| Chatterjee et al. [56] | Genital disc | Development 138:1099–1109 |
| Hartmann et al. [52] | Whole adult | RNA 17:453–68 |
| Graveley et al. [47] | Whole adult | Nature 471:473–9 |
Multiple genome-wide studies reveal that the gonads are the greatest source of sex-biased gene expression in Drosophila. This may be expected given their role in producing sex-specific gametes that are completely different in form. What is clear is that female and male gonads use distinct batteries of genes to produce their respective gamete. Testes are observed to express greater numbers of genes than ovaries and more genes with higher expression levels [34, 42, 51]. Furthermore, many of the testis-expressed genes appear unique to the testis [34]. Sex-biased transcript isoforms are also abundant in the gonad where hundreds of sex-specific or sex-biased transcript isoforms have been identified [45, 51, 52]. The greatest divergence of the male and female gonadal transcriptomes appears to occur when germ cells differentiate into eggs and sperm as populations of male and female germ stem cells and early dividing cysts in bam mutant gonads display higher transcriptome congruence (r = 0.716) than wild-type gonads (r = 0.182) [51]. Linking the unique batteries of expressed genes in male and female gonads to the somatic sex determination pathway may be especially challenging. Germ cell sex determination is achieved through nonautonomous action of the somatic sex determination pathway members in the somatic cells and intrinsic factors in the germ cells themselves [67]. dsx is not expressed in the germ cells and consistent with this observation, genetic data demonstrate that tra and dsx have no cell-autonomous function in germ cell sex determination [68–72]. It has been recently shown that Sxl is required intrinsically for female germ cell sex determination, revealing the existence of another branch from the selector trunk [73]. To date the only known signaling pathway under dsx control in the somatic gonadal cells affecting male germ cell sex-biased gene expression is the JAK/STAT pathway [74]. Identifying direct targets of SXL in embryonic germ cells and the signaling molecules under DSX control that emanate from the somatic cells to instruct germ cell sex determination is critical for making the connections between the sex-biased gene expression in the gonad—the largest source of sex-biased gene expression in the whole fly—to the pathway.
LOOKING FOR MINOR BRANCHES
Several studies have investigated the dependence of genome-wide sex-biased gene expression on members of the sex determination pathway [49, 50, 53, 59]. Expression profiles of wild-type sexed flies or tissues have been compared to those of tra and dsx mutants to identify genes under control of sex-determination pathway members. These studies suggest that genes with sex-biased gene expression may be regulated at all levels of the sex determination pathway: upstream of TRA, downstream of TRA and regulated by DSX or FRU, or downstream of TRA and not regulated by DSX or FRU; however, genetic and phenotypic confirmations of these observations are needed. There is also a physiological control of sex-biased expression as the status of the germline alters sex-biased expression in distant somatic tissues [66] and as tra and dsx mutations disrupt the germline some of the genes that appear to regulated upstream of tra or dsx may in fact be an indirect effect of the atrophied gonads of tra and dsx mutants. fit (female-specific independent of transformer) is an example of a gene with female-biased expression in the fat cells of the head that is regulated upstream of TRA (expression is associated only with XX individuals regardless of TRA function) and responsiveness to germline status [55, 66]. However, in fit’s case, lack of a germline has been shown to increase female-biased expression revealing negative feedback from the germline to the distant fat cells of the head and complexity of regulation for sex-biased gene expression [55, 66]. Genes potentially regulated upstream of TRA have been postulated to either be regulated directly by SXL or result from an effect of X chromosome composition [50, 53]. Evidence for this latter hypothesis is supported by the observation that genes demonstrating sex-biased expression in the Drosophila head and are not dependent on TRA are enriched on the X chromosome and may be associated with DCC entry sites [50, 53]. As a means of identifying genes that could be regulated directly by SXL, genes with sex-biased expression not dependent on TRA at the gene or transcript level were searched for matches to the SXL sequence-binding motif [50, 52]. These searches yielded few candidate SXL direct target genes (<10% of sex-biased transcript isoforms contained a SXL binding sequence in Chang et al. [50] and 9% of the sex-biased transcript isoforms in Hartmann et al. [52] when using a 100 bp window on either side of the splice site associated with sex-biased exon usage), although Chang et al. [50] caution that the SXL binding site has not been well characterized which would limit this analysis. A low yield also occurred when looking for TRA binding sites among the 362 genes in the head with sex-biased transcript isoforms where six genes were identified with a TRA binding site and these six included dsx, fru and Sxl [50]. Tissue specificity and developmental context should be considered in any future searches for direct SXL or TRA targets. The ability of ectopic Sxl expression to enable XY germ cells to enter the oogenesis pathway when transplanted into XX hosts depended on the developmental window of Sxl expression [73]. Sxl expressed in XY germ cells from embryonic stage 9 onwards was sufficient to allow oocyte production, whereas Sxl expression in XY germ cells beginning at embryonic stage 16/17 was not [73].
Transcriptome profiling experiments of dsx mutants have also shed light on DSXs transcriptional control of its targets [49, 53, 59]. For those genes identified as downstream of TRA and DSX, genome-wide transcriptome experiments have indicated that both DSX protein isoforms may be capable of either activation or repression of its targets [49, 53, 59]. In the genetic and biochemical characterization of DSX’s role in directly regulating transcriptional activity of the Yp1 and Yp2 genes at their shared enhancer, it was observed that both DSXF and DSXM directly bind the enhancer sequence and that DSXF activates Yp gene expression in females, whereas DSXM protein represses Yp gene expression in males [21, 30, 75]. The opposite transcriptional outcomes by DSX sex-specific isoform regulation of the same target gene became a model for how DSX may generally direct sexual differentiation through its target genes. This model holds true for another identified direct DSX target gene, bab1 [76]. However, comparison of expression profiles of wild-type, tra and dsx mutant tissues followed up with RNA in situ experiments suggest that DSX target genes may be regulated by DSX isoforms in diverse ways that can be classified in four different modes: (i) activation by DSXF and repression by DSXM as observed for the Yp genes; (ii) repression by DSXF and activation by DSXM; (iii) activation by both DSXF and DSXM; and (iv) repression by both DSXF and DSXM [49, 53, 59]. A solo mode exists as well: the DSX direct target gene Fad2/DesatF is activated only by DSXF protein and DSXM apparently plays no role in repressing its expression in males [77]. Identification of all DSX target genes and characterization of DSX’s role in their transcriptional activity are needed to understand the full spectrum of DSX’s function in transcriptional regulation.
Identifying the direct target genes of the transcription factors at the terminal end of the pathway is an essential step in understanding how sex-biased or sex-specific gene expression is produced and regulated. To this end, Luo et al. [78] have used the DamID protocol to identify the genome-wide targets of the DSXF protein. Among their list of genes associated with bound DSX protein is the already defined DSX target bab1, and bnl, a gene shown to be under DSX regulation in the genital disc [76, 79] suggesting that this data set will be a valuable asset for identifying direct DSX target genes and making connections between the sex determination pathway and genes with sex-biased gene expression.
CHALLENGES
The clear bottleneck for our understanding of sex determination and differentiation is in the canopy. We will need to develop a more refined map of the leaves, looking at individual tissues and cells through development, as well as the dependencies due to nonautonomous interactions among branches. We will also need to identify the direct targets of each branch of pathway, again in the context of space and time. This march toward the leaves will probably be iterative as more and more branch points are identified. Finally, we should not be overly enamored with hierarchical models, as sex determination intersects with multiple other pathways [76, 80, 81]. For example, to make sure that sex combs develop on the first leg only in males and that sex-specific pigmentation differences occur in the abdomen; sex-determining inputs must be coordinated with the HOX activators, selectors and realizators outlined by Garcia-Bellido.
Key point.
Genomics has given us an outstanding picture of transcription in the Drosophila sexes. The future challenge will be to link these changes to the key regulatory genes that were identified in the last century.
ACKNOWLEDGEMENTS
We thank our colleagues in the lab for many discussions and comments on the manuscript. This work supported by the Intramural Research Program of the NIH, NIDDK.
Biography
Emily Clough received her PhD degree working with Dr. Tulle Hazelrigg at Columbia University and is a postdoctoral fellow working on identifying DSX target genes.Brian Oliver received his PhD in the Antony Mahowald lab at Case Western Reserve University and postdoctoral training in the Bruce Baker lab at Stanford. He is currently chief, section of Developmental Genomics.
References
- 1.Garcia-Bellido A. Genetic control of wing disc development in Drosophila. Ciba Found Symp. 1975;29:161–82. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- 2.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan ET, Quon GT, Chua G, et al. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanut-Delalande H, Fernandes I, Roch F, et al. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 2006;4:e290. doi: 10.1371/journal.pbio.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busser BW, Bulyk ML, Michelson AM. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev. 2008;18:521–9. doi: 10.1016/j.gde.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986;6:3847–53. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturtevant A. A gene in Drosophila melanogaster that transforms females into males. Genetics. 1945;30:297–9. doi: 10.1093/genetics/30.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan TH, Redfield H, Morgan LV. Maintenance of a Drosophila Stock Center, in Connection with Investigations on the Constitution of the Germinal Material in Relation to Heredity. Vol. 42. Washington, DC: Yearbook of the Carnegie Institution; 1943. pp. 171–4. [Google Scholar]
- 9.Bridges CB. Triploid intersexes in Drosophila melanogaster. Science. 1921;54:252–4. doi: 10.1126/science.54.1394.252. [DOI] [PubMed] [Google Scholar]
- 10.Baker BS, Ridge KA. Sex and the single cell: I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- 12.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2012;13:123–34. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 13.Salz HK, Erickson JW. Sex determination in Drosophila: the view from the top. Fly. 2010;4:60–70. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell LR, Maine EM, Schedl P, et al. Sex-lethal, a Drosophila sex determinaton switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 1988;55:1037–46. doi: 10.1016/0092-8674(88)90248-6. [DOI] [PubMed] [Google Scholar]
- 15.McKeown M, Belote JM, Baker BS. A molecular analysis of transformer, a gene in Drosophila melanogaster that controls female sexual differentiation. Cell. 1987;48:489–99. doi: 10.1016/0092-8674(87)90199-1. [DOI] [PubMed] [Google Scholar]
- 16.Boggs RT, Gregor P, Idriss S, et al. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987;50:739–47. doi: 10.1016/0092-8674(87)90332-1. [DOI] [PubMed] [Google Scholar]
- 17.Nagoshi RN, McKeown M, Burtis KC, et al. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–36. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 18.Sosnowski BA, Belote JM, McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989;58:449–59. doi: 10.1016/0092-8674(89)90426-1. [DOI] [PubMed] [Google Scholar]
- 19.Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes Dev. 1988;2:477–89. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- 20.Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 1993;12:527–35. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burtis KC, Coschigano KT, Baker BS, et al. The Doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10:2577–82. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 23.Goodwin SF, Taylor BJ, Villella A, et al. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics. 2000;154:725–45. doi: 10.1093/genetics/154.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito H, Fujitani K, Usui K, et al. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci USA. 1996;93:9687–92. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryner LC, Goodwin SF, Castrillon DH, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–89. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 26.Heinrichs V, Ryner LC, Baker BS. Regulation of sex-specific selection of fruitless 5' splice sites by transformer and transformer-2. Mol Cell Biol. 1998;18:450–8. doi: 10.1128/mcb.18.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hildreth PE. Doublesex, a recessive gene that transforms both males and females of Drosophila into intersexes. Genetics. 1965;51:659–78. doi: 10.1093/genetics/51.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrett-Engele CM, Siegal ML, Manoli DS, et al. Intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development. 2002;129:4661–75. doi: 10.1242/dev.129.20.4661. [DOI] [PubMed] [Google Scholar]
- 29.Chase BA, Baker BS. A genetic analysis of intersex, a gene regulating sexual differentiation in Drosophila melanogaster females. Genetics. 1995;139:1649–61. doi: 10.1093/genetics/139.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterbury JA, Jackson LL, Schedl P. Analysis of the doublesex female protein in Drosophila melanogaster: role on sexual differentiation and behavior and dependence on intersex. Genetics. 1999;152:1653–67. doi: 10.1093/genetics/152.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Baker BS. Her, a gene required for sexual differentiation in Drosophila, encodes a zinc finger protein with characteristics of ZFY-like proteins and is expressed independently of the sex determination hierarchy. Development. 1998;125:225–35. doi: 10.1242/dev.125.2.225. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Baker BS. Hermaphrodite and doublesex function both dependently and independently to control various aspects of sexual differentiation in Drosophila. Development. 1998;125:2641–51. doi: 10.1242/dev.125.14.2641. [DOI] [PubMed] [Google Scholar]
- 33.Arbeitman MN, Furlong EE, Imam F, et al. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–5. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 34.Andrews J, Bouffard GG, Cheadle C, et al. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–43. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin W, Riley RM, Wolfinger RD, et al. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- 36.Ranz JM, Castillo-Davis CI, Meiklejohn CD, et al. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–5. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 37.Meiklejohn CD, Parsch J, Ranz JM, et al. Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci USA. 2003;100:9894–9. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayroles JF, Carbone MA, Stone EA, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker DA, Meadows LA, Wang J, et al. Variable sexually dimorphic gene expression in laboratory strains of Drosophila melanogaster. BMC Genomics. 2007;8:454. doi: 10.1186/1471-2164-8-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibson G, Riley-Berger R, Harshman L, et al. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics. 2004;167:1791–9. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIntyre LM, Bono LM, Genissel A, et al. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 2006;7:R79. doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parisi M, Nuttall R, Edwards P, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parisi M, Nuttall R, Naiman D, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stolc V, Gauhar Z, Mason C, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–60. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 45.Telonis-Scott M, Kopp A, Wayne ML, et al. Sex-specific splicing in Drosophila: widespread occurrence, tissue specificity and evolutionary conservation. Genetics. 2009;181:421–34. doi: 10.1534/genetics.108.096743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Sturgill D, Parisi M, et al. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–7. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graveley BR, Brooks AN, Carlson JW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–9. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang ZF, Machado CA. Evolution of sex-dependent gene expression in three recently diverged species of Drosophila. Genetics. 2009;183:1175–85. doi: 10.1534/genetics.109.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arbeitman MN, Fleming AA, Siegal ML, et al. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–21. doi: 10.1242/dev.01077. [DOI] [PubMed] [Google Scholar]
- 50.Chang PL, Dunham JP, Nuzhdin SV, et al. Somatic sex-specific transcriptome differences in Drosophila revealed by whole transcriptome sequencing. BMC Genomics. 2011;12:364. doi: 10.1186/1471-2164-12-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gan Q, Chepelev I, Wei G, et al. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010;20:763–83. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartmann B, Castelo R, Minana B, et al. Distinct regulatory programs establish widespread sex-specific alternative splicing in Drosophila melanogaster. RNA. 2011;17:453–68. doi: 10.1261/rna.2460411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldman TD, Arbeitman MN. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dauwalder B, Tsujimoto S, Moss J, et al. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002;16:2879–92. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujii S, Amrein H. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 2002;21:5353–63. doi: 10.1093/emboj/cdf556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatterjee SS, Uppendahl LD, Chowdhury MA, et al. The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development. 2011;138:1099–1109. doi: 10.1242/dev.055731. [DOI] [PubMed] [Google Scholar]
- 57.Barmina O, Gonzalo M, McIntyre LM, et al. Sex- and segment-specific modulation of gene expression profiles in Drosophila. Dev Biol. 2005;288:528–44. doi: 10.1016/j.ydbio.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 58.Xu A, Park SK, D'Mello S, et al. Novel genes expressed in subsets of chemosensory sensilla on the front legs of male Drosophila melanogaster. Cell Tissue Res. 2002;307:381–92. doi: 10.1007/s00441-002-0524-0. [DOI] [PubMed] [Google Scholar]
- 59.Lebo MS, Sanders LE, Sun F, et al. Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC Genomics. 2009;10:80. doi: 10.1186/1471-2164-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson WJ, Clark AG, Waldrip-Dail HM, et al. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA. 2001;98:7375–9. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casper AL, Van Doren M. The establishment of sexual identity in the Drosophila germline. Development. 2009;136:3821–30. doi: 10.1242/dev.042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–51. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 63.Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–9. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- 64.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 65.Baker DA, Nolan T, Fischer B, et al. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 2011;12:296. doi: 10.1186/1471-2164-12-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parisi MJ, Gupta V, Sturgill D, et al. Germline-dependent gene expression in distant non-gonadal somatic tissues of Drosophila. BMC Genomics. 2010;11:346. doi: 10.1186/1471-2164-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murray SM, Yang SY, Van Doren M. Germ cell sex determination: a collaboration between soma and germline. Curr Opin Cell Biol. 2010;22:722–9. doi: 10.1016/j.ceb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hempel LU, Oliver B. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Dev Biol. 2007;7:113. doi: 10.1186/1471-213X-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marsh JL, Wieschaus E. Is sex determination in germ line and soma controlled by separate genetic mechanisms? Nature. 1978;272:249–51. doi: 10.1038/272249a0. [DOI] [PubMed] [Google Scholar]
- 70.Rideout EJ, Dornan AJ, Neville MC, et al. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci. 2010;13:458–66. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinett CC, Vaughan AG, Knapp JM, et al. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schüpbach T. Autosomal mutations that interfere with sex determination in somatic cells of Drosophila have no direct effect on the germline. Dev Biol. 1982;89:117–27. doi: 10.1016/0012-1606(82)90300-1. [DOI] [PubMed] [Google Scholar]
- 73.Hashiyama K, Hayashi Y, Kobayashi S. Drosophila Sex lethal gene initiates female development in germline progenitors. Science. 2011;333:885–8. doi: 10.1126/science.1208146. [DOI] [PubMed] [Google Scholar]
- 74.Wawersik M, Milutinovich A, Casper AL, et al. Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature. 2005;436:563–7. doi: 10.1038/nature03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belote JM, Handler AM, Wolfner MF, et al. Sex-specific regulation of yolk protein gene expression in Drosophila. Cell. 1985;40:339–48. doi: 10.1016/0092-8674(85)90148-5. [DOI] [PubMed] [Google Scholar]
- 76.Williams TM, Selegue JE, Werner T, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–23. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shirangi TR, Dufour HD, Williams TM, et al. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo SD, Shi GW, Baker BS. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development. 2011;138:2761–71. doi: 10.1242/dev.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmad SM, Baker BS. Sex-specific deployment of FGF signaling in Drosophila recruits mesodermal cells into the male genital imaginal disc. Cell. 2002;109:651–61. doi: 10.1016/s0092-8674(02)00744-4. [DOI] [PubMed] [Google Scholar]
- 80.Tanaka K, Barmina O, Sanders LE, et al. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 2011;9:e1001131. doi: 10.1371/journal.pbio.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keisman EL, Baker BS. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshund sex-specifically in the genital imaginal disc. Development. 2001;128:1643–56. doi: 10.1242/dev.128.9.1643. [DOI] [PubMed] [Google Scholar]


