Abstract
The Drosophila dorsal-ventral (DV) axis is polarised when the oocyte nucleus migrates from the posterior to the anterior margin of the oocyte. Prior work suggested that dynein pulls the nucleus to the anterior side along a polarised microtubule cytoskeleton, but this mechanism has not been tested. By imaging live oocytes, we find that the nucleus migrates with a posterior indentation that correlates with its direction of movement. Furthermore, both nuclear movement and the indentation depend on microtubule polymerisation from centrosomes behind the nucleus. Thus, the nucleus is not pulled to the anterior, but is pushed by the force exerted by growing microtubules. Nuclear migration and DV axis formation therefore depend on centrosome positioning early in oogenesis and are independent of anterior-posterior axis formation.
The correct positioning of the nucleus is important for several developmental processes, such as cell migration, formation of the neuromuscular junction, and asymmetric cell divisions, whereas nuclear mislocalisation is a feature of neurological disorders, such as lissencephaly (1). Positioning of the nucleus plays an essential role in Drosophila axis formation, as the movement of the nucleus from the posterior of the oocyte to a point at its anterior circumference breaks radial symmetry to polarise the DV axis (2, 3). At stage 7 of oogenesis, an unknown signal from the posterior follicle cells induces a major reorganisation of the oocyte microtubule cytoskeleton. The posterior microtubule organising centre (MTOC) is disassembled, and microtubules are nucleated from the anterior-lateral cortex, resulting in an anterior-posterior (AP) gradient of microtubules that defines the AP axis (4). It is believed that dynein subsequently uses this polarised microtubule cytoskeleton to pull the nucleus to the oocyte anterior, making polarisation of the DV axis dependent on the prior polarisation of the AP axis (5-9).
The nucleus is pushed to the anterior by growing microtubules
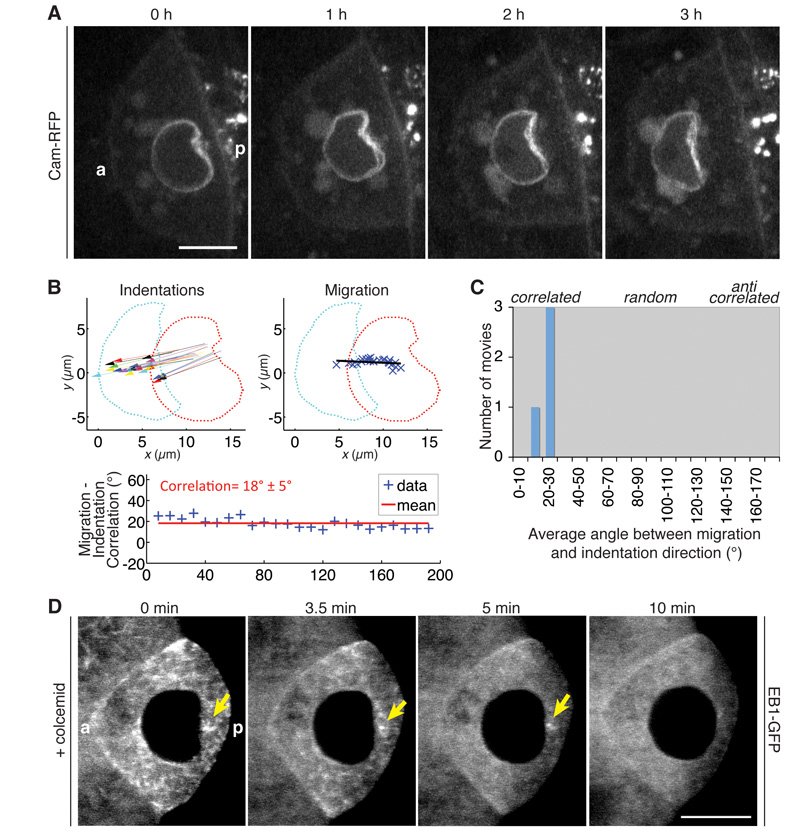
To investigate the mechanism of nuclear positioning directly, we imaged the movement of the nucleus in living oocytes. The nucleus migrates at a speed of 4.0 ± 0.7 μm/h (n=6), and takes 2-3 hours to move across the oocyte (Fig. 1). The trajectory of the nucleus is variable: sometimes it moves around the cortex of the oocyte directly to an anterior corner, but it often migrates up the centre of the oocyte and then turns to move along the anterior cortex (Fig. S1, Movie S1), confirming the random nature of this symmetry-breaking event. We observed that all migrating nuclei have large posterior indentations, suggesting that they are being pushed rather than pulled towards the anterior (Fig. 1A, Movie S2). This could reflect an intrinsic reorganisation of the nuclear architecture, or a deformation induced by an external force to the nucleus. In support of the latter view, the direction of the indentation correlates with the direction of migration, suggesting that the same force creates the indentation and moves the nucleus (Fig. 1B-C). This indentation is not an artefact of long-term imaging in oil, as egg chambers dissected directly into strong fixative have identical indentations (Fig. S2).
Fig. 1. Nuclear migration is driven by a posterior microtubule pushing force.
(A) Time course of a migrating nucleus. (B) Analysis of directions of nuclear indentations during migration (left), direction of overall migration (right), and the correlation between them, expressed as the angle between the directional vectors (bottom) (mean±s.d.). Red and cyan dots show the outline of the nucleus at the start and end of migration, respectively; blue Xs show the centroid of the nucleus during migration. (C) Mean angle between migration and indentation directions from four migrating nuclei. (D) Temporal merges of an EB1-GFP movie of a colcemid-treated egg chamber. Each image is a maximum projection of 5 time frames (equal to 10 s). Arrow, MTOC. (A, D) a, anterior; p, posterior; Scale bars, 10 μm.
This idea that the nucleus is pulled to the anterior by dynein is based on the finding that mutations in the dynein accessory factors, Lis1 and Bic-D, as well as disruption of the dynactin complex result in mislocalised nuclei at stage 10 (5-9). This is not compatible with the pushing model of nuclear movement, as motor proteins can only pull their cargoes. We therefore re-examined the role of the dynein complex by imaging the nucleus in Lis1 mutant egg chambers. Lis1 mutant oocytes are much smaller than normal because dynein is required for transport from the nurse cells into the oocyte (6). Nevertheless, the oocyte nucleus migrates normally with a prominent posterior indentation (Movie S3). Thus, dynein is presumably required for the anchoring of the nucleus once it has reached the anterior, rather than for its migration. Consistent with this, the nuclei are only rarely mispositioned in Lis1 and Bic-D mutant oocytes at stage 9, but are mislocalised much more frequently at later stages (Fig. S3).
Both actin and microtubule polymerisation can generate pushing forces that lead to cellular or organelle deformations (10). Two lines of evidence suggest that microtubules are responsible for the nuclear indentation: Firstly, depolymerisation of actin with latrunculin A or B does not affect nuclear positioning, whereas the microtubule-depolymerising drug, colcemid, induces mislocalised nuclei (11). Secondly, several microtubule-associated proteins become enriched on the posterior nuclear envelope during migration, including the Dynein light intermediate chain (Dlic), Calmodulin (Cam), and the Drosophila NuMA homologue Mushroom body defect, Mud (Fig. S4) (12-14).
To test the role of microtubules in the formation of the indentation, we added colcemid to living egg chambers expressing the +TIP protein, EB1-GFP (end binding-1), which forms a “comet” on the growing plus ends of microtubules (15). Colcemid takes 3.5 minutes to diffuse into the oocyte, as monitored by a decrease in the number of EB1 comets on growing microtubule plus ends. As soon as microtubule growth starts to decrease, the indentation diminishes in size (Fig. 1D, Movie S4). A focus of EB1-GFP persists posterior to the nucleus for several minutes, and as this disappears, the nucleus relaxes completely and becomes spherical. Thus, the nuclear indentation depends on microtubule polymerisation and its size is proportional to the number of growing microtubules.
The nuclear indentation depends on active posterior centrosomes
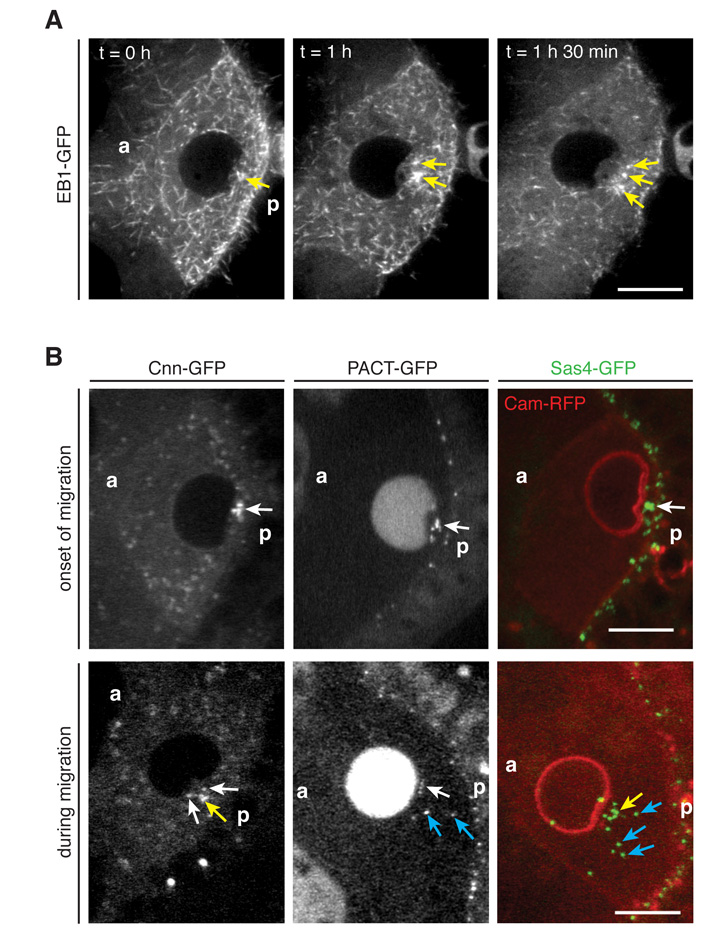
Using EB1-GFP to track the growing microtubule plus ends in time-lapse movies of nuclear migration revealed several strong foci of EB1-GFP behind the indentation, with growing microtubules emanating from them in all directions (Fig. 2A, Movie S5). This indicates that microtubules are nucleated from MTOCs behind the nucleus. These MTOCs resemble the centrosomes, which migrate from the nurse cells into the oocyte during early oogenesis in a dynein dependent manner, and localise to the posterior cortex as a result of the initial oocyte polarity (16-20). Indeed, the centriolar markers Sas4 and PACT, as well as a marker for pericentriolar material (PCM), Centrosomin (Cnn) (21), localise to the foci behind the nuclear indentation at the onset of migration (Fig. 2B). The centrosomes behave rather dynamically during migration and change reversibly from a dense cluster to a more dispersed distribution (Fig. 2B). Upon completion of nuclear migration, the centrosomes are recruited to the anterior-dorsal cortex of the oocyte, presumably as a consequence of the activation of the dynein-dependent anchoring mechanism that retains the nucleus in this position (Fig. S5, Movies S6-7). Active centrosomes are therefore positioned behind the nucleus before and during migration.
Fig. 2. Active centrosomes are localised behind the nuclear indentation.
(A) Temporal merges (20 time frames, equal to 10 s) of an EB1-GFP movie reveal active MTOCs (arrows) between the nuclear indentation and the posterior oocyte cortex. (B) The centriolar markers PACT-GFP and Sas-4-GFP and the PCM marker Cnn-GFP are localised behind the nucleus at the onset of migration (top). During migration, MTOCs can be clustered together (yellow arrows), associated with the nucleus (white arrows), or dispersed in the cytoplasm (blue arrows) (bottom). (A-B) a, anterior; p, posterior; Scale bars, 10 μm.
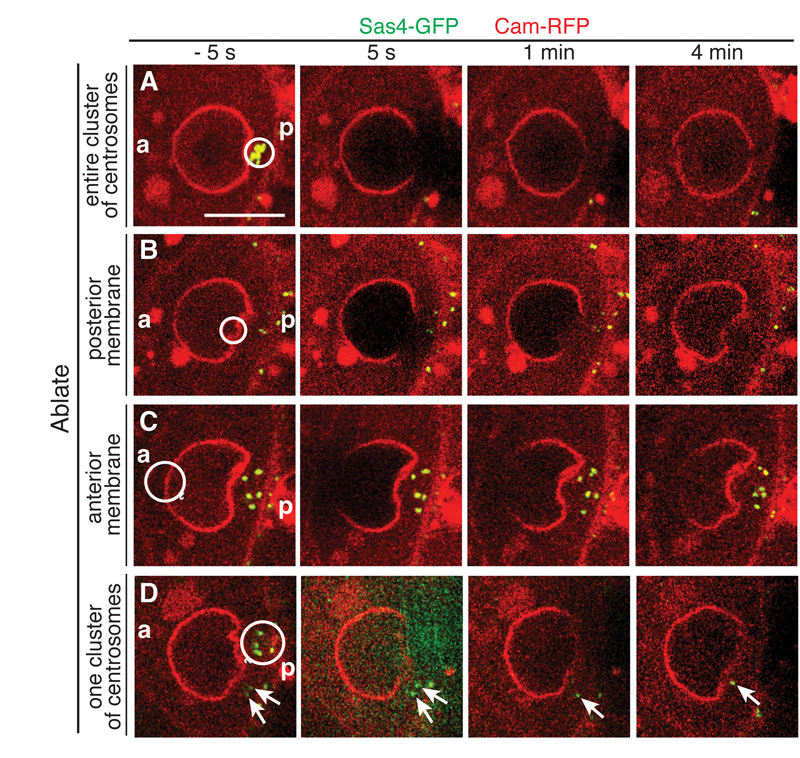
To test the role of the centrosomes in creating the nuclear indentation, we inactivated them by laser ablation. Upon ablation of the entire cluster of centrosomes, the indentation disappears and the nucleus becomes spherical within 1 min (Fig. 3A, Movie S8). This nuclear relaxation may occur more rapidly, as centrosome ablation causes local bleaching of the nuclear envelope, making it impossible to monitor nuclear shape during the first minute. Local laser ablation of the nuclear envelope at the site of the indentation has no effect, however, excluding the possibility that the disappearance of the indentation is a consequence of bleaching of the nuclear membrane (Fig. 3B, Movie S9). Furthermore, ablation of the anterior of the nucleus does not affect the indentation, arguing against any pulling force from the anterior (Fig. 3C, Movie S10). As described above, centrosomes are sometimes scattered behind the nucleus, causing multiple indentations. Ablating one cluster of centrosomes abolishes only the indentation facing them. The non-ablated centrosomes remain active and induce an indentation on the adjacent side of the nucleus (Fig. 3D, Movie S11). Thus, the nucleus is not a rigid structure and the growing microtubules from the centrosomes exert force on the nuclear envelope to induce its deformation.
Fig. 3. Laser ablation of the centrosomes abolishes the nuclear indentation.
(A, D) Clusters of centrosomes were bleached for 5 s. 1-4 min after ablation, the nuclear indentation facing the ablated centrosomes disappears. (B, C) When the nuclear membrane or the anterior of the nucleus is bleached, the indentation is maintained. (A-D) a, anterior; p, posterior; Circles, area of bleaching; Arrows, non-ablated, active centrosomes; Scale bar, 10 μm.
The centrosomes are dispensable for oogenesis (22). We therefore examined nuclear migration in DSas-4 mutant ovaries that lack centrosomes. Consistent with the previous study, all nuclei migrate to the anterior-dorsal corner (n=117) and show a posterior indentation during migration (Fig. S6, Movie S12). GFP-Cnn is still localised in foci behind the nucleus and EB1-GFP tracks reveal active posterior MTOCs (Fig. S6). Thus, acentrosomal MTOCs form in the absence of centrosomes and can provide the pushing force for nuclear migration.
Nuclear migration is independent of AP axis formation
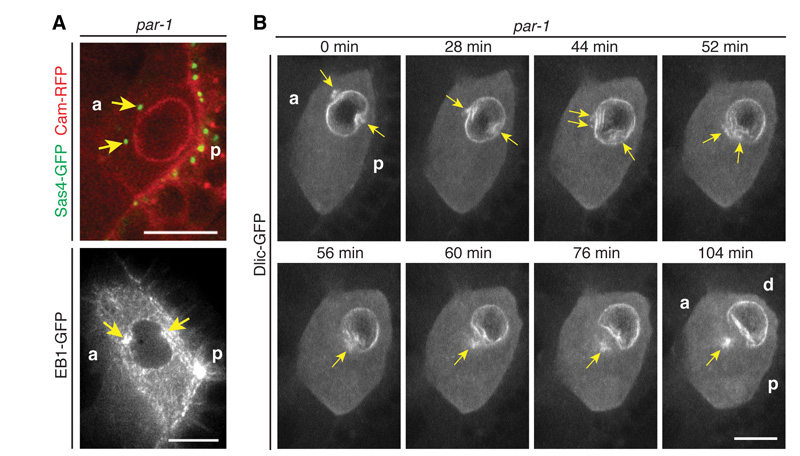
As a further test of the idea that the centrosomal microtubules push the nucleus to the anterior, we examined par-1 hypomorphs, in which some centrosomes fail to migrate to the posterior of the oocyte (19). These anterior centrosomes induce anterior nuclear indentations, leading to dumbbell-shaped nuclei, confirming the role of centrosomal microtubules in pushing the nucleus (Fig. 4A). These ectopic centrosomes eventually fuse with the posterior centrosomes to move the nucleus to the anterior-dorsal corner (Fig. 4B, Movie S13). This explains why the nucleus migrates normally in par-1 mutants, even though the anterior-posterior axis is not polarised (23). Consistent with this, the nucleus in wildtype oocytes can migrate to the anterior before the anterior-to-posterior microtubule gradient is established (Fig. 2A, Fig. S7).
Fig. 4. Mispositioned centrosomes induce ectopic nuclear indentations.
(A) Sas4-GFP (top) and a temporal merge of EB1-GFP (20 frames, equals 10 s) (bottom) reveal active, anterior centrosomes in par-16323/par-1W3 mutants, which induce an anterior indentation in the nucleus. (B) Nuclear migration in par-16323/par-1W3 mutants. At the onset of migration, the anterior centrosomes induce an ectopic anterior nuclear indentation. The anterior centrosomes eventually move around the nuclear membrane to cluster with the posterior centrosomes, inducing a broad nuclear indentation and rapid nuclear movement. (A-B) a, anterior; p, posterior; d, dorsal; Arrows, centrosomes; Scale bars, 10 μm.
Microtubule growth provides sufficient force to move the nucleus
Another documented example of nuclear positioning by microtubule pushing comes from S. pombe, where microtubule bundles push against the cell ends to maintain the nucleus in the cell centre (24). The oocyte nucleus moves a much greater distance, however, and appears to be pushed by the force exerted by single growing microtubules. To test the feasibility of this mechanism, we used Stoke’s law (F = 6πηrv) to estimate the drag force (F) exerted on the nucleus. Assuming a cytoplasmic viscosity (η) ≈ 100 Pas (25, 26) and the measured values of the nuclear radius (r) ≈ 5 μm and the velocity of migration (v) ≈ 4 μm/h yields a drag force ≈ 10 pN. We expect the actual drag force to be lower, because nuclear migration is so slow (1 nm/sec) that the cytoplasmic actin mesh will turn over ahead of the nucleus, decreasing the effective viscosity (27, 28). The longest microtubules can reach ≈ 10 μm between the posterior of the nucleus and the posterior oocyte cortex, resulting in a critical buckling force Fc ≈ 5 pN (29). This value is probably an underestimate, as microtubules embedded within an elastic cytoplasm in vivo have been reported to bear compressive loads a hundred times higher than in vitro (30). Each microtubule can therefore generate a pushing force of at least 5 pN. Thus, the force of only two microtubules pushing at any time should be sufficient to move the nucleus to the oocyte anterior.
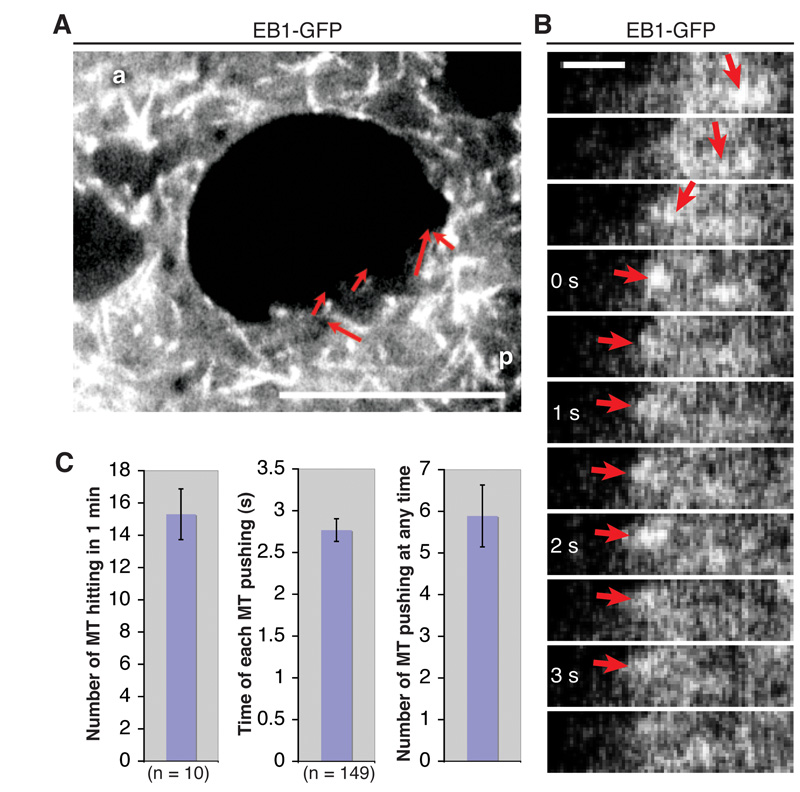
We measured the number of microtubules pushing the nucleus using EB1-GFP. 15.3 ± 1.6 microtubules hit the nuclear indentation per minute in one z-plane (n=10, 2 oocytes), and they continue growing and presumably exerting force on the nucleus for 2.77 ± 0.14 s (n=149) (Fig. 5A-C, Movie S14). Given the thickness of a confocal section (0.8 μm) and the radius of the indentation (4.3 ± 0.2 μm (n=10)), an average of 5.9 ± 0.7 microtubules are pushing the nucleus at any given time. Microtubule polymerisation can therefore provide sufficient pushing force to drive nuclear migration.
Fig. 5. The force of microtubule polymerisation is sufficient to move the nucleus.
Quantification of the number of microtubules hitting the posterior of the nucleus. (A) Temporal merge of 20 frames (equal to 10 s) of an EB1-GFP movie. Red arrows indicate tracked microtubules that hit the nuclear indentation. a, anterior; p, posterior; Scale bar, 10 μm. (B) Kymograph of a microtubule that pushes against the nuclear indentation for 3 s; arrows indicate the position of the EB1-GFP comet (plus end of a microtubule). Scale bar, 1 μm. (C) Quantification of the number of microtubules hitting the nuclear indentation, the time that each microtubule pushes, and the resulting average number of microtubules that are pushing against the indentation at any given time (error bars, ± s.e.m.).
Nuclear migration is triggered by release from a posterior anchor
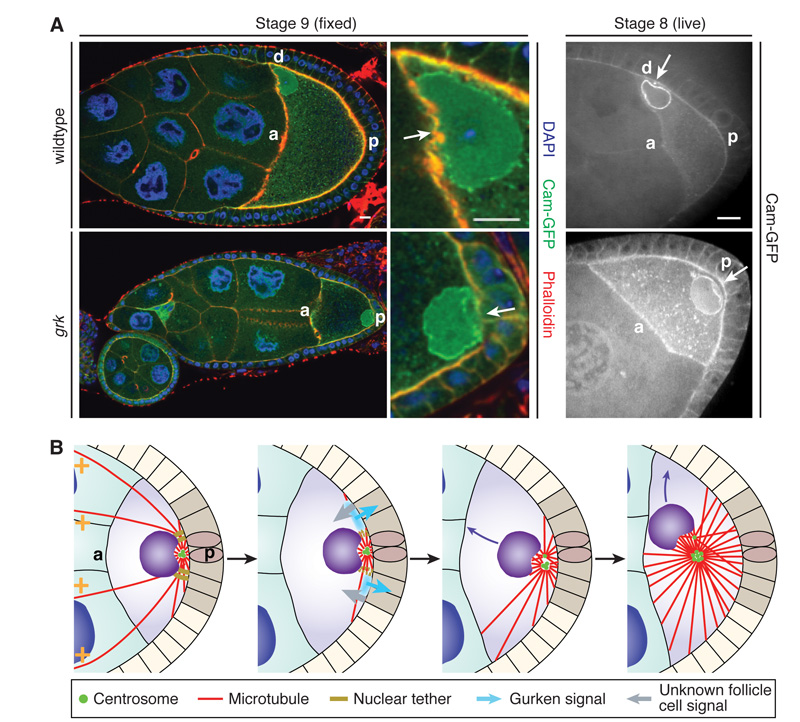
The migration of the nucleus is triggered by an unknown signal from the posterior follicle cells, which could act either by activating the centrosomes or by releasing the nucleus from a posterior tether. To address this question, we examined when the indentation appears during oogenesis. Active centrosomes are already localised behind the nucleus at stage 5 of oogenesis and induce a posterior indentation (Fig. S8A). This suggests that the centrosomes continually exert a pushing force on the nucleus, which is tethered to the posterior until it receives a signal for migration. The nucleus remains at the posterior in gurken (grk) mutants, which block follicle cell signalling to the oocyte (39% penetrance, n=70)(2, 3). These posterior nuclei still maintain a posterior indentation later in oogenesis (Fig. 6A), suggesting that they fail to migrate because they are not released from the posterior tether (Fig. S8B, Movie S15). Indeed, microtubules growing from active centrosomes probably always exert a pushing force on the nucleus that must be countered by an opposing pulling force or anchor to keep the nucleus in place. For example, a clear nuclear indentation is still visible adjacent to the centrosomes after the nucleus is anchored at the anterior (Fig. S5A).
Fig. 6. The nucleus is anchored to the posterior prior to migration.
(A) The nuclei often fail to migrate in grk2B6/grk2E12 mutants, but still have prominent posterior indentations (arrows), indicating that they are tethered at the posterior. a, anterior; p, posterior; d, dorsal; Scale bars, 10 μm. (B) A microtubule pushing model for nuclear migration. Before migration, the nucleus is tethered at the posterior with active centrosomes behind it (left panel). The posterior follicle cell signal induces the release of the nucleus from the tether, and growing microtubules then push the nucleus anteriorly (middle panels). This movement is essentially random and continues until the oocyte becomes wedged in an anterior corner (right panel).
Our results lead to a revised model for how the oocyte nucleus moves to break radial symmetry and polarise the Drosophila dorsal-ventral axis (Fig. 6B). This model explains the failure to recover mutants that specifically disrupt nuclear migration, since the driving force is provided solely by microtubule polymerisation. Furthermore, our results imply that migration is triggered by the release of the nucleus from a posterior anchor, rather than by microtubule reorganisation. Thus, polarisation of the dorsal-ventral axis is independent of the formation of the microtubule array that defines the anterior-posterior axis, as previously proposed.
Supplementary Material
Acknowledgments
We would like to thank J. Raff for providing the Dlic-GFP fly stock and centrosomal markers; H. Nash for providing the Mud antibody; S. Young for help with image analysis; and J. Guck, Y-C. Oei and P. K. Trong for help with the force calculations. This work was supported by a Wellcome Trust Principal Research Fellowship to D.St J., PhD scholarships from the FAZIT Stiftung, the Boehringer Ingelheim Fonds and the Cambridge European Trust to T.Z., the Gulbenkian PhD scholarship of the Fundação para a Ciência e Tecnologia to A.R., and core support from the Wellcome Trust and Cancer Research UK.
Footnotes
Supplementary Materials: www.sciencemag.org Materials and Methods Figures S1-S8 References (31-43) Movies S1-S15
References and Notes
- 1.Dupin I, Etienne-Manneville S. Int. J. Biochem. Cell Biol. 2011;43:1698. doi: 10.1016/j.biocel.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Reyes A, Elliott H, St Johnston D. Nature. 1995;375:654. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- 3.Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. Cell. 1995;81:967. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 4.Bastock R, St Johnston D. Curr. Biol. 2008;18:R1082. doi: 10.1016/j.cub.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Swan A, Suter B. Development. 1996;122:3577. doi: 10.1242/dev.122.11.3577. [DOI] [PubMed] [Google Scholar]
- 6.Swan A, Nguyen T, Suter B. Nat. Cell Biol. 1999;1:444. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- 7.Lei Y, Warrior R. Dev. Biol. 2000;226:57. doi: 10.1006/dbio.2000.9848. [DOI] [PubMed] [Google Scholar]
- 8.Duncan JE, Warrior R. Curr. Biol. 2002;12:1982. doi: 10.1016/s0960-9822(02)01303-9. [DOI] [PubMed] [Google Scholar]
- 9.Januschke J, et al. Curr. Biol. 2002;12:1971. doi: 10.1016/s0960-9822(02)01302-7. [DOI] [PubMed] [Google Scholar]
- 10.Mogilner A, Oster G. Curr. Biol. 2003;13:R721. doi: 10.1016/j.cub.2003.08.050. [DOI] [PubMed] [Google Scholar]
- 11.Koch EA, Spitzer RH. Cell Tissue Res. 1983;228:21. doi: 10.1007/BF00206261. [DOI] [PubMed] [Google Scholar]
- 12.Lee YC, Wolff J. J. Biol. Chem. 1982;257:6306. [PubMed] [Google Scholar]
- 13.Kardon J, Vale R. Nat. Rev. Mol. Cell Biol. 2009;10:854. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radulescu AE, Cleveland DW. Trends Cell Biol. 2010;20:214. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mimori-Kiyosue Y, Shiina N, Tsukita S. Curr. Biol. 2000;10:865. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 16.Mahowald AP, Strassheim JM. J. Cell Biol. 1970;45:306. doi: 10.1083/jcb.45.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grieder NC, de Cuevas M, Spradling AC. Development. 2000;127:4253. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- 18.Bolivar J, et al. Development. 2001;128:1889. doi: 10.1242/dev.128.10.1889. [DOI] [PubMed] [Google Scholar]
- 19.Huynh J, Shulman J, Benton R, St Johnston D. Development. 2001;128:1201. doi: 10.1242/dev.128.7.1201. [DOI] [PubMed] [Google Scholar]
- 20.Fichelson P, Jagut M, Lepanse S, Lepesant JA, Huynh JR. Development. 2010;137:815. doi: 10.1242/dev.045013. [DOI] [PubMed] [Google Scholar]
- 21.Rusan N, Rogers G. Traffic. 2009;10:472. doi: 10.1111/j.1600-0854.2009.00880.x. [DOI] [PubMed] [Google Scholar]
- 22.Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. Curr. Biol. 2007;17:1498. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulman JM, Benton R, St Johnston D. Cell. 2000;101:377. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- 24.Tran PT, Marsh L, Doye V, Inoue S, Chang F. J. Cell Biol. 2001;153:397. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bausch AR, Möller W, Sackmann E. Biophys. J. 1999;76:573. doi: 10.1016/S0006-3495(99)77225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hochmuth RM. J. Biomech. 2000;33:15. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- 27.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Dev. Cell. 2007;13:539. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe N. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 2010;86:62. doi: 10.2183/pjab.86.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dogterom M, Kerssemakers JW, Romet-Lemonne G, Janson ME. Curr. Opin. Cell Biol. 2005;17:67. doi: 10.1016/j.ceb.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Brangwynne CP, et al. J. Cell Biol. 2006;173:733. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin X, Daneman R, Zavortink M, Chia W. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15050. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada Y, Yonemura S, Ohkura H, Strutt D, Uemura T. Dev. Cell. 2006;10:209. doi: 10.1016/j.devcel.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Conduit PT, et al. Curr. Biol. 2010;20:2178. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. J. Cell Biol. 2004;165:673. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peel N, Stevens NR, Basto R, Raff JW. Curr. Biol. 2007;17:834. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jankovics F, Brunner D. Dev. Cell. 2006;11:375. doi: 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Basto R, et al. Cell. 2006;125:1375. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 38.Neuman-Silberberg FS, Schüpbach T. Cell. 1993;75:165. [PubMed] [Google Scholar]
- 39.Ran B, Bopp R, Suter B. Development. 1994;120:1233. doi: 10.1242/dev.120.5.1233. [DOI] [PubMed] [Google Scholar]
- 40.Golic KG. Science. 1991;252:958. doi: 10.1126/science.2035025. [DOI] [PubMed] [Google Scholar]
- 41.Chou T, Perrimon N. Genetics. 1996;144:1673. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu JX, Guan Z, Nash HA. Genetics. 2006;173:243. doi: 10.1534/genetics.105.051557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bianco A, et al. Nature. 2007;448:362. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.