Summary
The Hippo signaling pathway was initially defined by genetic studies in Drosophila to regulate tissue growth and organ size [1, 2]. This pathway is highly conserved in mammals and dysregulation of the Hippo pathway has been implicated in human cancer. Although the exact extracellular signal that controls the Hippo pathway is currently unknown, compelling evidence supports a critical role of the Hippo pathway in cell contact inhibition, which is a property commonly lost in cancer cells. Many molecules, such as the merlin tumor suppressor protein, have been identified as regulating the activity of the core Hippo pathway components [1, 2]. Acting downstream are two key transcription co-activators, YAP and TAZ, which mediate the major gene regulation and biological functions of the Hippo pathway. This article will focus on the physiological function and molecular regulation of YAP/TAZ and its Drosophila homolog Yki.
The Hippo pathway acts to restrict YAP and TAZ in mammals
The Hippo pathway is a newly discovered and evolutionally conserved signaling cascade. It regulates organ size control and stem cell property by governing cell proliferation and apoptosis. In vitro, it is a major regulatory mechanism in cell contact inhibition. Alterations of this pathway are increasingly recognized to be associated with cancer development. Components of the Hippo pathway were firstly discovered by functional genetic screens in Drosophila and shown to be evolutionally and functionally conserved in mammals. Basically, the Hippo pathway can be divided into three interlinked parts: the upstream regulatory components, the Hippo core kinase components, and the downstream transcriptional machinery. For the upstream regulatory components, much in depth discussion can been found in recent reviews [1, 2]. The Hippo core kinase cassette contains four proteins, two of which are kinases: Hpo and Wts in the fly and Mst1/2 and Lats1/2 in mammals. The other two proteins, Sav and Mats in the fly, and WW45 and Mob in mammals, act as adaptors/activators. In mammals, Mst1/2 in association with WW45 is activated by phosphorylation in response to upstream regulators. The activated Mst1/2-WW45 can phosphorylate and activate LATS1/2-Mob complex. The major target of the Hippo core kinase cascade is Yki transcription co-activator in the fly and YAP and TAZ which are the mammalian homologues. Phosphorylation of YAP and TAZ by the Hippo pathway leads to their sequestration in the cytoplasm by interaction with 14–3–3 proteins and ubiquitination-dependent proteosomal degradation. Therefore, the Hippo pathway acts to restrict the availability/functionality of YAP and TAZ in the nucleus by governing its distribution and protein levels. In the fly, Yki binds Scalloped (Sd) and activates transcription of downstream target genes like Diap, bantam and cycE. In mammals, YAP and TAZ interact primarily with transcriptional factors TEAD1–4 (TEADs) and activate expression of target genes such as CTGF, IGFBP3, ITGB2, Birc5/Survivin, Gli2, and Axl. In this review, we will focus on the discovery of YAP and TAZ, the demonstration of their function as transcriptional co-activators, their identification as targets of Hippo pathway, their involvement in cancer, and the transcriptional outcome of their complexes with TEAD transcriptional factors.
Identification of YAP and TAZ
YAP was originally identified in chicken as an interacting protein of Yes protein tyrosine kinase. However, the functional significance of this interaction is still not clear. Unlike the use of routine protein-protein interacting approaches, YAP was identified by generating anti-idiotypic antibodies against the N-terminal domain of the Yes protein. The interaction was defined to be mediated by the SH3 domain of Yes and Pro-rich region (PVKQPPPLAP) of YAP. Due to its size of 65 kDa, the chicken protein was referred to as YAP65 (Yes-associated protein of 65 kDa) [3]. The human and mouse homologues were identified by using the YAP65 cDNA to probe human and mouse cDNA libraries and the study was reported one year later [4]. During the course of sequence analysis by comparison of YAP with other proteins, a conserved module was noticed to be present in several proteins of various species such as human dystrophin, yeast Rsp5p, and mammalian Nedd-4. This was named the WW domain to reflect the sequence motif containing two conserved and consistently-positioned tryptophan (W) residues [5]. The WW domain was shown to bind PPXY motif around the same time [6]. Using a functional screen of a cDNA expression library, two proteins binding to the WW domain of YAP were identified as interacting partners and named as WBP-1 and WBP-2 (for WW domain Binding Protein). Sequence comparison between WBP1 and WBP2 followed by interaction assays demonstrated that the PPXY motif binds WW domain with relatively high affinity and specificity. The solution structure of YAP WW domain with PPXY motif was resolved in 1996 [7]. The human YAP gene, located at 11q13, can be transcribed into at least 4 isoforms based on annotation by NCBI. Isoform1, 2, 3 and 4 have 504, 450, 488, and 326 residues in length, respectively. Two consecutive WW domains were present in all isoforms except for isoform 2 which has only one WW domain. Isoform 3 containing 488 residues and two WW domains is most thoroughly studied and the cDNA clone encoding this isoform is most widely used for analysis in the scientific community. The schematic depiction of isoform 3 of YAP is shown in Figure 1A. The C-terminus contains a PZD-binding motif (TWL-COOH) for interaction with PDZ domain of other proteins such as ZO2 and NHERF2 [8, 9]
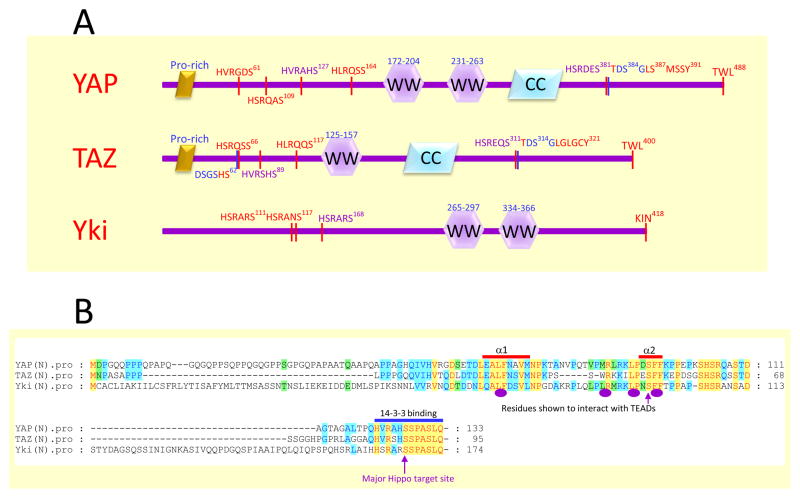
Figure 1.
Domain organization of YAP, TAZ and Yki.
A. The schematic drawing of YAP, TAZ and Yki. The key features are indicated such as the Pro-rich region, the WW domain, and coiled coil region. The HXRXXS motifs targeted by the Hippo pathway are also indicated with the major site in purple. The DSG-degron at the C-terminal part of YAP and TAZ is shown in blue, and the additional N-terminal DSGXXS degron of TAZ is also indicated.
B. Alignment of the N-terminal region of YAP, TAZ and Yki. Residues identical in all three proteins are indicated in red with yellow background. The residues experimentally demonstrated to be involved in interaction with TEADs are indicated with purple ovals (using TAZ in the study) and arrow (using YAP in the analysis) below the alignment. These residues are located within the alpha1 and alpha2 helices (indicated above the alignment), which were revealed structurally to contact TEADs directly. The major Hippo target site whose phosphorylation created a binding site for 14–3–3 proteins is also highlighted.
TAZ (transcriptional co-activator with PDZ-binding motif) is also referred to as WWTR1 (WW domain containing transcription regulator 1). It was originally identified in 2000 as a 14–3–3 binding protein using immobilized 14–3–3 proteins to pull down interacting proteins derived from in vitro translation reactions [8]. 14–3–3 proteins are a family of 7 homologous proteins having the ability to bind phosphorylated serine with certain sequence motifs and are thus involved in diverse cellular functions including differentiation, cell cycle progression and apoptosis through their ability to interact with diverse intracellular phosphoproteins involved in signal transduction network. Although NCBI annotation has indicated that the TAZ gene may be transcribed into three variants, they all have the same coding region for a protein having 400 amino acids in length. Phosphorylation of S89 was shown to be important for interaction with 14–3–3 proteins. TAZ is homologous to YAP with 46% amino acid sequence identify (with YAP isoform 3) and displaying similar domain organization but having only one WW domain (Figure 1A). Biochemically, TAZ was shown to display transcriptional co-activator function via interaction with PPXY-containing transcriptional factors through its WW domain. The C-terminal region (amino acids 165–395) was shown to be responsible for the transcriptional co-activation property [8]. Several transcriptional factors such as Runx/PEBP2, AP2, C/EBP, c-Jun, Krox-20, Krox-24, MEF2B, NF-E2, Oct-4 and p73 contain proline-rich PPXY motif, are speculated to be interacting transcriptional factors for TAZ (and possibly for YAP) (reviewed in [1]). Similar to YAP, the TAZ C-terminus has a PDZ-binding motif (TWL-COOH), which has been shown to mediate interaction with the first PDZ domain of NHERF-2 [8].
Both YAP and TAZ are homologous to fly Yki, which was identified as a downstream target of the Hippo pathway in 2005 [9]. The identification of Yki and the demonstration of its sequence homology with YAP and TAZ have two important implications. Firstly, like Yki, YAP and TAZ may serve as downstream targets of potential mammalian Hippo pathway. Secondly, like YAP and TAZ, Yki may function as a transcriptional co-activator to regulate the transcriptional outcome of the Hippo pathway. Importantly, YAP can functionally substitute Yki in Drosophila, indicating it is a true Yki ortholog. Like YAP, Yki contains two WW domains (depicted in Figure 1A). The N-terminal region of Yki is most homologous to YAP and TAZ. The sequence alignment of the N-terminal regions of YAP, TAZ and Yki is shown in Figure 1B, in which the residues responsible for interaction with TEAD transcriptional factors in mammals and Scalloped in the fly, respectively, are indicated (see below).
Both YAP and TAZ genes have been studied using knockout mice and revealed to have different physiological functions. Knockout of the YAP gene in mice leads to early embryonic lethality, suggesting an essential role in development [10]. Three independent knockout studies of the TAZ gene suggest an important role of TAZ in the kidney and lung as TAZ−/− mice developed renal cysts characteristic of polycystic kidney disease (PKD) [11–13]. The cellular role of TAZ may be to maintain the integrity of renal cilia to ensure the structural integrity of the kidney [11]. Furthermore, levels of calcium-permeable cation channel protein polycystin 2 (PC2) were increased in TAZ−/− kidney and TAZ was shown to link PC2 to the beta-Trcp of SCF (beta-Trcp) E3 ubiquitin ligase pathway [12]. Glis3 is recently shown to interact with TAZ to maintain the normal development and architecture of the kidney [14]. The P/LPXY motif in the C terminus of Glis3 may mediate the interaction with TAZ so that TAZ is able to function as a co-activator of Glis3-mediated gene transcription. Localization to the primary cilium and interaction with TAZ may be involved in the Glis3 signaling pathway. In addition to a role in the kidney, TAZ knockouts also exhibited defects in the lung characteristic of pulmonary emphysema [13].
YAP and TAZ as transcriptional co-activators
Although YAP was identified in 1995, its biochemical function remained elusive until a study 4 years later showed that YAP possesses transcriptional co-activator activity [15]. Several transcription factors such as c-Jun, AP-2, NF-E2, C/EBPalpha and PEBP2/CBF, Runx/PEBP2, Krox-20, Krox-24, MEF2B, Oct-4 and p73 contain the PPXY motif and could be potential target for YAP (reviewed in [1]). The PY motif in the transcription factor PEBP2 was investigated and shown to be important for transcriptional activation [15]. Yeast two-hybrid screens then identified YAP as an interacting protein with the PPXY motif. The WW domains of YAP were revealed to interact with the PPXY motif of PEBP2. The C-terminal region (residues 276 and 472) of YAP exhibited strong transactivation property. Over-expression of YAP also conferred transcription-stimulating activity on PEBP2alpha, suggesting that YAP may potentiate the transcriptional outcome of interacting transcriptional factors. This study established YAP as a transcriptional co-activator. The subsequent identification of homologous TAZ and the demonstration that it also functions as a transcriptional co-activator further supported the notion that YAP and TAZ are transcriptional co-activators that may interact with transcriptional factor with PPXY motifs. However, whether PPXY-containing transcriptional factors are major physiological partners for YAP, TAZ and Yki remains to be confirmed.
An interesting twist in the study of YAP/TAZ/Yki cellular function was the identification of YAP as an interacting protein for TEAD2 transcriptional factor, which does not have the PPXY motif found in other YAP interacting transcription factors [16]. Flag and HA-tagged TEAD2 and its potential interacting proteins were affinity-purified by double sequential immuno-precipitation. Among 12 candidate interacting proteins, only one protein segregated tightly with TEAD2 during sedimentation analysis by glycerol gradient and this protein was identified as YAP. The C-terminal half of TEAD2 was mapped to be responsible for interaction with YAP. In view of the fact that TEAD2, together with TEAD1, TEAD3 and TEAD4 form a homologous TEAD family, YAP was shown to interact also with TEAD1, TEAD3 and TEAD4. The N-terminal region of YAP (residue 32–121, Figure 1B) was shown to possess full interacting potential as the full-length YAP [16]. Importantly, TEAD-mediated transcription was dependent on YAP and YAP-TEAD complex can bind DNA containing TEAD-binding elements. This study suggests that YAP is a general transcriptional co-activator for TEAD transcription factors. Since the TEAD-interacting N-terminal region is most highly conserved among YAP, TAZ and Yki (Figure 1B), the fly TEAD homology Scalloped (Sd) could be a physiological target of Yki in Drosophila. When Drosophila Yki was identified in 2005 as the downstream target of the Hippo tumor suppressor [9], studies on YAP shed new light on the context of potentially conserved Hippo pathway in mammalian cells and its cellular and physiological role in cell proliferation and apoptosis.
YAP, TAZ and Yki as downstream targets of the Hippo pathway
Genetic screens in Drosophila have identified Wts, Hpo, Sav and Mats as tumor suppressors regulating tissue growth by controlling cell proliferation and apoptosis. These 4 proteins are structurally conserved in mammals [17–22]. Wts-Mats in the fly and LATS1/2-Mob1 in human act as protein kinase complexes downstream of the Hpo/Sav and Mst/Sav, respectively. Yeast two-hybrid screen was performed to identify interacting proteins using the N-terminal region of Wts as the bait. Three independent clones encoding for a novel protein were isolated and named (Yki) because its mutation resulted in reduced organ size. Interestingly, Yki is homologous to YAP [9]. Functionally, over-expression of Yki caused massive tissue overgrowth phenocopying loss of function of Hpo, Sav, or Wts. The tissue overgrowth was due to increased cell proliferation and decreased apoptosis (a net increase of cell number). The levels of cell-death inhibitor Diap1 and cell-cycle regulator cyclinE were increased in response to Yki over-expression. Consistently, inactivation of Yki through mutations caused tissue atrophy. Mechanistically, Yki is phosphorylated and inactivated by Wts. Thus the Hippo core components can negatively regulate Yki through direct phosphorylation. The observed tissue overgrowth due to mutations of the Hippo core components can be explained by increased Yki activity as the upstream negative regulatory pathway is compromised. Functional conservation of YAP was established because YAP was able to rescue Yki mutation in the fly. This study establishes a linear signaling pathway (Hpo/Sav to Wts/Mats to Yki to Diap1/CyclinE) in the fly to regulate cell proliferation and apoptosis.
Three landmark studies in 2007 [23–25] have conceptualized the paradigm of the conserved Hippo pathway as the major signaling cascade regulating organ size in vivo and cell contact inhibition in vitro [26]. These studies also elucidated the molecular mechanism through which YAP/Yki is regulated by the Hippo pathway [23–25]. The first study observed that the subcellular location of YAP is dependent on cell density. YAP is primarily present within the nucleus in sparsely growing cells, whereas upon confluence when contact inhibition comes into play, YAP accumulates in the cytoplasm, thereby rendering it spatially unable to function as a transcriptional co-activator. This cell-density regulated cytoplasmic sequestration of YAP correlates with its increased phosphorylation. Mechanistically, Mst2 and LATS2 were shown to act coordinately to phosphorylate YAP at HXRXXS motifs, which is defined as a Lats recognition motif (Figure 1B), with the S127-containing motif being the major site whose phosphorylation created a binding site for 14–3–3 proteins and resulted in cytoplasmic localization. TAZ and Yki have a corresponding motif with S89 and S168 as the major site, respectively (Figure 1B). In sparsely cultured cells, over-expression of LATS2 is able to force YAP S127 phosphorylation and its cytoplasmic sequestration, but mutation of the S127 into Ala abrogates this cytoplasmic shift. Circumventing the regulation of YAP and Yki by LATS2 and Wts respectively through the S127A/S168A mutation also results in enhanced growth-promoting activity. Interaction with 14–3–3 proteins is the basis for cytoplasmic sequestration of S127-phosphoryalted YAP. Earlier studies showed similar interaction of S127-phosphorylated YAP and S89-phosphorylated TAZ with 14–3–3 proteins [8, 27]. It was concluded that the cell-cell interactions upon confluence would trigger a cascade of signaling events that activate the Hippo pathway, which in turn phosphorylates YAP leading to enhanced interactions with 14–3–3 proteins and cytoplasmic sequestration.
This model is supported by genetic data in Drosophila. In searching for suppressors of the small wing phenotype caused by Hpo over-expression, three suppressors were isolated and named as dumbo in accordance with increased wing size [23]. Interestingly, all three suppressor mutations were found to be in the Yki gene. Two mutations were due to substitution of H164 in the Wts recognition motif in the Yki whereas the third mutation was due to substitution of P170 that is essential for Yki to bind 14–3–3. Moreover, expression of the phosphorylation defective YAP-S127A mutant caused a much stronger overgrowth phenotype in Drosophila than the wild type YAP. These studies unequivocally established the functional importance of YAP phosphorylation by Lats and 14–3–3 binding in relaying the biological function of the Hippo pathway, and the mechanistic conservation of this regulation in the fly and mammals.
Independently, study in the fly showed that Hippo pathway caused cytoplasmic sequestration of Yki and that Yki S168 is the major site targeted by Wts, leading to growth suppression. Consistently, loss of Hippo signaling due to mutation of Hippo or Wts results in nuclear accumulation of Yki accompanied by increased tissue overgrowth. This study also extended into the mammalian system in which it was revealed that S127 of YAP is phosphorylated by LATS1/2. More significant was the demonstration that variations of YAP levels can overcome organ size control in vivo in transgenic mice; an induced increase of YAP in the liver led to its enlargement due largely to increased cell numbers. Continued YAP over-expression can expand liver mass from 5% to about 25% of body weight, yet this effect is reversible as the enlarged liver reverts to almost normal size when over-expression of YAP is restrained for a sufficient period of time. This dramatic and reversible manipulation of liver size through YAP changes alone positions the Hippo pathway as the major mechanism controlling liver size in mammals. Microarray studies showed that several growth-promoting and/or anti-apoptotic genes are up-regulated by YAP such as c-Myc, Sox4, BIRC5/survivin and BIRC2/cIAP1. Increased level of BIRC5/survivin is necessary for YAP to induce anchorage-independent growth in vitro. Finally, sustained and prolonged high level of YAP expression in the liver of transgenic mice leads eventually to tumorigenesis characteristic of hepatocellular carcinoma [24]. The tumor inducing effect of YAP is particularly profound when YAP is over-expressed at early stages of development.
Another independent study also showed that YAP is sufficient for inducible and reversible liver enlargement in transgenic mice [25]. For the first time, this study also linked YAP expression to stem/progenitor cells in the intestine. YAP is primarily expressed in the crypt compartment where these cells reside. YAP overexpression in the intestine of transgenic mice correlates with elevated levels of cyclin D and BclXL, and causes dysplasia due to over proliferation of the crypt stem/progenitor cells. Interestingly, this parallels the correlation between YAP expression levels and enhanced levels of cyclin D and BclXL in human colon cancers. This study thus implicates YAP as a critical link between stem/progenitor cells and colon cancer cells [25]. These three seminal studies in 2007 firmly established that the conserved Hippo pathway regulates the function of transcriptional co-activator YAP and Yki in mammals and fly, respectively, and that over-expression of YAP or its over-activation due to intrinsic Hippo pathway mutations is able to abrogate cell contact inhibition in vitro and organ size control in vivo to promote tissue overgrowth and cancer development.
After its identification, TAZ has been shown to function as a transcriptional co-activator implicated in modulating mesenchymal stem cell differentiation [8]. By co-activating Runx2-dependent transcription while repressing PPARγ-dependent transcription, TAZ is able to promote differentiation into osteoblasts and to suppress differentiation to adipocytes [28]. Subsequent studies revealed that TAZ is also targeted by the Hippo pathway and capable of promoting oncogenic transformation in vitro [29, 30]. In one study, it was shown that TAZ is able to promote cell proliferation and epithelial-mesenchymal transition (EMT) and the Hippo pathway acts to restrict TAZ activity. Several serine residues in the conserved HXRXXS motifs of TAZ were phosphorylated by Hippo pathway with S89 phosphorylation creating the 14–3–3 binding site, leading to cytoplasmic retention that result from interaction with 14–3–3 proteins. In another study, it was shown that TAZ promotes migration, invasion, and tumorigenesis of breast cancer cells. The protein levels of TAZ in a panel of breast cancer cell lines correlate with the invasiveness of cancer cells. Over-expression of TAZ in low-expressing MCF10A cells not only causes morphologic changes characteristic of cell transformation and EMT but also promotes cell migration and invasion. Knockdown of TAZ expression in MCF7 and Hs-578T cells reduced cell migration and invasion and also inhibited anchorage-independent growth in soft agar and tumorigenesis in nude mice of MCF7 cells. TAZ protein was seen to be over-expressed in a fraction of breast cancer samples, suggesting that TAZ is an oncogene and this study represents the first link of TAZ over-expression with invasive property and cancer development.
In addition to regulating the cytoplasmic retention through phosphorylation of YAP S127, TAZ S89 and Yki 168 to create a binding site for 14–3–3 proteins, there are other sites (4, 3, and 2 additional sites, see Figure 1A, for YAP, TAZ and Yki, respectively) that are targeted by the Hippo core kinases. Multiple phosphorylation sites in Yki are found to contribute to Yki functional inactivation [31]. Moreover, among these sites, the functional consequence has been established for YAP S381 and TAZ S311 [32, 33]. Phosphorylation of YAP S381 and TAZ S311 facilitate the phosphorylation of S384 of YAP and S314 of TAZ by casein kinase 1, leading to recruitment of SCF E3 ubiquitin ligase and ubiquitination of YAP and TAZ followed by proteosomal degradation. Therefore, in addition to transiently regulating cytoplasmic sequestration, Hippo pathway can also prime permanent proteosomal degradation via phosphorylating YAP S381 and TAZ S311. These duel regulations enable Hippo pathway to tightly restrict the functionality of YAP and TAZ to prevent unwanted growth. Whether Yki is also under similar regulation is not yet defined.
A recent study revealed a role of YAP/TAZ in mechanotransduction that may be regulated independent of the Hippo pathway [34]. It is increasingly being recognized that physical and mechanical cues in microenvironment such as extracellular matrix (ECM) stiffness may regulate cell behaviour. Through mechanotransduction, cells translate these external stimuli into intracellular biochemical signals controlling cell growth, differentiation and cancer malignant progression. YAP and TAZ were identified as nuclear relays of mechanical signals exerted by ECM rigidity and cell shape. The regulation of YAP and TAZ was shown to require Rho GTPase activity and tension of the actomyosin cytoskeleton, but is independent of the Hippo pathway. Functionally, YAP and TAZ are required for differentiation of mesenchymal stem cells induced by ECM stiffness and for survival of endothelial cells regulated by cell geometry. Over-expression of activated YAP transcends physical constraints in dictating cell behaviour, suggesting that YAP and TAZ act as sensors and mediators of mechanical cues instructed by the cellular microenvironment. Several recent studies have also implicated the function of actin cytoskeletal organization in regulating YAP/Yki phosphorylation although the precise mechanism remains to be elucidated [35–38]. One may speculate that the actin cytoskeleton, cell morphology, cell attachment, and mechanosensing act through a common mechanism to regulate YAP/TAZ/Yki activity.
TEAD transcriptional factors as major mediators of YAP, TAZ and Yki in transcription and functional outcome
As briefly mentioned above, YAP was identified as a tight and major interacting protein for TEAD2 and proposed to function as a general transcriptional co-activator for the TEAD transcriptional factors [16]. TEAD1 was originally identified more than 20 years ago as transcriptional enhancer factor 1 (TEF-1) for SV40 virus [39] and it is also involved in transcriptional enhancement of E6 and E7 expression of papillomavirus 16 [40]. There are 4 related family members (TEAD1–4) in mammals and the fly has one highly homologous protein (with 68% amino acid identity with TEAD1) called Scalloped (Sd), which was cloned as a transcriptional factor required for sensory organ differentiation and wing morphogenesis in the fly [41]. The N-terminal region of TEADs and Sd contains a conserved TEA domain involved in recognizing DNA elements such as GGAATG in the promoter region of target genes [42]. The crystal structure of the TEA domain has been resolved and revealed a three-helix bundle with a homeodomain fold [42]. Interestingly, the helix 3 also contains a bipartite nuclear localization signal (NLS) [43] (Figure 2). The C-terminal region of TEADs and Sd is involved in interaction with YAP/TAZ/Yki [16] (Figure 2). Several recent studies establish that TEADs in mammals and Sd in the fly are the major transcriptional factors mediating the biological outcome of YAP/TAZ and Yki, respectively, which is governed by the Hippo pathway.
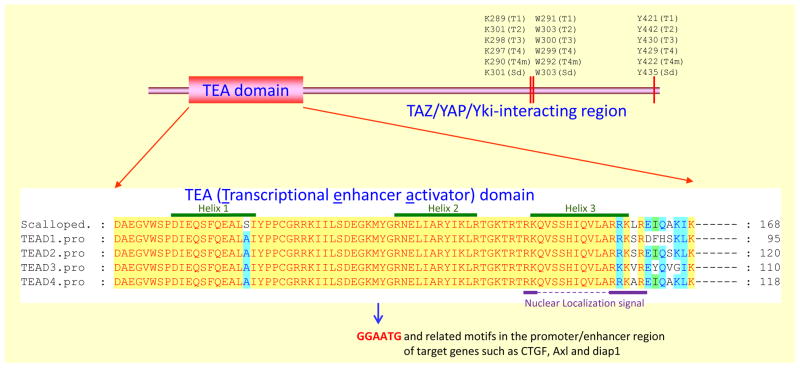
Figure 2.
Domain organization of TEADs and scalloped (Sd). The N-terminal region of TEADs and Sd contains the highly conserved TEA domain responsible for interaction with DNA elements such as GGAATG in the promoter region of target genes (such as CTGF and Axl). The amino acid alignment of the TEA domain of TEAD1–4 and Sd is also shown. The TEA domain forms a 3-helix bundle and the regions for the three helices are indicated on top. The nuclear localization signal is indicated below the alignment. The C-terminal region of TEADs and Sd is responsible for interaction with YAP/TAZ/Yki. Three residues conserved in TEADs and Sd that were experimentally demonstrated (using mouse TEAD4 in the study) to be important for interaction with YAP/TAZ/Yki (using mouse YAP as an interacting partner) are indicated (T1–T4 for TEAD1–4; T4m for mouse TEAD4, Sd for fly scalloped).
In one study, TEADs were shown to be essential in mediating YAP-dependent gene expression and functional outcome such as cell growth, oncogenic transformation, and EMT [44]. S94 in YAP is important for interaction with TEADs and mutation of this residue abolished most if not all property of YAP in promoting cellular transformation and mediating the transcriptional outcome as assessed by microarray, suggesting that interaction with TEADs is the major functional pathway of YAP. Congruent with this, YAP and TEAD bind to a common set of genomic targets. In addition, CTGF was identified as a direct target gene of TEAD-YAP complex and the promoter region of CTGF gene contains several GGAATG motifs for TEAD-binding. The functional conservation was also demonstrated for the interaction of fly Yki and Sd in promoting cell proliferation and tissue outgrowth [44–46]. In one study, the Yki-induced gene diap1 was revealed to contain a minimal Sd-binding element that is responsive to the Hippo pathway. Similarly, Yki mutant defective in interaction with Sd inactivated Yki function in inducing tissue overgrowth in vivo [46]. Furthermore, Sd was shown to promote Yki nuclear localization as well as recruit Yki to the promoter/enhancer region of the diap1 gene. A constitutively active Sd promotes tissue overgrowth, suggesting that Yki acts as a transcriptional co-activator to activate Sd [45].
Similar functional relationship between TEAD and TAZ is also implicated, TEADs were shown to be the major interacting partners for TAZ and this interaction is important for TAZ to mediate cellular oncogenesis as assessed by anchorage-independent soft agar growth as well as for target gene transcription [47, 48]. Mechanistically, interaction with TEADs is essential for TAZ to accumulate in the nucleus, because mutant analysis revealed a strict correlation between the ability of TAZ to interact with TEADs, to transform cells and to be accumulated in the nucleus. The TAZ residues (together with the corresponding residue of YAP S94) shown to be essential for TEAD-interaction and transformation are well conserved in YAP and Yki and they are indicated in Figure 1B.
Further verifying the functional importance of YAP-TEAD interaction is the co-crystal structure of YAP-TEAD complex [49, 50]. The structural analyses have validated previous biochemical studies that had identified key residues important for interaction between TEAD and YAP or TAZ [47, 48]. The structural basis for the YAP/TEAD interaction will be explored in detail by Sudol et al in this issue of Semin Cell Dev Biol. It is worth noting that Y421 in TEAD1 is mutated in human Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) [51] and is essential for TEAD to interact with YAP/TAZ. These observations underscore the in vivo importance of the partnership between YAP/TAZ and TEAD and provide a molecular basis for the pathogenesis of Sveinsson’s chorioretinal atrophy.
Angiomotin family and Wbp2 as novel regulator of YAP and TAZ
Proteomics approach was employed by several labs to identify novel interacting proteins for YAP/TAZ/Yki. In addition to transcription factors and 14–3–3 proteins, there are other proteins co-immunoprecipitated with YAP and TAZ. Among them were Wbp2, Angiomotin (Amot), Angiomotin-like 1 (Amot-L1), and Angiomotin-like 2 (Amot-L2). Wbp2 was earlier identified as an interacting protein for YAP [52, 53], but the functional relevance was not addressed. Two recent studies suggest that Wbp2 is a positive regulator of YAP/TAZ in mammals and Yki in the fly, respectively, in YAP/TAZ/Yki-mediated gene expression, cell proliferation and tissue growth [54, 55]. The sole WW domain of TAZ is important to interact with PPXY motifs of Wbp2. The mammalian Wbp2 has three PPXY motifs but the second one is most important for interaction with TAZ. Knockdown of endogenous Wbp2 suppresses, whereas over-expression of Wbp2 enhances, TAZ-driven cell transformation and gene expression [54]. The fly Wbp2 was similarly shown to promote Yki-dependent tissue growth and this is mediated by direct interaction of the WW domain of Yki and the PPXY motif of Wbp2 [55]. Therefore, Wbp2 is a downstream component of the Hippo pathway positively regulating YAP-TEAD complex in promoting cell proliferation. Interestingly, a recent study showed that Wbp2 is tyrosine-phosphorylated at Y192 and Y231 by c-Src and c-Yes kinases. Functionally, over-expression of Wbp2 and its phospho-mimetic mutant in MCF7 cells promoted tumor formation in xenograft model in nude mice [56]. Y192 is only 5 residues apart from the second PPXY motif; it will be interesting to investigate whether the interaction of Wbp2 with YAP/TAZ/Yki is regulated by Tyr-phosphorylation.
Three independent studies have recently identified Angiomotin (Amot), Angiomotin-like 1 (AmotL1) and Angiomotin-like 2 (AmotL2) as negative regulators of YAP and TAZ [57–59]. All three studies demonstrated direct interaction of YAP/TAZ with Amot, AmotL1 and/or AmotL2 and the interaction is mediated by the sole WW domain of TAZ and the first but not the second WW domain of YAP with the PPXY motif in the N terminus of Amot family proteins. Over-expression of Amot family proteins caused cytoplasmic retention of TAZ and YAP and suppressed its transcriptional outcome such as the expression of CTGF and Cyr61. Conversely, knockdown of Amot family proteins in MDCK cells caused activation of YAP/TAZ and loss of contact inhibition. Interestingly, the Hippo refractory TAZ mutant (S89A) is also negatively regulated by Amot and AmotL1, supporting a phosphorylation independent regulation of YAP/TAZ by AMOT. However, AMOT knockdown also decreased Lats kinase activity and YAP phosphorylation although the molecular mechanism of AMOT in Lats regulation is currently unknown. Since Amot family proteins are integral components of cell junction complexes [60], their interaction with YAP and TAZ may play a key role in regulating the Hippo pathway in response to cell contact in vitro and organ size control in vivo. The interacting proteins for TAZ, YAP and Yki are schematically summarized in Figure 3.
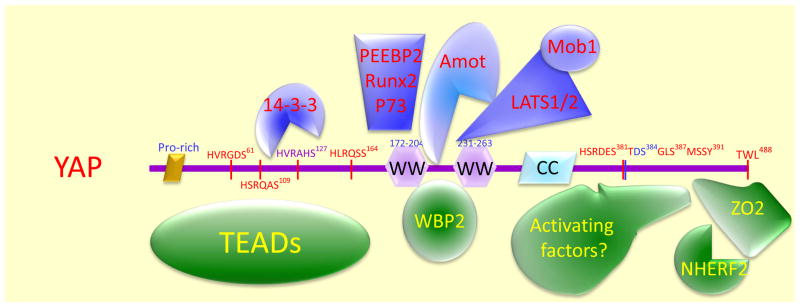
Figure 3.
Summary of proteins shown to interact with YAP/TAZ/Yki. Amot family proteins are present only in mammalian cells but the drosophila expanded has been shown to interact with WW domain of fly Yki. The potential factors that interact with the C-terminal transactivation domain to promote transcriptional program are yet to be defined. The C-terminal PZD-binding motif is specific for YAP and TAZ but not present in Yki and has been shown to interact with ZO2 and NHERF2. Not all proteins interact with YAP at the same time. In fact, some interaction may be mutually exclusive. For example, the 14–3–3 associated YAP would be in cytoplasm whereas the TEAD associated YAP should be nuclear.
α-catenin is an actin binding protein that bridges the cytoskeleton to cell surface adhesion molecule cadherin. α-catenin is also found to interact with and inhibit YAP [61]. α-catenin accomplishes these functions by modulating interaction with 14–3–3 and PP2A phosphatase. Therefore, α-catenin acts in a manner similar to AMOT to inhibit YAP/TAZ. Given the fact that α-catenin is associated with adherent junction and AMOT is associated with tight junction, these two proteins may relay the cell-cell contact signals to inhibit cell growth via modulating YAP/TAZ.
The role of YAP and TAZ in cancer development
Increasing evidence supports that YAP and TAZ are oncogenes in mammalian cells [1, 62]. Genome-wide analysis using mouse model of liver cancer showed chromosome amplification at 9qA1, which is syntenic to human chromosome region 11q22. Among genes in the amplified region are YAP and cIAP1, the latter being a dIAP1-related protein. YAP and cIAP1 are individually oncogenic, but they can cooperate to accelerate tumor growth [63]. In an independent study, an amplification of a smaller chromosomal region within 9qA1 was identified in mouse mammary tumors and YAP is the only gene within this narrower region. Amplification of 11q22 was also observed in several human cancers. YAP gene is also amplified in human intracranial ependymomas, oral squamous cell carcinomas, and medulloblastomas [64–67], demonstrating a critical role of YAP in human cancer. Moreover, YAP over-expression has been found in many human tumors, such as approximately 45% liver cancers [23]. High YAP expression is also associated with poor prognosis of several human cancers, including esophageal squamous cell carcinomas, hepatocellular carcinomas, non-small cell lung cancer, and ovarian cancer [68–71].
Among genes induced by YAP, CTGF and Axl are direct targets of YAP-TEAD complex. Knockdown of CTGF or Axl partially impaired the oncogenic property of YAP [44, 72]. In addition, amphiregulin (AREG) has been implicated as an effector of YAP in conferring growth factor-independent growth, but whether TEADs are involved is not clear [73]. YAP is also implicated in stem cell property of neuroprogenitor cells [74] and embryonic stem (ES) cells [75]. These observations suggest that the ability of YAP to promote stem cell property is part of the mechanism responsible for its oncogenic behavior. Since EMT is also associated with stem cell property [76, 77], the ability of YAP to promote EMT is consistent with this hypothesis.
The first link of TAZ with human cancer was instigated by the observation that the protein level is higher in invasive breast cancer cell lines and a fraction of primary breast cancer samples [29]. Screening over 40 human cancer cell lines led to the finding that TAZ levels are higher in invasive Hs578t, BT549 and MDA-MB-231 breast cancer cells while its levels are lower in noninvasive MCF7 and ZR75.1 cells. Over-expression and knockdown approaches were used to demonstrate that TAZ is able to promote cell migration and invasion and is important for the transformed phenotype of breast cancer cells [29]. TAZ not only promotes cell proliferation and EMT but also causes oncogenic transformation when over-expressed [30]. The oncogenic property of TAZ is greatly enhanced when S89 is mutated into alanine, confirming its negative regulation by the Hippo pathway through phosphorylation at S89. Furthermore, along with TEAD4, TAZ mRNA level was increased in basal type and triple negative breast cancers [78, 79]. A recent study has shown that TAZ may confer resistance to Taxol (paclitaxel) through its target genes Cyr61 and CTGF, as knockdown of Cyr61 and CTGF reversed TAZ-induced Taxol resistance in breast cancer cells [80].
Supporting the concept that TAZ is an oncogene, TAZ was recently linked to cancer stem cell-related traits in breast cancer cells [81]. TAZ is required to sustain self-renewal and tumor-initiation capacities in breast CSCs. Importantly, TAZ level/activity is elevated in CSCs and in poorly differentiated human tumors and have prognostic value. TAZ is sufficient to endow self-renewal capacity to non-CSCs. Mechanistically, TAZ forms a complex with the cell-polarity determinant Scribble, and loss of Scribble, or induction of EMT, uncouples the inhibition of TAZ by the Hippo core kinases [81]. Genomic analysis of breast cancer patients revealed that TAZ is among the genes that have increased de novo mutations in the metastatic cells as compared with the primary cancer cells in the same patients. F229V was detected but the functional consequence on proliferation and metastatic property is yet to be examined [82].
In addition to breast cancer, TAZ is over-expressed in non-small cell lung cancer (NSCLC), while TAZ knockdown in NSCLC cells suppressed proliferation and oncogenic properties [83]. Furthermore, TAZ may contribute to malignant glioma through regulating mesenchymal differentiation [84]. Gene expression profiling of glioblastoma (GBM) showed that patients with a mesenchymal (MES) gene expression signature exhibit poor overall survival and treatment resistance. TAZ was revealed to be highly associated with the MES network. Functionally, TAZ knockdown in MES glioma stem cells (GSCs) decreased expression of MES markers, invasion, self-renewal, and tumor formation. Conversely, overexpression of TAZ induced MES marker expression and aberrant osteoblastic and chondrocytic differentiation in a TEAD-dependent fashion. Interacting with TEAD2 was shown to be important and therefore TAZ and TEAD may drive the MES differentiation of malignant glioma.
Conclusions
Rapid progresses in the Hippo pathway studies have established the important function of this signaling pathway in cell proliferation, cell death, organ size control, and tumor development. Convincing genetic and biochemical data have shown that YAP/TAZ are the major downstream effectors of the Hippo pathway directly phosphorylated by Lats though it is likely that Lats may have additional substrates to exert its full biological functions. Moreover, TEADs have emerged as the key transcription factors that partner with and mediate the transcription response of YAP/TAZ whereas the functional significance of other YAP interacting transcription factors require further investigation. Many components have been identified to act upstream of the Hippo core kinase cascade. However, key questions, such as the mechanism of MST activation and extracellular signals regulating the Hippo-YAP pathway, remain to be addressed.
Highlights.
The Hippo pathway plays a key role in organ size control and dysregulation of the pathway contributes to tumorigenesis
The YAP/TAZ transcription co-activators are the majn downstream effectors of the Hippo pathway
YAP/TAZ are phosphorylated and inhibited by the Lats kinase of the Hippo pathway
The TEAD family proteins are the major target transcription factors mediating gene expression by YAP/TAZ
Acknowledgments
We would like to express our sincere thanks to Dr Siew Wee Chan and Alice Tay for their valuable help in preparing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan D. The hippo signaling pathway in development and cancer. DevCell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–52. [PubMed] [Google Scholar]
- 4.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, et al. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–41. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 5.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–3. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–23. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, et al. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–9. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 8.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14–3–3 and PDZ domain proteins. Embo J. 2000;19:6778–91. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A. 2007;104:1631–6. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, et al. TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol. 2007;27:6383–95. doi: 10.1128/MCB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. AmJ Physiol Renal Physiol. 2008;294:F542–F53. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- 14.Kang HS, Beak JY, Kim YS, Herbert R, Jetten AM. Glis3 is associated with primary cilia and Wwtr1/TAZ and implicated in polycystic kidney disease. Mol Cell Biol. 2009;29:2556–69. doi: 10.1128/MCB.01620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. Embo J. 1999;18:2551–62. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–41. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–63. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 18.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–46. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 19.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–67. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–56. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 21.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–78. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 22.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–85. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 23.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–33. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–60. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 26.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–92. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14–3–3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 28.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–8. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 29.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–8. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 30.Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–36. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–27. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu CY, Zha ZY, Zhou X, Zhang H, Huang W, Zhao D, et al. The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem. 2010;285:37159–69. doi: 10.1074/jbc.M110.152942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrighton KH. Mechanotransduction: YAP and TAZ feel the force. Nat Rev Mol Cell Biol. 2011;12:404. doi: 10.1038/nrm3136. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, Janody F. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138:2337–46. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 36.Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. Embo J. 2011;30:2325–35. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–14. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 38.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–68. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 40.Ishiji T, Lace MJ, Parkkinen S, Anderson RD, Haugen TH, Cripe TP, et al. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. Embo J. 1992;11:2271–81. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992;6:367–79. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- 42.Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103:17225–30. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magico AC, Bell JB. Identification of a classical bipartite nuclear localization signal in the Drosophila TEA/ATTS protein scalloped. PLoS One. 2011;6:e21431. doi: 10.1371/journal.pone.0021431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–98. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–87. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–58. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. JBiolChem. 2009;284:13355–62. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Chan SW, Zhang X, Walsh M, Lim CJ, Hong W, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, et al. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–40. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, et al. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) HumMolGenet. 2004;13:975–81. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 52.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, et al. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–7. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 53.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, et al. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–41. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 54.Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–10. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X, Milton CC, Poon CL, Hong W, Harvey KF. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011;18:1346–55. doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim SK, Orhant-Prioux M, Toy W, Tan KY, Lim YP. Tyrosine phosphorylation of transcriptional coactivator WW-domain binding protein 2 regulates estrogen receptor alpha function in breast cancer via the Wnt pathway. Faseb J. 2011;25:3004–18. doi: 10.1096/fj.10-169136. [DOI] [PubMed] [Google Scholar]
- 57.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–26. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Huang J, Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J Biol Chem. 2011;286:4364–70. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, et al. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–48. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 61.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, et al. The Hippo pathway in biological control and cancer development. J Cell Physiol. 2011;226:928–39. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 63.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baldwin C, Garnis C, Zhang L, Rosin MP, Lam WL. Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 2005;65:7561–7. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez L, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–41. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Modena P, Lualdi E, Facchinetti F, Veltman J, Reid JF, Minardi S, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24:5223–33. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 67.Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–42. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 68.Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–85. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010 doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, et al. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–22. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 71.Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, et al. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2011;32:389–98. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- 72.Xu MZ, Chan SW, Liu AM, Wong KF, Fan ST, Chen J, et al. AXL receptor kinase is a mediator of YAP-dependent oncogenic functions in hepatocellular carcinoma. Oncogene. 2011;30:1229–40. doi: 10.1038/onc.2010.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. NatCell Biol. 2009;11:1444–50. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–34. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–18. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 77.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 79.Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, et al. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes ChromosomesCancer. 2008;47:490–9. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 80.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71:2728–38. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 81.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–72. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 82.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou Z, Hao Y, Liu N, Raptis L, Tsao MS, Yang X. TAZ is a novel oncogene in non-small cell lung cancer. Oncogene. 2011;30:2181–6. doi: 10.1038/onc.2010.606. [DOI] [PubMed] [Google Scholar]
- 84.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25:2594–609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]