Summary
Successful targets for anti-cancer drugs such as clofarabine and gemcitabine, ribonucleotide reductases (RNRs) provide the precursors for DNA biosynthesis and repair. Recently, we reported that dATP inhibits E. coli class Ia RNR by driving formation of RNR subunits into α4β4 rings. Here, we present the first X-ray structure of gemcitabine-inhibited E. coli RNR and show that the previously described α4β4 rings can interlock to form an unprecedented (α4β4)2 megacomplex. This complex is also seen in a higher-resolution dATP-inhibited RNR structure presented here, which employs a distinct crystal lattice from that observed in the gemcitabine-inhibited case. With few reported examples of protein catenanes, we use data from small-angle X-ray scattering and electron microscopy to both understand the solution conditions that contribute to concatenation in RNR as well as present a mechanism for the formation of these unusual structures.
Introduction
Essential for DNA biosynthesis and repair, the prototyptic class Ia ribonucleotide reductase (RNR) from E. coli employs two subunits α2 and β2 that perform radical-based chemistry to convert ribonucleotides to deoxyribonucleotides. The β2 subunit houses the diferric-tyrosyl radical cofactor used to initiate radical chemistry, while α2 contains catalytic and regulatory sites (Fig. 1). Prior to each round of catalysis, a radical transfer occurs in which the radical moves an unprecedented 35 Å from β2 to the catalytic site in the α2 subunit, generating the essential catalytic thiyl radical (Uhlin et al., 1994; Seyedsayamdost et al., 2007; Licht et al., 1996). Although RNRs are successful targets for anti-cancer drugs such as clofarabine and gemcitabine (Hertel et al., 1990; Huang et al., 1991; Plunkett et al., 1997; Bonate et al., 2006; Zhenchuk et al., 2009; Gandhi et al., 2006), there is surprisingly little structural information depicting holoRNRs (containing both subunits) with or without these anti-cancer drugs. Here we present two crystal structures of inhibited forms of holoRNR from E. coli: the first structure of a gemcitabine-diphosphate (F2CDP) inactivated holocomplex to 4.45 Å resolution and the highest resolution structure of a dATP-inhibited RNR, improving the resolution from 5.65 Å (Ando et al., 2011) to 3.95 Å.
Figure 1.
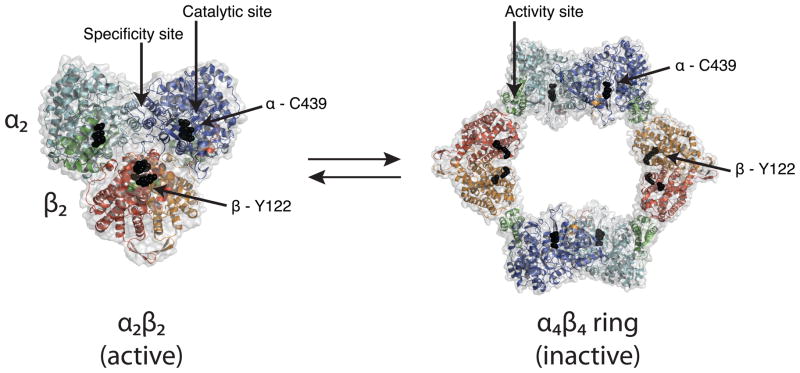
Structures of E. coli class Ia RNR. Left, a pseudo atomic model of the active α2β2 state of E. coli RNR based on SAXS (Ando et al., 2011). Right, crystal structure of the dATP-inactivated α4β4 form of RNR (Ando et al., 2011). α2 subunits are in cyan/blue with N-terminal domain in green and β2 in red/orange. Residues along the proposed radical path between the diiron center (green spheres) and Y122 in β and C439 in α are shown as black spheres, and highlight how the proposed radical transfer pathway in the active enzyme is disrupted in the α4β4 ring. Also labeled are the two types of allosteric effector binding sites: a specificity site and an activity site. The specificity site, found at the α dimer interface, responds to intracellular levels of deoxyribonucleoside 5′-triphosphates (dATP, TTP, dGTP) and ATP such that balanced pools of deoxyribonucleotides are maintained (Reichard, 1988). The activity site, found at the N-terminal domain controls the overall enzymatic rate, binding ATP to activate the enzyme or dATP to turn it off (Larsson et al., 1966a; Larsson et al., 1966b; Brown et al., 1969).
Gemcitabine-diphosphate (F2CDP) is a mechanism-based inhibitor of both E. coli and human RNR that binds and reacts at the catalytic site and irreversibly inhibits α2 using only one equivalent of F2CDP per dimer. After reaction with the inhibitor, one α per α2 dimer becomes covalently modified by the sugar of F2CDP (van der Donk et al., 1998; Wang et al., 2007). Size exclusion experiments indicated that reaction with the drug results in a stabilized complex of the two subunits not seen in the presence of other substrates or effectors (Wang et al, 2007), motivating our use of this inhibitor in crystallization experiments. In contrast to the chemotherapeutic agent F2CDP, dATP is a physiological and reversible inhibitor of RNR, responsible for down-regulating activity in the presence of excess deoxynucleotides (Larsson et al., 1966a; Larsson et al., 1966b; Brown et al., 1969). dATP exerts its effect by binding to the activity site on α2 and shifting the conformational equilibrium away from an active α2β2 state toward an inactive α4β4 state (Ando et al., 2011; Rofougaran et al., 2008) (Fig. 1). Accompanying this oligomeric state change is a dramatic movement of the β2 subunit which swings from a position docked with α2 to a position almost 40 Å away, forming connections with another α2 subunit and leading to formation of a ring-like α4β4 structure (Fig. 1). In the resulting α4β4 structure, the radical transfer pathway is completely abrogated (Fig. 1) (Ando et al., 2011).
Amazingly, we find that for both the F2CDP and dATP inhibited RNR structures, two α4β4 rings interlock to form concatenated (α4β4)2 rings. The observation of concatenated rings was completely unexpected – the structure is literally tied in knots. Although concatenated enzyme structures have been reported in the literature (Lee et al., 2004; Cao et al., 2005), previously it was not clear if these structures were more than crystallographic space group artifacts. To address this question, we compare RNR X-ray structures solved in different space groups, and use small angle X-ray scattering (SAXS) to explore whether concatenated rings can form in solution. We also examine what role, if any, F2CDP plays in the formation of RNR interlocked rings. Finally, using electron microscopy (EM), we obtain snapshots of partially formed RNR rings, providing a glimpse into the ring assembly process. With so little known about the structures of holoRNRs and about enzyme concatenation, these data are of broad interest.
Results
F2CDP-inhibited and dATP-inhibited Structures of E. coli Class Ia RNR
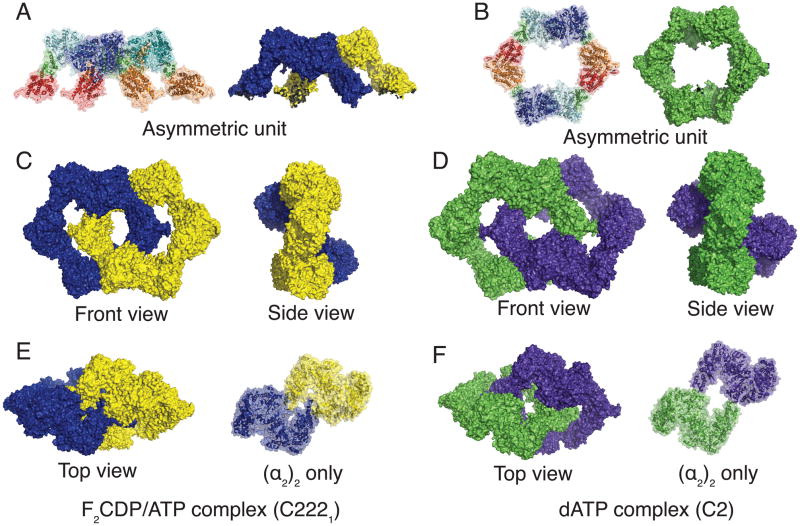
Crystallization using F2CDP treated α2 and β2 subunits in the presence of ATP, has yielded a 4.45 Å resolution structure of this complex with Rwork/Rfree values of 23.3/26.7 (Table 1). In this C2221 crystal form, the asymmetric unit contains two α2 dimers and four β monomers (Fig. 2A). Applying crystallographic symmetry operations generates four physiological β2 dimers and results in a (α4β4)2 complex comprising two asymmetric units (Fig. 2C). The α2 dimers are packed near one another, burying a total surface area of 1200 Å2 between the two dimers (Fig. 2E). Each α monomer also makes contacts to two distinct β2 molecules, one from its own ring, burying ~1500 Å2 total, and one from the interlocked ring, burying an additional 160 Å2 (Fig. S1).
Table 1.
X-ray data collection and refinement statistics.
| F2CDP/ATP-RNR | dATP-RNR | |
|---|---|---|
| Data collection | ||
| Space group | C2221 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 128.7, 398.9, 319.4 | 280.5, 155.7, 166.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 119.1, 90.0 |
| Wavelength (Å) | 0.9769 | 0.9794 |
| Resolution (Å) a | 50.0 – 4.45 (4.61-4.45) | 30.0 - 3.95 (4.09-3.95) |
| Rsym a | 11.5 (35.7) | 16.1 (45.7) |
| <I/σI> a | 8.5 (2.2) | 5.0 (2.3) |
| Completeness (%)a | 91.1 (70.6) | 96.5 (96.5) |
| Redundancy a | 3.8 (2.0) | 2.9 (2.9) |
| Refinement | ||
| Resolution (Å) | 49.0 – 4.45 | 30.0 - 3.95 |
| Number of reflections | 41827 | 52934 |
| Rwork/Rfree (%) | 23.3/26.7 | 25.9/28.4 |
| Number of atoms | ||
| Proteinb | 35053 | 34976 |
| Nucleotides | 124 | 348 |
| Fe | 8 | 8 |
| Water | 8 | 8 |
| Average B-factors (Å2) | ||
| Protein | 147.8 | 109.0 |
| Nucleotidesc | 171.4 | 130.0 |
| Fe | 121.6 | 74.8 |
| Water | 109.3 | 89.3 |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.008 | 0.02 |
| Bond angles (°) | 1.01 | 0.74 |
| PDB ID | 4ERP | 4ERM |
Figure 2.
Multiple crystal forms display interlocked α4β4 rings for E. coli RNR. Each α4β4 ring is colored separately with F2CDP/ATP-RNR complex in blue/yellow and dATP-RNR complex in green/purple. (A,B) One asymmetric unit for each complex is shown as ribbons with transparent surface and with full surface, for F2CDP/ATP- and dATP-RNR complexes, respectively. (C,D) The full (α4β4)2 RNR complex is shown from front and side orientations for each complex. (E,F) A top down view of each RNR complex shows the packing of two α2-dimers (left) and close-up of interactions of just the α2 dimers in an identical orientation (right). Other known structures with interlocked rings are shown in Fig. S3.
The concatenation of two α4β4 rings to make an (α4β4)2 complex is also seen in the 3.95 Å resolution structure of dATP-inhibited E. coli class Ia RNR (Rwork/Rfree of 25.9/28.4). In this C2 crystal form (Table 1), one α4β4 complex is present in the asymmetric unit (Fig. 2B), however, if two asymmetric units are shown, a second ring is found to be interlocked with the first (Fig. 2C). While both the F2CDP/ATP structures and dATP-inhibited structures show two concatenated α4β4 rings, the relative orientation of the rings with respect to each other is different (Fig. 2C, D). As a consequence, the packing of the two α2 subunits is not as close in the dATP-inhibited structure with only 230 Å2 of total buried surface area between the two α2 dimers (Fig. 2F compared to 2E), and each ring is slightly more asymmetric (Fig. S1). Again, each α monomer makes contacts to two distinct β2 molecules, one from its own ring (Fig. 3), burying ~1500 Å2 total, and one from an interlocked ring, burying an additional 400 Å2. Overall, the (α4β4)2 structures are strikingly similar given the different space groups and inhibitor/effector combinations used, indicating that these concatenated rings are formed reproducibly under a variety of conditions. To test the necessity of a small molecule inhibitor such as F2CDP or dATP on the formation of the interlocked ring structure, crystallization was attempted under the same conditions as the F2CDP/ATP-RNR structure, but with CDP in place of F2CDP and with an inactive form of β2 (met-β2). When nucleated with F2CDP/ATP-RNR microcrystals, these crystallization conditions yield the same C2221 crystal form, suggesting that small molecule inhibitors are not essential for this arrangement of molecules in the crystal lattice (data not shown).
Figure 3.
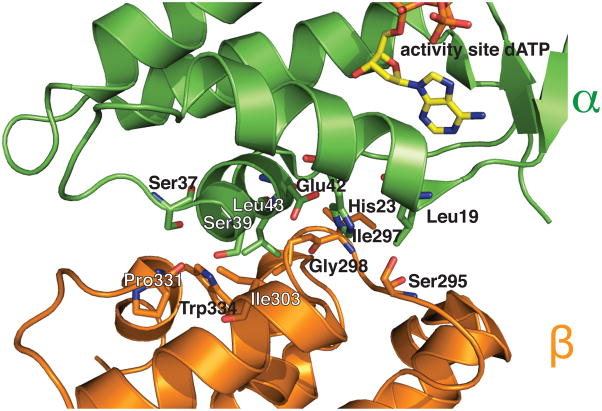
Detailed view of the α-β interface within an α4β4 ring. Protein is shown in ribbons with residues found near the interface shown in sticks. Residue names and numbers are labeled. α is colored in green and β in orange (carbons same color as ribbon, oxygen in red, nitrogen in blue) with dATP bound at the activity site in sticks (carbon in yellow, oxygen in red, nitrogen in blue, phosphorus in orange). Additional close-up views of interfaces and distances between α2 and β2 subunits are shown in Fig. S1.
Solution Studies of Concatenated Ring Formation
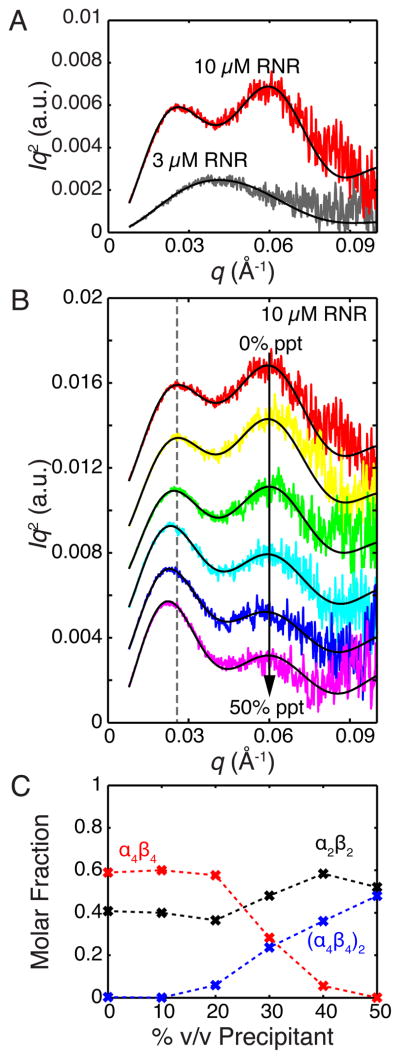
Previously, we showed that dATP drives the formation of an α4β4 ring that is stable over a wide range of protein concentrations (2 – 30 μM) in solution (Ando et al., 2011). Here, we used SAXS to investigate the effect of F2CDP on the solution conformation of E. coli RNR. Scattering profiles were obtained from RNR in the presence of 4 molar equivalents of F2CDP per α2 and are shown in Kratky representation (Iq2 vs. q) in Fig. 4A. At 10 μM α2 and β2, the Kratky plot is multimodal with a significant peak observed at q~0.06 Å−1 (Fig. 4A, red), a signature feature of the non-globular α4β4 ring (Ando et al., 2011). Decreasing the protein concentration to 3 μM α2 and β2 leads to a monomodal Kratky plot, indicative of a globular shape (Fig. 4A, dark grey). A 270 kDa molecular weight estimation obtained by a Porod invariant method (Fischer et al., 2010), suggests that this 3 μM sample can be described as a globular α2β2 consistent with the model in Fig. 1, a species previously observed by SAXS in the absence of dATP (Ando et al., 2011). Linear combinations of theoretical profiles calculated from the α2β2 and α4β4 models shown in Fig. 1 yield good fits to the experimental profiles (Fig. 4A, black curves). Consistent with our previous observations (Ando et al., 2011), these data indicate that an increase in protein concentration favors a shift in equilibrium from predominantly α2β2 to predominantly α4β4.
Figure 4.
SAXS studies on F2CDP-treated E. coli RNR reveal conditions important for oligomeric state changes. (A) The Kratky plots of 3 μM (dark grey) and 10 μM (red) F2CDP- treated E. coli RNR. The presence of one large peak at 3 μM RNR indicates a globular species, such as α2β2, while the presence of two peaks at 10 μM RNR indicates a non-globular species, such as the α4β4-ring. A two-component fit to the data (black line) indicates that the solution at 3 μM RNR is 94% α2β2 and 6% α4β4, while the solution at 10 μM RNR is 43% α2β2 and 57% α4β4. (B) The Kratky representation of 10 μM F2CDP-treated RNR over the course of a titration with 0–50% v/v crystallization precipitant indicates a shift from a less globular species, such as a α4β4-ring, to a more globular species, such as (α4β4)2 The movement of the first peak from the dotted line to lower q is consistent with an increase in overall radius of gyration. A three-component fit to the data (black curves) yields (C) the fractions of α2β2, α4β4, and (α4β4)2 species present at the indicated percentages of precipitant. Data suggest that precipitant favors the formation of (α4β4)2 from α4β4.
In a second SAXS experiment, the crystallization precipitant for the F2CDP/ATP RNR structure (25% PEG 3350, 100 mM HEPES pH 7.5, 200 mM NH4(CH3COO), 5% glycerol) was titrated into 10 μM F2CDP-treated RNR. Upon addition of precipitant, the shape of the Kratky plot alters dramatically (Fig. 4B). The signature peak of the α4β4 ring at q~0.06 Å−1 decreases relative to the first peak at lower q, indicative of increasing globularity with increasing precipitant. This change in shape is concomitant with an increase in overall radius of gyration from 69 to 77 Å, exceeding the previously determined value for the α4β4 ring (71.1 Å) (Ando et al., 2011). Unsurprisingly, two-component linear combinations of α2β2 and α4β4 species described above were insufficient to describe the data. Good fits were obtained by the inclusion of an (α4β4)2 species in the linear combinations (Fig. 4B, black curves) yielding the fractions of each species (Fig. 4C). Here, the relative orientations of the rings seen in the dATP-inhibited structure provide a better fit to the solution data than those observed in the F2CDP/ATP structure.
Because experimental scattering profiles are obtained at finite protein concentrations and therefore are not devoid of excluded volume effects, molar fractions extracted from fitting theoretical profiles are not precise quantities. Thus, the observation of a trend across the titration series is essential. The trends observed in Fig. 4C provide strong support that the α4β4 species are used to form the (α4β4)2 species. Together, these data are consistent with the addition of crystallization precipitant facilitating the conversion of non-globular ring species to interlocked rings that are larger in size and more globular.
EM Analysis Shows Population of Partial Ring Intermediates
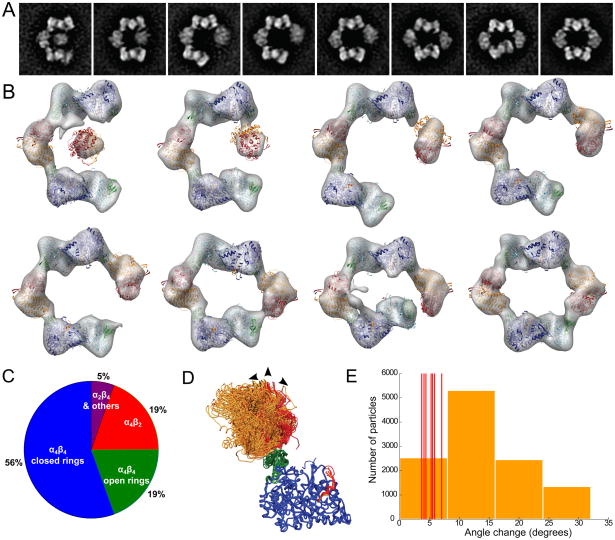
The formation of (α4β4)2 species from α4β4 rings, suggests that these RNR rings must be flexible or come apart. To explore this issue, we analyzed electron microscopy images of α2 and β2 (150 nM) in the presence of inhibitor dATP (50 μM). At these low protein concentrations, no (α4β4 )2 species are expected and we find that the complexes are a mixture of completely closed α4β4 rings and partially formed species. Previously, we focused our analysis of these EM images on investigating the structure of the dominant species, the fully assembled dATP-inhibited α4β4 RNR ring (Ando et al., 2011), while here we were interested in extending the analysis to include the partially assembled rings. EM images of RNR oligomers were aligned and clustered by similarity, resulting in 33 classes of RNR complexes with different composition and structure. Approximately 56% of particles (in 14 of 33 classes) form completely closed α4β4 rings in the presence of 50 μM dATP while other class averages reveal broken rings in which one connection between adjacent α2 and β2 dimers has been severed (19% of particles) and partial rings with a missing β2 subunit (19% of particles) (Fig. 5 and Fig. S2). The open and partial rings may represent assembly or disassembly intermediates and, together with the asymmetry observed in many of the closed rings, suggest that the connections between α2 and β2 can be readily formed and broken.
Figure 5.
Electron microscopy analysis of dATP-inhibited RNR reveals flexibility in the α4β4 ring. (A) Class averages of a closed α4β4 ring (right) and α4β4 in opened-ring conformations with severed interaction between the α2 and β2 subunits. (B) 3D reconstructions for each class with fit atomic structures (PDB ID 4R1R and 1RIB) illustrate the flexibility in the α2 - β2 interface (also see Movie S1). (C) The conformational distribution of particles based on the entire set of 33 class averages (Fig. S2). (D) Interacting α-β pairs from the fitted models in (B) and from dATP and F2CDP crystal structures, aligned using the α subunits (blue with N-terminal domain in green). Rotations of the β subunits from the crystal structures (red) or from the EM structures (orange) were measured relative to chain E from the dATP crystal structure (arrows). (E) Red lines indicate the rotation measured for each α-β pair from the crystal structures. The histogram (orange bars) presents the angular distribution of α-β interactions observed in the EM reconstructions.
Comparison between the class averages suggests a hinge motion in the α-β interaction. By overlaying the class variance images and the class averages, additional heterogeneity is apparent within each class, particularly those classes with severed α-β connections (Fig. S2). Thus the conformations captured by EM class averages appear to represent samples along a continuum of motion in the α-β interface. To assess the degree of flexibility in the α-β interaction, atomic models of α2 and β2 were fitted into 3D maps of the structures represented by the severed-ring EM classes. Each interacting α-β pair from both the EM reconstructions (Fig. 5B) and from the two crystal structures (Fig. 2) was aligned and compared. This analysis reveals that the α-β orientation within the closed α4β4 rings varies by up to 7°, and that α-β interfaces in the less constrained open conformations can readily accommodate motions of 10–25° (Fig. 5D–E). Such elasticity of the α4β4 ring would seem to be required both for conversion of an α4β4 inactive state into an active α2β2 species of RNR as well as for the formation of the (α4β4)2 interlocked rings.
Discussion
Despite the significant interest in RNRs due to their sophisticated allosteric regulation and success as drug targets, the interactions between the α2 and β2 subunits of the class Ia enzymes have been challenging to visualize. Here we present two X-ray structures of the prototypic E. coli RNR enzyme that display interactions between both subunits. These structures represent inhibited forms of RNR, one inhibited with chemotherapeutic agent gemcitabine-diphosphate (F2CDP) and the other bound with the negative allosteric regulator dATP. Unexpectedly, structure data show that the subunits are arranged into two interlocked α4β4 rings. Although rare, two concatenated protein structures have been previously described (Lee et al., 2004; Cao et al., 2005). In particular, RecR, an enzyme involved in homologous recombinational DNA repair, forms interlocked tetrameric-ring structures in its crystal lattice (Fig. S3). Since DNA is proposed to bind in the center of one RecR tetrameric ring, the presence of the second interlocked ring is not thought to be physiologically relevant (Lee et al, 2004). However, the ability of RecR tetramers to interlock may indicate a propensity of these rings to open and close, an important aspect of its DNA binding function. Thus two of the three concatenated protein structures involve tetrameric ring structures that are either known or proposed to open and close as part of their cellular activities.
The other example of interlocked protein rings is found with bovine mitochondrial peroxiredoxin III, which forms magnificent concatenated dodecameric structures (Cao et al., 2005) (Fig. S3). Here it is not known which state of the enzyme is active or why such a multimeric complex might form. For both RecR and peroxiredoxin III, data are not available to firmly establish if these interlocked structures form in solution, although the authors of the peroxiredoxin studies (Cao et al., 2005) mention preliminary analytical ultracentrifugation data that suggest some formation of concatenated rings in solution. Also, while mechanisms of protein ring concatenation have been considered for both RecR and peroxiredoxin III, experimental support is unavailable (Lee et al., 2004; Cao et al., 2005).
To address the conditions under which interlocked protein rings can form in a crystal lattice, we investigated whether F2CDP was required in the E. coli RNR crystallization solution for the formation of the C2221 crystal arrangement of concatenated rings. We find that it is not required, consistent with the mode of F2CDP inhibition necessitating active enzyme (the α2β2 state). Instead, the requisite high protein concentrations for crystallization can explain the formation of α4β4 rings in the C2221 cell. Structural data also show that the (α4β4)2 state can form in a C2 crystal form in the presence of inhibitor dATP, known to form the α4β4 ring. Thus despite the different crystallization conditions and inhibitors used, E. coli RNR preferentially forms interlocked rings in the crystalline state.
The solution-based technique, SAXS, was employed to examine whether interlocked rings form in solution. Consistent with the above observations from crystallography, the presence of F2CDP is not the determinant for concatenation of E. coli RNR rings. Instead, the primary determinant is the presence of ingredients from the crystallization precipitant (25% PEG 3350, 100 mM HEPES pH 7.5, 200 mM NH4(CH3COO), 5% glycerol). Interestingly, one ingredient, PEG, is a well-known molecular crowding agent (Ellis, 2001; Minton, 2000; Zhou et al., 2008) that often favors high order oligomerization (Hamiaux et al., 2000). A dramatic shift from α4β4 to (α4β4)2 is observed upon addition of precipitant (Fig. 4B), providing the first unambiguous experimental support that concatenated rings can form in solution.
It is intriguing that the subunits of the α4β4 ring do not rearrange into compact assemblies in the presence of a crowding agent such as PEG, but rather the ring retains its hole (with a diameter of ~80 Å) and that hole is filled with a second doughnut-like ring. The preference to respond to crowding with concatenation of individual α4β4 rings rather than by subunit rearrangement is surprising in that RNRs from other species, such as yeast, will form larger complexes (α6β2) (Fairman et al., 2011). Despite the similarity in the structure of the yeast and E. coli α2 subunits (Uhlin et al., 1994; Xu et al., 2006), the E. coli enzyme does not form α-hexamers. The only complexes larger than the active α2β2 state that E. coli RNR appears to form are α4β4 or when crowded (α4β4)2.
A significant remaining question is how concatenated rings form. Three mechanisms can be considered (Lee et al., 2004; Cao et al., 2005). In the first, one ring is formed and the second ring is assembled around the first ring. In the second mechanism, two rings simultaneously become interlocked due to association of monomers from distinct rings before the assembly process is complete. In the third, a ring would open, embrace a second ring, and reclose. Our SAXS data favors the third mechanism. In particular, the SAXS data show that in the presence of precipitant, the fraction of α2β2 remains fairly constant while the fraction of α4β4 decreases significantly as the (α4β4)2 species develops (Fig. 4C). This observation indicates that interlocked rings form from α2 and β2 subunits that were previously part of single rings. While rings could come apart completely, with subunits reassembling around an intact ring or assembling simultaneously to form interlocked rings, it is hard to explain why the amount of α2β2 species would not be more dramatically affected in such a case. Thus, this SAXS result is more consistent with mechanism three in which rings open and reclose. In addition, EM images of E. coli RNR with inhibitor dATP show that ~19% of the particles are α4β4 rings that have opened to varying degrees (Fig. 5), depicting the flexibility necessary for this mechanism of concatenation (Movie S1).
It is also interesting to consider whether there might be a benefit to the formation of E. coli RNR concatenated rings in vivo, or phrased another way, could the formation of interlocked rings be a further means of cellular RNR activity modulation? While the production of deoxynucleotides by RNR is essential for DNA biosynthesis and repair, overproduction can be detrimental (Wheeler et al., 2005), requiring all organisms including E. coli to adopt cellular processes to both up and down regulate RNR activity (Elledge et al., 1993). The modulation of E. coli RNR activity by ATP and dATP levels, which involves the binding of ATP/dATP to the activity sites on α2 and the subsequent equilibrium shift between active α2β2 and inactive α4β4 states, provides a sensitive and efficient method for both up and down regulation of RNR function (Brown et al., 1969; Rofougaran et al., 2008; Ando et al., 2011). The higher resolution structure of dATP-inhibited RNR presented here allows us to identify residues at this key α-β interface near the activity site within the α4β4 complex (Fig. 3). Although polar residues such as Trp, Ser, Glu and His line these surfaces, providing some potential for hydrogen bonding interactions, the overall contact area is small (Fig. 3), consistent with the ability of E. coli RNR to efficiently sever these interactions and convert to an active state in response to ATP. While this small contact area allows for a facile conversion from an inactive state, there could be cellular conditions in which it would be desirable to maintain RNR in an inactive state that does not directly interconvert with active enzyme. One can imagine a scenario in which an abundance of α4β4 rings formed under high dATP cellular conditions might lead to interlocked (α4β4)2 rings in a crowded cellular milieu. The sequestration of α2 and β2 into these interlocked rings would limit their dissociation, slowing their transition back to an active enzyme state. The additional disassembly of (α4β4)2 into single α4β4 rings before conversion back to active α2β2 would then lead to a more gradual increase in the levels of deoxynucleotides. Whether such a mechanism of regulation exists and what would drive such a transformation in vivo requires further study.
Finally, with recent 6.6 Å resolution X-ray data for yeast RNR revealing hexameric α subunit ring-like structures in the presence of dATP (Fairman et al., 2011), it is apparent that attention must be paid to the propensity of multiple RNRs to form rings. While the contributions of particular residues to the stability of the protein-protein interfaces within these rings remains to be determined, it is interesting to note that the residues on the E. coli α nearest the α-β interface (Fig. 3) are not conserved, even within bacterial species. Exploiting differences in ring structures may offer new strategies to inhibit RNRs in a species-specific manner. Clearly the ability of RNR subunits to form and break interactions at interfaces is key to allosteric regulation of activity, and we now find that this subunit flexibility can even lead to the formation of interlocked rings. In particular, the work presented here demonstrates that the α4β4 rings formed by the E. coli enzyme have the proclivity to concatenate in the presence of crowding agents. With conclusive evidence for formation of these interlocked rings in solution, our SAXS and EM data also provide some of the first experimental data to address the mechanism of protein ring concatenation. While beautiful to observe, these concatenated rings appear to be more than a crystallization magic trick. We now understand the relationship between the previously determined α4β4 dATP-inhibited state of E. coli RNR and this interlocked ring system, with information about when and how these concatenated rings can form.
Methods
Reagent Preparation
Cytidine 5′-diphosphate was purchased from Sigma-Aldrich and dissolved into assay buffer (50 mM HEPES, pH 7.6, 15 mM MgCl2, 1 mM EDTA). The pH was slowly adjusted to 7–8 with NaOH and the concentration was determined spectroscopically using ε271 of 9.1 mM−1 cm−1. High purity solutions of ATP, and dATP were purchased from USB Corporation. F2C was a gift to the group of J.S. from Eli Lilly. F2CDP was prepared as described (Artin et al., 2009).
Protein Preparation
The α2 and β2 proteins were prepared as described (Salowe et al., 1986; Salowe et al., 1987). The concentrations of α2 and β2 were determined using ε280 of 189 and 131 mM−1cm−1, respectively; all molar concentrations are dimer concentrations (i.e., α2 or β2).
Crystallization of F2CDP-inactivated RNR
The F2CDP-inactivation reaction was carried out immediately before crystallization by supplementing a protein mixture (31 μM α2 and 62 μM β2 in 50 mM HEPES, 15 mM MgCl2 1 mM EDTA, pH 7.6) with 3 mM ATP and 5 mM DTT and 120 μM F2CDP, and incubating in a 25°C water bath for ten minutes prior to crystallization. The F2CDP inactivated complex was crystallized by adding 2 μL of the protein mixture to 4 μL of precipitant solution (25% PEG 3350, 100 mM HEPES pH 7.5, 200 mM NH4(CH3COO), 5% glycerol) and equilibrating over a 500 μL reservoir of precipitant solution at room temperature. To grow crystals large enough for data collection, drops were streak seeded from crushed crystals grown in similar conditions. Crystals appeared after one week and continued to grow for several weeks. Crystals were cryo-cooled by adding 5 μL of solution containing 25% PEG 3350, 100 mM HEPES pH 7.5, 200 mM NH4(CH3COO), and 20% glycerol to the drop containing crystals then looping out of this drop after 1–3 minutes and plunging directly in liquid N2. Diffraction data were collected at the Cornell High Energy Synchrotron Source (CHESS) beamline A1 at 100 K and processed using HKL2000 (Otwinowski et al., 1997) (Table 1).
Crystallization of dATP-inactivated RNR
For the dATP structure, hydroxyurea-inactivated β2 (met-β2) was used in place of active β2. Met-β2 was prepared from purified active β2 as described (Ando et al., 2011). Crystals were grown by adding 1 μL of protein (25 μM α2 and 50 μM met-β2 in 50 mM HEPES, 15 mM MgCl2 1 mM EDTA pH 7.6 supplemented with 10 mM dATP and 5 mM DTT) to 1 μL of precipitant solution (10% PEG 3350, 100 mM MOPS pH 7.5, 200 mM Mg(CH3COO)2, 25 mM MgCl2, 6 mM n-Nonyl-β-D-maltopyranoside (Hampton Research) and 5% glycerol) and equilibrating over a 500 μL reservoir of precipitant without n-Nonyl-β-D-maltopyranoside at room temperature. Crystals were cryo-cooled after one week by looping thorough solutions of 13% PEG 3350, 100 mM MOPS pH 7.5, 200 mM Mg(CH3COO)2, 40 mM MgCl2, 5 mM DTT, 10 mM dATP and successive concentrations of 10, 15, and 20% glycerol and plunged directly in liquid N2. Diffraction data were collected at the Advanced Photon Source at Argonne National Labs on beamline 24ID-C at 100 K and processed using HKL2000 (Otwinowski et al., 1997) (Table 1).
Structure Solution and Refinement
The initial F2CDP-RNR structure was solved with a lower resolution data set by molecular replacement in the program PHASER (McCoy et al., 2007) using previously solved structures of α and β monomers with no ligands included as search models (PDB ID 1RLR (Uhlin et al, 1994) and 1PFR (Logan et al., 1996) respectively). After higher resolution data was obtained, initial refinement of the F2CDP structure was done using Fourier synthesis methods. The dATP-RNR structure was solved using Fourier synthesis methods from the previously published 5.65 Å structure (Ando et al., 2011). For both structures, iterative rounds of data refinement and model building were performed in CNS 1.3 (Brünger et al., 1998) and COOT (Emsley et al., 2010) respectively. Refinement consisted of rigid body, low temperature (300K) simulated annealing using deformable elastic network restraints (Schroeder et al., 2010) and group B-factor refinement with two B-factors per residue, one for main-chain atoms and one for sidechain atoms. During all rounds of refinement, loose non-crystallographic symmetry (NCS) restraints were used.
Final Model of F2CDP/ATP-RNR
At 4.45 Å resolution, secondary structure can be unambiguously assigned while side chain positions are not well defined. Side chains are included in the model based on their positions in previously solved high resolution structures however their high B-factors (average of 147.8) indicate the positions are not precisely determined. In the F2CDP-RNR structure, there is some positive density in all four catalytic sites where the inhibitor would be expected to bind and react (Fig. S4). Because we do not know the identity of the reaction product and the resolution of the data is too poor to distinguish possible products, nothing has been modeled in these sites. The catalytic site of the α subunit contains two redox active cysteine residues that are oxidized to a disulfide during the reaction (Thelander, 1974; Uhlin et al., 1994). All four catalytic site disulfide pairs appear to be reduced. All residues present in the model are listed in Table S1 with refinement statistics in Table 1. In addition the model contains ATP at all four activity sites as well as 2 Fe3+ atoms with a bridging oxygen and 2 Fe ligated H2Os in each β chain. Positive difference density at the di-iron center could be accounted for by modeling the bridging oxygen and iron ligated water molecules based on their location in previously solved high resolution structures of the β subunit. After including them in the model all difference density was accounted for showing that even at this relatively low resolution we can detect water bound at the di-iron center. In the Ramachandran plot, 90.3%, 9.6%, 0.1% and 0.0% of residues are in favored, allowed, generously allowed, and disallowed regions respectively.
Final Model of dATP-RNR
All residues present in the 3.95 Å resolution dATP model are listed in Table S1 with refinement statistics in Table 1. The model contains dATP at all four specificity sites and all four activity sites (Fig. S4). In addition, density was seen in all four catalytic sites. Only diphosphates bind at the catalytic site but no diphosphates were added to the crystallization so dADP, a likely contaminant of dATP and product of RNR, was modeled in this site (Fig. S4). In addition 2 Fe3+ atoms with a bridging oxygen and 2 Fe ligated H2Os are present in each β chain, modeled as described for the F2CDP-RNR structure. In the Ramachandran plot, 90.2%, 9.8%, 0.0% and 0.0% of residues are in favored, allowed, generously allowed, and disallowed regions respectively.
Small-angle X-ray Scattering of F2CDP-inactivated RNR
The F2CDP-inactivation reaction was carried out immediately prior to SAXS data collection. A protein mixture (30 μM α2 and 30 μM β2 in assay buffer (50 mM HEPES, 15 mM MgCl2 1 mM EDTA pH 7.6)) was supplemented with 3 mM ATP, 5 mM DTT, 120 μM F2CDP (final concentrations), and incubated in a 25°C water bath for ten minutes. In the precipitant titration experiment, in order to mimic crystallization conditions, the 30 μM inactivated mixture was diluted 3-fold with precipitant mixtures (25% PEG 3350, 100 mM HEPES pH 7.5, 200 mM NH4(CH3COO), 5% glycerol) that were diluted with water to give the total v/v precipitant concentration indicated. To obtain data at 3 μM RNR, the 30 μM stock solution was diluted 10-fold with assay buffer containing 3 mM ATP and 5 mM DTT.
SAXS was performed at the Cornell High Energy Synchrotron Source (CHESS) G1 station using a 9.5 keV 250 mm square X-ray beam with a flux of several 1012 photons/s/mm2. Data were collected on a custom 1024×1024 pixel CCD detector (Tate et al., 1995) with a sample-to-detector distance of ~2 m. The transmitted intensity was measured on a PIN diode beamstop (Ando et al, 2008). Protein solutions and reference solutions were prepared such that the buffer contents matched.
Samples were contained in 2 mm path length acrylic cells (ALine Inc.) (Ando et al, 2008) with 7.5 μm kapton windows (Chemplex). The same cell was used for acquisition of protein solution and background scattering pairs. Several 1 and 2 s exposures were taken per sample separated by 10 s pauses. The scattering images were corrected and integrated about the beam center following previously described image correction procedures (Ando et al, 2008). Exposures that did not display apparent radiation-induced changes were averaged and normalized by the transmitted intensities. Background scattering was subtracted from the protein solution scattering to produce the one-dimensional protein scattering profile, I(q), as a function of q, where q = 4π/λ sin θ; 2θ is the scattering angle, and λ is the X-ray wavelength.
Theoretical scattering curves were calculated from atomic coordinates using CRYSOL (Svergun et al., 1995). Here, the resulting forward scattering intensities (I(q) at q=0) are proportional to the square of the molecular weights. Linear combinations of theoretical scattering curves, Ifit(q), were fitted to SAXS data using least-squares fitting in MATLAB:
where Ii(q) and fi are the scattering curve and molar fraction for the i-th species, N is the number of species, and c is a constant scaling factor.
The average globularity of the species in solution was investigated by Kratky analysis (Glatter et al., 1982). Here, the scattering profile is plotted as Iq2 vs. q to emphasize the power-law dependence in the mid-q range. I(q) of globular, folded species display a q−4 dependence in this range, giving rise to a single prominent peak in the Kratky plot. Multiple distinct peaks in a Kratky plot are observed in the case of proteins with multiple domains in a non-globular arrangement (Bernadó et al., 2010) as in the inhibited α4β4 RNR structure (Ando et al., 2011).
Electron Microscopy (EM) of dATP-inhibited RNR
For EM studies, met-β2 was used in place of active β2. 150 nM α2 and met-β2 were combined with 1 mM CDP and 50 μM dATP in assay buffer. 5 μl of the mixture was applied to a carbon coated EM grid that had been glow discharged immediately before use. The grid was washed 3 times with a stain solution containing 2% uranyl acetate (Ted Pella) and 0.2% trehalose and then a second carbon layer was applied (Tischendorf et al., 1974). Eighty-nine pairs of CCD images were collected of the −55° tilted and untilted specimen at 50,000x magnification and 120 kV on a Tecnai F20 microscope (FEI) operated manually using Leginon (Suloway et al., 2005) to store the images. CCD frames have a pixel size of 2.18 Å at the specimen level.
Ring and C-shaped particles were selected from the untilted images using EMAN2’s e2boxer.py (Tang et al., 2007) and TiltPicker (Voss et al., 2009) was modified to import and transform the coordinates onto the tilted images. 14,055 pairs of particles were windowed and 160 particles were removed from the dataset after an initial reference-free alignment and classification as members of 4 poor quality classes. An iterative alignment and supervised classification routine was then repeated on the remaining 13,895 particles using SPIDER as described previously (Brignole et al., 2009) resulting in 33 classes. 3D density maps were reconstructed for each class using CTF-corrected tilted particles (Radermacher et al., 1987; Radermacher et al., 2001) supplemented with a random selection of 10% of the untilted particles (Fairman et al., 2011). Each reconstruction was low-pass filtered at its resolution as determined by the Fourier shell correlation at 0.5.
The reconstructions were interpreted by iterative rigid fitting crystal structures of α2 (4R1R) (Eriksson et al., 1997) and β2 (1RIB) (Nordlund et al., 1993) using UCSF Chimera’s “fitmap” command (Goddard et al., 2007). When compared with the fit structures and our α4β4 crystal structures, the EM density maps appeared compressed in the dimension perpendicular to the carbon support film by a factor of ~1/3, a well-documented artifact of specimen preservation in negative stain (Cheng et al., 2006; Kellenberger et al., 1982). Despite the flattening, contours of the crystal structures could be clearly recognized on the upper and lower surfaces of the EM reconstructions, indicating that the features had been merely compressed in the z-dimension. To compensate for the compression artifact, density maps were stretched to voxel dimensions of 4.36 × 4.36 × 6.54 Å and the fitting of individual subunits was refined. Interacting α-β pairs were extracted from the fitted models and from the α4β4 crystal structures and then the α-subunits of each pair were superimposed in Chimera. Relative motions of the β subunits were calculated with Chimera’s “measure rotation” command (Goddard et al., 2007). Chain E of the dATP crystal structure was chosen as the frame of reference because this chain has an intermediate conformation among the β subunits extracted from the α4β4 crystal structures.
An animation that illustrates the observed conformational flexibility was generated using the “Morph Conformations” tool in Chimera to interpolate between the coordinates of models shown in Fig. 5 (Movie S1).
Supplementary Material
HIGHLIGHTS.
Two structures of inactivated RNRs show two concatenated α4β4 rings in the crystal
Solution scattering shows rings interlock in the presence of crystallization solution
Electron microscopy shows open and incomplete states of ring formation
Mechanism of protein ring concatenation by ring opening and closing is proposed
Acknowledgments
We thank Jun Wang for help with initial crystallization experiments. For assistance with SAXS and crystallographic data collection at CHESS, we thank Dr. Mackenzie Firer-Sherwood and Rebekah Bjork, and CHESS scientists, Drs. Chae Un Kim, Arthur Woll and Richard Gillilan. We thank Prof. Sol Gruner (Cornell) for access to SAXS equipment. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-0936384, and the MacCHESS resource is supported by NIH/NCRR award RR-01646. Crystallographic data were also collected at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by NIH/NCRR award RR-15301. The APS is also supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Electron microscopy was performed at the National Resource for Automated Molecular Microscopy, which is supported by the NIH though the National Center for Research Resources P41 program (RR017573). This work was supported by the National Institutes of Health grants T32GM08334 (C.M.Z.), F32GM904862 (N.A.), F32DK080622 (E.J.B.), GM67167 (F.J.A.), P30-ES002109 (to C.L.D.), and GM29595 (J.S.). C.L.D. is a Howard Hughes Medical Institute Investigator. C.M.Z. performed crystallization and crystal structure determination and analysis. N.A. performed SAXS experiments and data analysis. E.J.B. performed EM experiments and image analysis. F.J.A., J.S., and C.L.D. were involved in study design and data analysis. The manuscript was written and/or edited by C.M.Z., N.A, E.J.B., F.J.A., J.S., and C.L.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ando N, Chenevier P, Novak M, Tate MW, Gruner SM. High hydrostatic pressure small-angle X-ray scattering cell for protein solution studies featuring diamond windows and disposable sample cells. J Appl Crystallog. 2008;41:167–175. [Google Scholar]
- Ando N, Brignole EJ, Zimanyi CM, Funk MA, Yokoyama K, Asturias FJ, Stubbe J, Drennan CL. Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc Natl Acad Sci. 2011;108:21046–21051. doi: 10.1073/pnas.1112715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artin E, Wang J, Lohman GJ, Yokoyama K, Yu G, Griffin RG, Bar G, Stubbe J. Insight into the Mechanism of Inactivation of Ribonucleotide Reductase by Gemcitabine 5′-Diphosphate in the Presence or Absence of Reductant. Biochemistry. 2009;48:11622–11629. doi: 10.1021/bi901590q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadó P. Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. Eur Biophys J. 2010;39:769–780. doi: 10.1007/s00249-009-0549-3. [DOI] [PubMed] [Google Scholar]
- Bonate PL, Arthaud L, Cantrell WR, Jr, Stephenson K, Secrist JA, Weitman S. Discovery and development of clofarabine: A nucleoside analogue for treating cancer. Nat Rev Drug Discov. 2006;5:855–863. doi: 10.1038/nrd2055. [DOI] [PubMed] [Google Scholar]
- Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat Sturct Mol Biol. 2009;16:190–197. doi: 10.1038/nsmb.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969;46:39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR System (CNS), A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cao Z, Roszak AW, Gourlay LJ, Lindsay JG, Isaacs NW. Bovine Mitochondrial Peroxiredoxin III Forms a Two-Ring Catenane. Structure. 2005;13:1661–1664. doi: 10.1016/j.str.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bio Essays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: Obvious but Underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and Developmet of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Uhlin U, Ramaswamy S, Ekberg M, Regnström K, Sjöberg BM, Eklund H. Binding of allosteric effectors to ribonucleotide reductase protein R1: reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- Fairman JW, Wijerathna SR, Ahmad MF, Xu H, Nakano R, Jha S, Prendergast J, Welin RM, Flodin S, Roos A, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, de Oliverira Neto M, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J Appl Crystallog. 2010;43:101–109. [Google Scholar]
- Gandhi V, Plunkett W. Clofarabine: Mechanisms of action, pharmacology and clinical investigations. In: Peters GJ, editor. Cancer Drug Discovery and Development: Deoxynucleoside Analogs in Cancer Therapy. Humana Press; Totowa, NJ: 2006. pp. 153–171. [Google Scholar]
- Glatter O, Kratky O. Small angle X-ray scattering. Academic Press; London; New York: 1982. [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Pérez J, Prangé T, Veesler S, Riès-Kautt M, Vachette P. The BPTI decamer observed in acidic pH crystal forms pre-exists as a stable species in solution. J Mol Biol. 2000;297:697–712. doi: 10.1006/jmbi.2000.3584. [DOI] [PubMed] [Google Scholar]
- Hertel LW, Boder GB, Kroin JS, Rinzel SM, Poore GA, Todd GC, Grindey GB. Evaluation of the antitumor-activity of gemcitabine (2′,2′-difluoro-2′-deoxycytidine) Cancer Res. 1990;50:4417–4422. [PubMed] [Google Scholar]
- Huang P, Chubb S, Hertel LW, Grindey GB, Plunkett W. Action of 2′,2′-difluorodeoxycytidine on DNA-synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- Kashlan OB, Scott CP, Lear JD, Cooperman BS. A Comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- Kellenberger E, Häner M, Wurtz M. The wrapping phenomenon in air-dried and negatively stained preparations. Ultramicroscopy. 1982;9:139–150. doi: 10.1016/0304-3991(82)90236-4. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallog. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A, Reichard P. Enzymatic synthesis of deoxyribonucleotides. IX Allosteric effects in the reduction of pyrimidine ribonucleotides by the ribonucleoside diphosphate reductase system of Escherichia coli. J Biol Chem. 1966a;241:2533–2539. [PubMed] [Google Scholar]
- Larsson A, Reichard P. Enzymatic synthesis of deoxyribonucleotides. X Reduction of purine ribonucleotides; allosteric behavior and substrate specificity of the enzyme system from Escherichia coli. J Biol Chem. 1966b;241:2540–2549. [PubMed] [Google Scholar]
- Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht S, Gerfen GJ, Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271:477–481. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- Logan DT, Su XD, Aberg A, Regnström K, Hajdu J, Eklund H, Nordlund P. Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site. Structure. 1996;15:1053–1064. doi: 10.1016/s0969-2126(96)00112-8. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A. Implications of Macromolecular Crowding for Protein Assembly. Curr Opin Struct Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- Mowa MB, Warner DF, Kaplan G, Kana BD, Mizrahi V. Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J Bacteriol. 2008;191:985–995. doi: 10.1128/JB.01409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Silva JC. Ribonucleotide reductase as a target to control apicomplexan diseases. Curr Issues Mol Biol. 2011;14:9–26. [PubMed] [Google Scholar]
- Nordlund P, Eklund P. Structure and Function of the Escherichia coli Ribonucleotide Reductase Protein R2. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Plunkett W, Huang P, Gandhi V. Gemcitabine: actions and interactions. Nucleosides Nucleotides Nucleic Acids. 1997;16:1261–1270. [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Ruiz T, Wieczorek H, Gruber G. The structure of the V(1)-ATPase determined by three-dimensional electron microscopy of single particles. J Struct Biol. 2001;135:26–37. doi: 10.1006/jsbi.2001.4395. [DOI] [PubMed] [Google Scholar]
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an α6β2 octamer. J Biol Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- Rofougaran R, Crona M, Vodnala M, Sjöberg BM, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J Biol Chem. 2008;283:35310–353118. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- Salowe SP, Stubbe J. Cloning, overproduction, and purification of the B2 subunit of ribonucleoside-diphosphate reductase. J Bacteriol. 1986;165:363–366. doi: 10.1128/jb.165.2.363-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salowe SP, Ator MA, Stubbe J. Products of the inactivation of ribonucleoside diphosphate reductase from Escherichia coli with 2′-azido-2′-deoxyuridine 5′-diphosphate. Biochemistry. 1987;26:3408–3416. doi: 10.1021/bi00386a024. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Chan CT, Mugnaini V, Stubbe J, Bennati M. PELDOR spectroscopy with DOPA-β2 and NH2Y-α2s: distance measurements between residues involved in the radical propagation pathway of E. coli ribonucleotide reductase. J Am Chem Soc. 2007;129:15748–15749. doi: 10.1021/ja076459b. [DOI] [PubMed] [Google Scholar]
- Schroeder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Crystallog. 1995;28:768–773. [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tate MW, Eikenberry EF, Barna SL, Wall ME, Lowrance JL, Gruner SM. A Large-Format High-Resolution Area X-ray Detector Based on a Fiber-Optically Bonded Charge-Coupled Device (CCD) J Appl Crystallog. 1995;28:196–205. [Google Scholar]
- Thelander L. Reaction mechanism of ribonucleotide diphosphate reductase from Escherichia coli. J Biol Chem. 1974;249:4858–4862. [PubMed] [Google Scholar]
- Tischendorf GW, Zeichhardt H, Stoffler G. Determination of the location of proteins L14, L17, L18, L19, L22, L23 on the surface of the 50S ribosomal subunit of Escherichia coli by immune electron microscopy. Mol Gen Genet. 1974;134:187–208. doi: 10.1007/BF00267715. [DOI] [PubMed] [Google Scholar]
- Torrents E, Sahlin M, Biglino D, Gräslund A, Sjöberg BM. Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical. Proc Nat Acad Sci. 2005;102:17946–17951. doi: 10.1073/pnas.0506410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- Van der Donk WA, Yu G, Pérez L, Sanchez RJ, Stubbe J, Samano V, Robins MJ. Detection of a new substrate-derived radical during inactivation of ribonucleotide reductase from Escherichia coli by gemcitabine 5′-diphosphate. Biochemistry. 1998;37:6419–6426. doi: 10.1021/bi9729357. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine 5′-diphosphate inhibit ribonucleotide reductases. Proc Natl Acad Sci. 2007;104:14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler LJ, Rajagopal I, Mathews CK. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 2005;4:1450–1456. doi: 10.1016/j.dnarep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Xu H, Faber C, Uchiki T, Fairman JW, Racca J, Dealwis C. Structure of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation. Proc Natl Acad Sci. 2006;103:4022–4027. doi: 10.1073/pnas.0600443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhenchuk A, Lotfi K, Juliusson G, Albertioni F. Mechanisms of anti-cancer action pharmacology of clofarabine. Biochem Pharmacol. 2009;78:1351–1359. doi: 10.1016/j.bcp.2009.06.094. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu Rev Biochem. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.