Abstract
Reports of nuclear tRNA aminoacylation and its role in tRNA nuclear export (Lund and Dahlberg, 1998; Sarkar et al., 1999; Grosshans et al., 2000a) have led to the prediction that there should be nuclear pools of aminoacyl-tRNA synthetases. We report that in budding yeast there are nuclear pools of tyrosyl-tRNA synthetase, Tys1p. By sequence alignments we predicted a Tys1p nuclear localization sequence and showed it to be sufficient for nuclear location of a passenger protein. Mutations of this nuclear localization sequence in endogenous Tys1p reduce nuclear Tys1p pools, indicating that the motif is also important for nucleus location. The mutations do not significantly affect catalytic activity, but they do cause defects in export of tRNAs to the cytosol. Despite export defects, the cells are viable, indicating that nuclear tRNA aminoacylation is not required for all tRNA nuclear export paths. Because the tRNA nuclear exportin, Los1p, is also unessential, we tested whether tRNA aminoacylation and Los1p operate in alternative tRNA nuclear export paths. No genetic interactions between aminoacyl-tRNA synthetases and Los1p were detected, indicating that tRNA nuclear aminoacylation and Los1p operate in the same export pathway or there are more than two pathways for tRNA nuclear export.
INTRODUCTION
In eukaryotic cells most RNAs are transcribed in the nucleus but perform their cellular functions in the cytosol. Export of RNA from the nucleus occurs across nuclear pores, channels connecting the nuclear and cytosolic compartments (for review, see Görlich and Kutay, 1999). Transport of macromolecules across the nuclear boundary requires a functional small GTPase, Ran, its regulators, and members of the family of Ran-binding proteins known as importins and exportins. It has been proposed that RanGTP, primarily located in the nucleus, and RanGDP, primarily located in the cytosol, establish a gradient across the nuclear boundary that determines the directionality of macromolecular flow (Izaurralde et al., 1997). Nuclear RanGTP forms a trimeric complex with a Ran-binding exportin and a cargo, and the complex is exported to the cytosol, where RanGAP activates hydrolyzsis of RanGTP to RanGDP, thereby releasing the cargo (for review, see Görlich and Kutay, 1999).
There are 14 members in Saccharomyces cerevisiae of the importin/exportin family and >20 members in higher eukaryotes, and the various family members have distinct specificities for transport cargo (for review, see Görlich and Kutay, 1999). Exportin-t has been shown to bind tRNA directly (Arts et al., 1998a; Hellmuth et al., 1998; Kutay et al., 1998). In vertebrate cells excess nuclear exportin-t facilitates tRNA nuclear export (Arts et al., 1998a; Kutay et al., 1998), and ablation of nuclear exportin-t via nuclear injection of anti-exportin-t inhibits tRNA nuclear export (Lipowsky et al., 1999), supporting the model that exportin-t is the β-importin family member responsible for export of tRNA to the cytosol. In yeast, the β-importin member closest in sequence to vertebrate exportin-t is Los1p. Los1p appears to be the exportin-t homologue because Los1p binds tRNA and los1 mutants accumulate nuclear pools of tRNA (Sarkar and Hopper, 1998; Grosshans et al., 2000a). However, LOS1 is not an essential gene in yeast (Hurt et al., 1987), leading to the idea that there are alternative nuclear export mechanisms, at least for yeast (Sarkar et al., 1999; for review, see Grosshans et al., 2000b).
It was recently proposed that nuclear tRNA aminoacylation functions in a proofreading step that monitors tRNA processing and structure before nuclear export of tRNA to the cytosolic protein synthesis machinery (Lund and Dahlberg, 1998). Lund and Dahlberg (1998) showed that tRNAs are aminoacylated in Xenopus nuclei and that prevention of tRNA aminoacylation inhibits export of tRNAs to the cytosol. It appears that such a proofreading system is conserved because in the yeast, S. cerevisiae, tRNAs are aminoacylated while in the nucleus (Sarkar et al., 1999), and inhibition of aminoacylation causes tRNA nuclear accumulation (Sarkar et al., 1999; Grosshans et al., 2000a).
Although aminoacylation has been implicated in tRNA export from the nucleus to the cytosol, most tRNA aminoacylation occurs in the cytosol. This is because aminoacylation is an integral part of the cytosolic protein synthesis system. Nevertheless, there have been previous reports describing detection by microscopy of nuclear pools of aminoacyl-tRNA synthetases in mammalian cells (Popenko et al., 1994; Barbarese et al., 1995) and quite recently a report of cofractionation of several aminoacyl-tRNA synthetase activities with nuclei from rodent cells (Nathanson and Deutscher, 2000). The goal of our work was to determine whether there are nuclear pools of aminoacyl-tRNA synthetases in yeast and, if so, to determine the consequences of depleting the pools on tRNA nuclear export.
We previously showed that a temperature-sensitive (ts) allele of TYS1 (tys1-1), encoding a defective tyrosyl-tRNA synthetase, Tys1-1p, caused tRNA nuclear accumulation in yeast (Sarkar et al., 1999). If indeed the defect in tRNA nuclear export in this mutant is due to lack of aminoacylation of tRNATyr in the nucleus, then there normally should be a nuclear pool of Tys1p. We report that nuclear pools of tyrosyl-tRNA synthetase exist, and alterations of TYS1 that inhibit delivery of Tys1p to the nucleus result in tRNA nuclear accumulation. However, aminoacylation of nuclear tRNA appears not to be absolutely required for tRNA nuclear export because depletion of the nuclear synthetase pool appears not to affect cell growth. Moreover, no genetic interactions between alterations of aminoacyl-tRNA synthetases and Los1p could be detected. Thus, it appears that either nuclear aminoacylation of tRNA and Los1p operate in series in a single path or they operate in parallel, and there are more than two pathways for the export of tRNA from the nucleus to the cytosol.
MATERIALS AND METHODS
Strains and Media
The following yeast strains were used: ts2 (MATa ade2-101 his3Δ200 tyr1 ura3-52 tys1-1; Sarkar et al., 1999); SSS708 (MATa ade2-101 his3Δ200 tyr1 ura3-52 tys1-1 los1::Kanr; see below for construction); SS328 (MATα ade2-101 his3Δ200 lys2-801 ura3-52; Vijayraghavan et al., 1989); SSS706 (MATα ade2-101 his3Δ200 lys2-801 ura3-52 los1::Kanr; see below for construction); SJ17 (MATα ura3–52 leu2-3,-112 gal4; from J.E. Hopper); A364a (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1; Hartwell and McLaughlin, 1968); SSS707 (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1 los1::Kanr; see below for construction); ts341 (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1 ils1-1; Hartwell and McLaughlin, 1968); SSS705 (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1 ils1-1 los1::Kanr; see below for construction); ts19:3:4 (MATa ade1 leu2 his5 lys11 gal1 gal2 mes1-1; McLaughlin and Hartwell, 1969); SSS703 (MATa ade1 leu2 his5 lys11 gal1 gal2 mes1-1 los1::Kanr; see below for construction); X2316-3C (MATα SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1; Hopper et al., 1980); 201-1–5 (MATα los1-1 SUP4 ade2-1 can1-100 lys1-1 his5-2 trp5-48 ura3-1; Hopper et al., 1980); BJ2168 (MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 gal2; Jones, 1991). Yeast strains were maintained on YEPD medium or synthetic defined media lacking appropriate nutritional ingredients. For growth assays, cells were grown in liquid media to select for plasmids, and then 10 μl of serial dilutions of cells were applied to solid media and incubated for ∼3 days at the designated temperatures.
Escherichia coli RR1 and DH5α were used for propagation of recombinant DNA constructs. E. coli cells were maintained on YT media or YT media containing the appropriate antibiotic to select for plasmid expression.
Construction of Plasmids/Strains
TYS1 Constructs
To construct YCpTYS1-GFP, TYS1 was amplified by polymerase chain reaction (PCR) using primers az 32 (CCCGGGTAGCTATTCTTCAAC) and az 33 (CCCGGGCAATTTGGTTTC CTC) containing SmaI sites and a plasmid containing a 9.7- kilobase pair insert encoding TYS1 (YCpAzC5; Sarkar et al., 1999) as the template. The amplified DNA contained 226 bp upstream of the TYS1 open reading frame. The amplified DNA was cloned into pGEM-T (Promega, Madison, WI) to generate pGEM-T-TYS1. TYS1 was released from pGEM-T by SmaI and ligated into pRS416 at the SmaI site. To generate an in-frame fusion of TYS1 to green fluorescence protein (GFP), GFP was released from pRS415-myo-GFP (kindly provided by Dr. R. Li, Harvard Medical School, Boston, MA) by BamHI/SacII digestion. The GFP DNA was ligated into YCpTYS1 at the BamHI/SacII sites located at the 3′-end of TYS1. YCpTYS1-myc was constructed by replacing the GFP fragment in plasmid YCpTYS1-GFP with a BamHI/SacII fragment from pRS415-myo-myc (kindly provided by Dr. R. Li) encoding six repeats of a myc epitope.
To mutate the nuclear localization sequence (NLS) of TYS1 we used reverse PCR (Hemsley et al., 1998) with pGEM-T-TYS1 as the template and primers az 36 (GCTGAGGAACCAAAGAATAAGGG TACTAAG) and az 37 (CTCTTCCGACTTTTGTGGTGTGGCAAC). In the resulting plasmid, pGEM-TYS1-nls1, TYS1 lysine (K) residues K364, K365, K367, and K368 were changed to glutamic acid (E). TYS1-nls1 was released from pGEM-TYS1-nls1 by SmaI and used to replace TYS1 in YCpTYS1-myc generating YCpTYS1-nls1-myc. The same procedure also generated pGEM-TYS1-nls2 that contained a 1 base addition before amino acid 360 and a 1 base deletion at codon 365, resulting in the following changes: TYS1, TPQKSKKAKKPK (359–370); TYS1-nls1, TPQKSEEAEEPK; TYS1-nls2, TPTKVGRAEEPK. TYS1-nls2 was released from pGEM-T-TYS1-nls2 by SmaI and used to replace TYS1 in YCpTYS1-myc generating YCpTYS1-nls2-myc.
TYS1-lacZ Constructions
TYS1 regions were amplified by PCR using primers with terminal BamHI sites and cloned into pGEM-T. The TYS1 primers used were: az 38 and az 39 (codons 1–105), az 30 and az 31 (codons 106–169), az 40 and az 41 (codons 170–231), az 28 and az 29 (codons 232–264), az 42 and az 43 (codons 265–351), and az 26 and az 27 (codons 352–384). Subcloned fragments were released from pGEM-T by BamHI digestion and were inserted at the BamHI site of pFB1–7a (Moreland et al., 1987) generating in-frame fusions of parts of TYS1 with LacZ. All plasmids, except the one encoding amino acids 170–231, expressed β-galactosidase in yeast.
Construction of los1::Kanr:
The LOS1 gene in ts2, SS328, A364a, ts341, and ts19:3:4 was replaced with a los1::Kanr cassette by a one-step gene disruption method (Guldener et al., 1996). The los1::Kanr cassette was generated by PCR amplification using oligonucleotide primers SRIM09 (CTGCGCCTGAAAGCTATTGACCTTGCTTTAAAACAGAAAGTGGAT CTGATATCACCTA) and SRIM27 (CGAGGAATGCTAGAACGGATTCAG CAGCTGGTAA ATGCACAGGTCGACAACCCTTAAT) and plasmid pUG6 containing the Kanr cassette as the template (Guldener et al., 1996). The resulting strains were verified using primers SRIM30 (AGGTTACTCATTGTGGGATC) and SRIM31 (ATTCTACGCTACCGATTGGC).
Microscopy
To determine the cellular location of GFP-tagged Tys1p in living yeast cells, cells containing YCpTYS1-GFP were grown overnight in media lacking uracil and containing 10 ng/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). Cells were harvested and washed once in phosphate-buffered saline and resuspended in phosphate-buffered saline. The cells were visualized through the fluorescein isothiocyanate (FITC) channel of a Microphot-FX microscope Nikon, Melville, NY). Images were captured with a SenSys charge-coupled device camera (Photometrics, Tucson, AZ) using QED software (QED Imaging, Inc., Pittsburgh, PA). Immunofluorescence experiments were performed as previously described (Hopper et al., 1990). Yeast strain SJ17 harboring plasmid (pFB1-7a) containing sequences of TYS1 fused with lacZ were grown in selective media and fixed for 1 h with formaldehyde. β-Galactosidase antigen was detected by rabbit anti-β-gal antibody at a dilution of 1:400 followed by secondary FITC-conjugated goat anti-rabbit (Jackson Immunoresearch, West Grove, PA) at a dilution of 1:600.
Cell Fractionation
Yeast strain BJ2168 was transformed with YCpTYS1-myc, YCpTYS1-nls1-myc, or YCpTYS1-nls2-myc and used for fractionation according to a previously published procedure (Dove et al., 1998; Peng and Hopper, 2000). Yeast cells were grown in 1 l of defined media lacking uracil at 30°C to OD600 = 0.6–0.7. The cells were harvested by centrifugation and washed with ice-cold water. For each gram wet weight of cells, 4 ml of 100 mM Tris-HCl (pH 9.0), 50 mM dithiothreitol, 5 mM EDTA (pH 9.0) buffer was added and incubated for 10 min at room temperature. The cells were washed with ice-cold 1.1 M sorbitol, and cell walls were removed by addition of 1.5 ml of Glusulase (New England Nuclear, DuPont, Boston, MA) and 0.3 ml of 10 mg/ml Zymolase 20T (ICN, Costa Mesa, CA) in 25 ml of sorbitol per 1011 cells. Digestion proceeded at 30°C for 1.5–2 h. Spheroplasts collected by centrifugation at 4°C were washed once with 25 ml of ice-cold 1.1 M sorbitol and resuspended in 20 ml of 1.1 M sorbitol. The spheroplasts were centrifuged through a cushion (22% sorbitol, 5% Ficoll 400, containing a cocktail of protease inhibitors) at 4000 × g for 10 min at 4°C and resuspended in 25 ml of lysis buffer (20% Ficoll in PM buffer [3.75 mM K2HPO4, 8.25 mM KH2PO4, 0.6 mM MgCl2, pH 6.5]) containing a cocktail of protease inhibitors and immediately lysed by five quick strokes in a Dounce homogenizer. Cell walls and debris were removed by centrifugation at 2°C, and aliquots of supernatant were used as the source for total protein. The supernatant was centrifuged at 13,000 × g for 10 min at 2°C, and aliquots of supernatant were used as the source of cytosolic protein. The remainder of the supernatant was transferred to a step gradient containing 30, 40, and 50% Ficoll, each in PM buffer. The gradients were centrifuged at 58,400 × g for 2 h at 2°C. Nuclei located primarily in the 40% layer were collected by aspiration. Protein was recovered from nuclei by trichloroacetic acid (TCA) precipitation.
Proteins (20 μg) from each fraction were resolved on a 10% SDS polyacrylamide gel. The separated proteins were transferred to nylon membranes and probed with mouse anti-myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:1000 followed by horseradish peroxidase-conjugated secondary anti-mouse antibody (Amersham, Arlington Heights, IL) at a dilution of 1:5000. Proteins were detected by enhanced chemiluminescence (ECL) according to the manufacturer's instructions (Amersham). For detection of Nsp1p, mouse anti-Nsp1 (Tolerico et al., 1999) was used at a dilution of 1:20,000 followed by secondary horseradish peroxidase-conjugated anti-mouse antibody at a dilution of 1:5,000. Detection of Rna1p was with rabbit anti-Rna1p (6142, Hopper et al., 1990) at a dilution of 1:20,000 followed by anti-rabbit horseradish peroxidase-conjugated secondary antibody at a dilution of 1:5,000. Fluorograms were scanned (UMAX, UMAX Data Systems, Hsinchyu, Taiwan), and the relative amount of cross-reacting protein was determined using algorithms provided by NIH-IMAGE (http://rsb.info.nih.gov/nih-image/Default.html).
In Situ Hybridization
The method used was a modification of published procedures (Amberg et al., 1992; Kadowaki et al., 1992; Sarkar and Hopper, 1998). Each mutant was grown at 23°C and then shifted to 37°C for 2 h followed by fixation with freshly prepared 4% paraformaldehyde in 0.1 M potassium phosphate (pH 6.5), 5 mM MgCl2 buffer for 3 h at room temperature. After the cells were washed three times with phosphate buffer, they were treated with Zymolase-20T (ICN) in phosphate buffer containing 1.2 M sorbitol and 25 mM dithiothreitol for 10–15 min at 37°C. Spheroplasted cells were applied to wells of Teflon-faced slides coated with poly-l-Lysine. Cells were dehydrated by treatment for 5 min each with 70, 90, and 100% ethanol and dried. The cells were incubated with prehybridization buffer (4× SSC; (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 1× Denhardt's solution (Denhardt, 1966), 500 μg/ml single-stranded sonicated salmon sperm DNA, 125 μg/ml E. coli tRNA, 10% dextran sulfate) at 37°C for 3 h. Hybridization was performed with the same buffer containing 400–500 pg/μl digoxigenin-labeled oligonucleotides for 12–16 h at 37°C in a humidified chamber. Both prehybridization and hybridization buffers contained RNasin (Promega). Oligonucleotides were end-labeled with digoxigenin-11-dUTP (Boehringer Mannheim, Indianapolis, IN) using terminal transferase (Life Technologies, Rockville, MD). Probes specific for tRNATyr, tRNAMet, and tRNAIle were previously described (Sarkar et al., 1999). After hybridization, cells were washed three times each in 2× SSC and 1× SSC at 42°C for tRNA probes and 37°C for the poly(A) RNA probe (10 min/wash). The cells were then briefly rinsed with 4× SSC, and 0.1% Triton-X-100 and incubated with 4× SSC and 1% bovine serum albumin (BSA) for 2 h at room temperature. Cells were incubated with 1:30 dilution of FITC-labeled anti-digoxigenin antibody (Boehringer Mannheim) in 4× SSC and 1% BSA for 2 h at room temperature. Unbound antibody was removed by washing two times in 4× SSC and twice in 4× SSC and 0.1% Triton X-100 at room temperature for 10 min each. The cells were counterstained with 1 μg/ml DAPI and mounting medium was applied. Images were obtained as described above.
tRNA Aminoacylation
Tyrosyl-tRNA synthetase activity in crude cell extracts was assayed by a modification of a published procedure (Natsoulis et al., 1986). Yeast were grown in defined media at 23°C to a density of 1 × 107 cells/ml, shifted to 37°C for 2 h, and harvested and resuspended in breaking buffer (50 mM Tris-HCl, pH 7.5, 10 mm EDTA) containing a cocktail of protease inhibitors and glass beads. Cells were broken by vortexing. The extracts were clarified by centrifugation, and the supernatant was stored at −70°C in 50% glycerol and used as a source of aminoacyl-tRNA synthetase. The reaction mixtures contained 100 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 50 mM KCl, 0.5 mM EDTA, 2.5 mM ATP, 300 μg tRNA (type X from baker's yeast, Sigma), 150 μg of protein from the extracts and radioactive tyrosine. Single-label reaction mixtures contained 10 μCi of [3H]tyrosine (New England Nuclear) and were incubated at 25°C. Double-label experiments contained 1.0 μCi of a mixture of 14C amino acids (New England Nuclear) in addition to 10 μCi [3H]tyrosine and were incubated at 30°C. The reactions were terminated at 0, 5, 10, and 20 min by addition of cold 10% TCA. Labeled tRNAs were collected onto filters (HA type, Millipore, Bedford, MA) preincubated with unlabeled amino acids, washed five times with 5% TCA, and dried, and the radioactivity was determined. Incorporation of radioactivity into tRNA was determined by subtracting values from a BSA mock reaction from values obtained with extracts.
Sequence Alignments
Alignments were performed as described previously (Stanford et al., 2000). The BLAST server (Altschul et al., 1997) at National Center for Biotechnology Information was utilized to search for similar proteins. Clustal X (Thompson et al., 1997) was used to do multiple alignments, and Gene Doc (Nicholas et al., 1997) was used to shade the alignments. Shaded alignments were used as the basis for schematic block diagrams.
RESULTS
The Vast Majority of Tys1p Is Located in the Cytosol
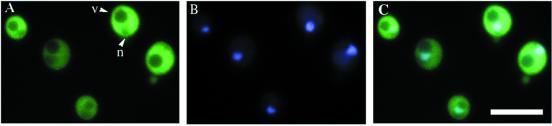
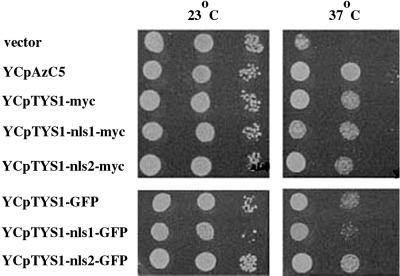
We used fluorescence microscopy to determine the cellular location of a GFP-tagged Tys1p (Figure 1). TYS1-GFP includes 226 bp upstream of TYS1, and it is tagged with GFP at the last codon. The encoded fusion protein complemented the tys1-1 ts growth defect when expressed from a centromere-containing vector (YCpTYS1-GFP). However, at the nonpermissive temperature (37°C) the cells with YCpTYS1-GFP grew a little slower than cells possessing a centromere vector containing a 9.7-kb genomic DNA encoding Tys1p (YCpAzC5; Figure 2). In live cells, the vast majority of Tys1p-GFP is cytosolic. Two organelles within yeast cells appear to possess less GFP signal than the cytosol: the vacuole (v, Figure 1A) and the nucleus (n, Figure 1A) identified by its colocalization with DNA-specific DAPI staining (Figure 1, B and C). The vacuole appears more depleted for the GFP signal than the nucleus. This could be due to the fact that the vacuole is larger and excludes surrounding cytosol or to the presence of a small pool of Tys1p in the nucleus and its absence in the vacuole.
Figure 1.
Cellular location of Tys1p-GFP. The location of Tys1p-GFP encoded by YCpTys1-GFP in ts2 cells was determined by autofluorescence. (A) Detection of GFP. v, identifies a vacuole; n, identifies a nucleus. (B) DAPI staining of the DNA in the same cells. (C) Overlap of images A and B. Bar, 10 μm.
Figure 2.
Ability of variant TYS1 constructs to complement the ts phenotype of the tys1-1 mutation. ts2 mutant cells with the indicated plasmids were grown and serially diluted, and aliquots were spotted onto complete minus Ura media. Plates were incubated at 23 or 37°C, as indicated, for 3 days.
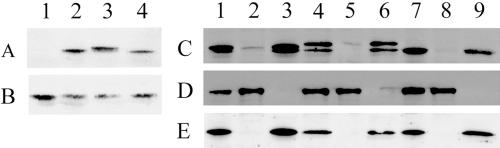
To determine whether there is a small nuclear Tys1p pool, we also used cell fractionation. YCpTYS1-GFP was modified by replacing GFP with six myc epitopes to generate YCpTYS1-myc. Tys1p-myc encoded by YCpTYS1-myc complemented the tys1-1 ts growth defect, but as for the TYS1-GFP construct, the cells containing YCpTYS1-myc grew somewhat slower at the nonpermissive temperature than cells containing YCpAzC5 (Figure 2). Total cell extracts from a yeast strain carrying YCpTYS1-myc possess a protein of the appropriate mobility that cross-reacts with monoclonal anti-myc (Figure 3A, lane 2), whereas extracts from cells possessing vector alone do not produce such a protein (Figure 3A, lane 1). The cellular extracts were fractionated into nuclear and cytosol-enriched fractions, and the location of Tys1p-myc was determined by Western analysis. Figure 3 shows the results of one of three independent cell fractionations. Monoclonal anti-Nsp1p (32D6; Tolerico et al., 1999) was used to detect Nsp1p, a nucleoporin (Hurt, 1988; Figure 3D), and polyclonal anti-Rna1p was used to detect the primarily cytosolic RanGAP (Hopper et al., 1990; Feng et al., 1999; Figure 3E). The cytosolic fraction lacked detectable nuclear proteins because little or no Nsp1p was apparent in this fraction. The nuclear fraction appeared to be largely free of cytosolic proteins because there was no detectable Rna1p in this fraction. The vast majority of Tys1p-myc cofractionated with the cytosol (Figure 3C, lanes 1–3). Nevertheless, a small Tys1p signal cofractionating with the nuclear-enriched fraction was reproducibly obtained. Judging from the enrichment and yield of Nsp1p, we estimate that ∼1.5% of the total yeast Tys1p fractionates with nuclei. Interestingly, similar percentages of aminoacyl synthetase activities copurify with rodent nuclei (Nathanson and Deutscher, 2000).
Figure 3.
Location of Tys1p by subcellular fractionation. (A) Western analysis to show the specificity of anti-myc to identify Tys1p-myc proteins. Lane 1, ts2 cells containing vector alone; lane 2, ts2 cells with YCpTYS1-myc; lane 3, ts2 cells with YCpTYS1-nls1-myc; lane 4, ts2 cells with YCpTYS1-nls2-myc. (B) Same blot as for A using antisera 6142 to identify Rna1p. (C) Location of myc-tagged proteins in cell fractions. Lanes 1–3, extracts from cells with YCpTYS1-myc; lanes 4–6, extracts from cells with YCpTYS1-nls1-myc; lanes 7–9, extracts from cells with YCpTYS1-nls2-myc. Lanes 1,4, and 7 have total cell extracts. Lanes 2, 5, and 8 have nuclear-enriched fractions. Lanes 3, 6 and 9 have cytosolic-enriched fractions. (D) The same blot as in C incubated with 32D6 monoclonal anti-Nsp1p to assess the fractionation of an authentic nuclear protein. (E) The same blot as in C incubated with anti-sera 6142 anti-Rna1p to assess the fractionation of a protein primarily located in the cytosol.
Tys1p Possesses Sequences Sufficient and Important for Nuclear Import
Both microscopic and biochemical fractionation studies indicated that the majority of the Tys1p pool is cytosolic. However, neither method ruled out a nuclear Tys1p pool and, in fact, the data could be interpreted to support the presence of a small nuclear pool of this protein. To address this issue in another manner, we attempted to learn whether Tys1p contains information for nuclear import. We recently described an approach to identify motifs specifying cell biological information. The premise is that enzyme activities broadly distributed among the eubacterial, archaeal, and eukaryotic kingdoms should have sequence conservation for catalytic and substrate recognition domains. In contrast, protein domains devoted to determining location in eukaryotic cells should not be conserved and, perhaps, may be absent from the eubacterial and archaeal homologues. We found that eukaryotic sorting isozymes have peptide domains that their archaeal and eubacterial counterparts lack. Protein regions known to specify subcellular location were included in all the eukaryotic counterparts but were absent from all the archaeal/eubacterial counterparts. We named the eukaryotic additions ADEPTs for additional domains in eukaryotes for protein targeting (Tolerico et al., 1999; Stanford et al., 2000).
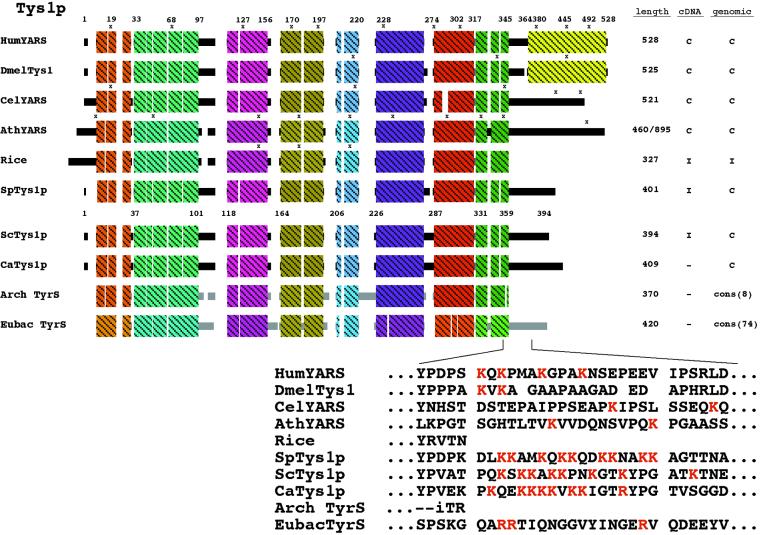
To assess whether yeast Tys1p possesses an ADEPT that can serve as an NLS, we generated a sequence alignment of tyrosyl-tRNA synthetases (Figure 4). Similar types of phylogenetic sequence comparisons led Schimmel and Wang (1999) to predict NLS motifs in numerous aminoacyl-tRNA synthetases. Generation of the tyrosyl-tRNA synthetase alignment was somewhat complicated because there appear to be two different tyrosyl-tRNA synthetase families. The eubacterial family, consisting of two branches, is more similar to eukaryotic organellar enzymes than to either archaeal or eukaryotic cytoplasmic enzymes. The other family, composed of eukaryotic cytoplasmic tyrosyl-tRNA synthetases and the archaeal counterparts, are more similar to each other than to the eubacterial enzymes. Also, several tyrosyl-tRNA synthetases are fusions between the synthetase and a cytokine, EMAPII (for review, see Schimmel and Ribas De Pouplana, 2000; C-terminal block of the human and Drosophila synthetases, Figure 4). Nevertheless, comparisons of the alignments of a consensus of the archaeal tyrosyl-tRNA synthetases to several eukaryotic proteins showed that the eukaryotic tyrosyl-tRNA synthetases generally possess a C-terminal region lacking in archaeal tyrosyl-tRNA synthetases (Figure 4). However, some plants lack the C-terminal extension and have an amino terminal addition instead, as depicted by the rice homologue (Figure 4). Inspection of the potential C-terminal ADEPT indicated that the S. cerevisiae, Schizosaccharomyces pombe, and Candida albicans fungal sequences contain regions similar to the simple basic NLS motif (Dingwall and Laskey, 1991).
Figure 4.
Sequence alignment of tyrosyl-tRNA synthetases. Schematic diagram showing alignment of selected eukaryotic cytosolic tyrosyl synthetases with consensus block diagrams of archaeal and eubacterial counterparts. The shaded boxes represent blocks of similarity and do not infer any structural information. The eubacterial shading is deliberately different to reflect dissimilarity from the eukaryotic and archaeal Tys1 proteins. The sequence of a putative ADEPT near the carboxyl terminus is shown below the alignments. Columns to the right indicate the length of the proteins in amino acids and whether cDNA and genomic DNA sequence information is complete (C) or incomplete (I). Cons (n) indicates that a consensus sequence from designated number of organisms was used to generate the block diagram. An x marks the locations of introns. Accession numbers: human (HumYARS), 2665519; Drosophila melanogaster (DmelTys1), 7294108; Caenorhabditis elegans (CelYARS), 7320764; Arabidopsis thaliana (AthYARS), 6560759; rice, expressed sequence tags: AU081543, AU077843, AU081542, AU077844, AQ365016, AQ865733; S. pombe (SpTys1p) 3183174; S. cerevisiae, (ScTys1p), 6321624; C. albicans (CaTys1p), Con4-2782.
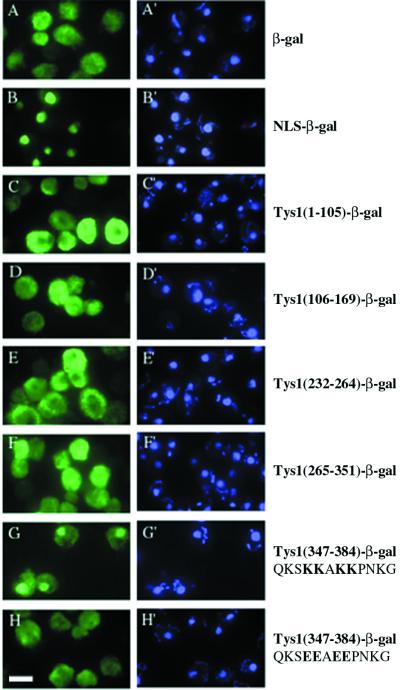
To determine whether the yeast C-terminal potential ADEPT contained functional nuclear targeting information, we amplified a TYS1 region containing this extension and fused it to a gene encoding a cytosolic β-galactosidase (Moreland et al., 1987). We also amplified other TYS1 regions and inserted them in the same site of the vector and transformed each plasmid into yeast. The cellular location of β-galactosidase encoded by each plasmid was determined by indirect immunofluorescence using anti-β-galactosidase (Figure 5). Cells harboring the parent vector generated cytosolic β-galactosidase (Figure 5A), whereas cells harboring a similar vector encoding the histone H2B NLS fused to β-galactosidase (Moreland et al., 1987) generated a nuclear signal (Figure 5B). All regions of yeast TYS1 (except amino acids 170–231, which did not yield a fusion protein that cross-reacts with anti-β-galactosidase) shared with the archaeal consensus sequence generated β-galactosidase signals located in the cytosol (Figure 5, C–F). In contrast, the potential ADEPT located at the C terminus fused to the β-galactosidase caused it to be located in the nucleus (Figure 5G). The data indicate that the C-terminal Tys1p extension is able to function as an NLS.
Figure 5.
Cellular location of Tys1-β-galactosidase fusion proteins. The location of β-galactosidase signal in yeast cells containing plasmids encoding portions of Tys1p fused in-frame to β-galactosidase was determined by indirect immunofluorescence. Left, FITC detection of anti-β-galactosidase. Right, DAPI staining of DNA. (A and A′) Yeast cells containing the parent pFB1-7a vector encoding a cytosolic β-galactosidase. (B and B′) The plasmid pFB1-67a encodes a fusion of the H2B NLS to β-galactosidase. (C and C′ to G and G′) Plasmids encode the indicated wild-type Tys1p amino acids fused in-frame to β-galactosidase. (H and H′) The plasmid encodes changes of four basic amino acids to four acidic amino acids as indicated by bold letters. Bar, 5μm.
If the Tys1p ADEPT is an NLS, then its activity should depend on consensus amino acids necessary for activities of other authentic NLS motifs. The activity of the simple basic NLS motif is dependent on the presence of the basic amino acids (Dingwall and Laskey, 1991). We mutated four of the closely spaced lysines to glutamic acids (Figure 5, G and H, bold letters) and assessed the consequences on the subcellular location of the β-galactosidase reporter protein. The amino acid changes resulted in a dramatic redistribution of the reporter from mostly nuclear to mostly cytosolic (Figure 5H). Therefore, the activity of the ADEPT is dependent on NLS consensus amino acids, bolstering the notion that it is an authentic NLS.
The studies utilizing the β-galactosidase show that the C-terminal ADEPT is sufficient for locating a reporter protein to the nucleus, but they do not prove that these sequences act as an NLS in the endogenous Tys1p. To learn whether these amino acids function in this manner, we attempted to alter the same basic amino acids to acidic amino acids in YCpTYS1-myc. Using reverse PCR (Hemsley et al., 1998) we obtained variant TYS1 genes. DNA sequence analyses of two variants showed one (YCpTYS1-nls1-myc) to have the intended four K to E changes. A second construct (YCpTYS1-nls2-myc) contained a deletion and an insertion in the vicinity of the lysine residues, resulting in a change from QKSKKAKKPK to TKVGRAEEPK.
We assessed the consequences of alterations of the putative NLS motif on the subcellular distribution of Tys1p-myc. As discussed above, a small pool of wild-type Tys1p cofractionated with nuclei (Figure 3C, lanes 1–3). The same method was used to locate Tys1p-nls1-myc and Tys1p-nls2-myc. Each mutant protein was produced in approximately equivalent quantities in yeast (compare lanes 2–4 in Figure 3A with lanes 1, 4, and 7 in Figure 3C), but the nls mutant proteins displayed slightly different electrophoretic mobilities. Tys1p-nls1-myc migrated slightly slower than the wild-type protein and usually appeared as a doublet (Figure 3, A, lanes 2 and 3, and C, lanes 1 and 4), whereas Tys1p-nls2-myc migrated somewhat faster than the wild-type counterpart (Figure 3, A, lanes 2 and 4, and C, lanes 1 and 7). The locations of mutant proteins were determined by separation of cell extracts into nuclear- and cytosol-enriched fractions followed by Western analyses using anti-myc (Figure 3C, lanes 4–9). In each case the cytosolic and nuclear marker proteins were in the anticipated cellular fractions (Figure 3, D and E, lanes 4–9). In independent subcellular fractionations the amount of Tys1p-nls1-myc cofractionating with nuclei was reduced to 40% of the amount of wild-type Tys1p-myc cofractionating with nuclei (in Figure 3C, compare lane 2 with lane 5). In contrast, the amount of Tysp-nls2-myc protein that cofractionated with nuclei was reduced to <20% of the amount of wild-type Tys1p-myc cofractionating with nuclei (in Figure 3C, compare lane 2 with lane 8). The data bolster the notion that the ADEPT contains an NLS because it is sufficient to deliver a passenger protein to the nucleus and it is also important for establishing a small nuclear pool for the endogenous protein.
Mutations of the Putative TYS1 NLS Motif Cause Defects in Nuclear Export of tRNA
To determine the consequences of the NLS mutations on the catalytic activity of Tys1p we assessed enzyme activity in vitro. Initially, we compared the levels of tyrosyl-tRNA synthetase activity from wild-type TYS1 cells to tys1-1 cells. Crude extracts were prepared from wild-type TYS1 or tys1-1 cells grown at the permissive temperature and incubated at the nonpermissive temperature for tys1-1 (37°C) for 2 h before preparing extracts. We defined the amount of [3H]Tyr incorporated into tRNA obtained from wild-type extracts as 100% and determined the relative amount of activity in the extracts from the mutant cells. By these assays, mutant tys1-1 cells had little, if any (2 ± 2%) tyrosyl synthetase activity compared with extracts from wild-type cells (Azad, Stanford, Sarkar, and Hopper, unpublished results). The same procedure showed that cells with YCpTYS1-myc, YCpTYS1-nls1-myc, and YCpTYS1-nls2-myc had approximately equivalent tyrosyl-tRNA synthetase activities (Azad, Stanford, Sarkar, and Hopper, unpublished results). However, tyrosyl-tRNA synthetase activity using these crude extracts was variable, and attempts to purify synthetase activity from the extracts were unsuccessful.
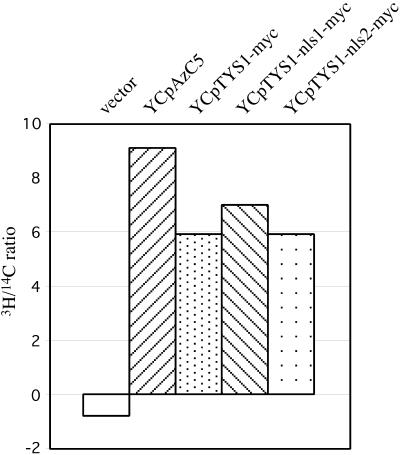
To reduce variability encountered by comparing enzyme activities from independent crude extracts to each other, we provided each extract with an internal control and then determined the relative amount of tyrosyl-tRNA synthetase to total aminoacyl-tRNA synthetases in each cellular extract. Reactions contained a mixture of 14C-amino acids in addition to [3H]tyrosine and the ratio of 3H/14C incorporation into tRNA for each time point was determined (Figure 6). As expected, tys1-1 cells with vector alone showed poor incorporation of 3H into tRNA compared with 14C incorporation; in fact, [3H]tyrosine incorporation into tRNA was lower than for the BSA negative control. Enzyme activity levels in the various yeast strains correlated well with their growth rate (Figure 2). For example, the 3H/14C ratio of extracts from cells with YCpAzC5 was greater than cells with myc-tagged Tys1p (Figure 6). The ratios of 3H/14C amino acid incorporation into tRNA from cells with Tys1p-myc, Tys1p-nls1-myc, and Tys1p-nls2-myc were equivalent to each other. Thus, alteration of the C-terminal ADEPT does not significantly affect Tys1p catalytic activity.
Figure 6.
Tyrosyl-tRNA synthetase activity in cells with various TYS1 alleles. Extracts were prepared from ts2 mutant cells containing plasmids with various TYS1 alleles and were assayed for the incorporation of [3H]tyrosine and 14C-amino acids into tRNA. The 3H/14C ratio of an average of two independent assays at the 10-min time point is shown.
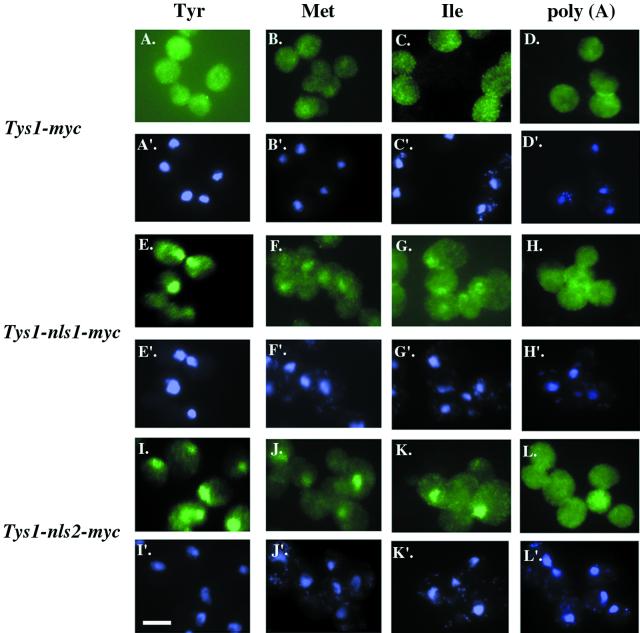
If the detected nuclear Tys1p pool functions in tRNA export from the nucleus to the cytosol, then alteration of the distribution of Tys1p that reduces its nuclear pool should affect tRNA nuclear export. To test this, we assessed tRNA nuclear export using in situ hybridization (Figure 7). We previously showed that cells with the tys1-1 mutation had substantial nuclear accumulation of tRNATyr, tRNAMet, and tRNAIle but not poly(A) RNA when incubated at the nonpermissive temperature for 2 h before preparing the cells for in situ analysis (Sarkar et al., 1999). Mutant tys1-1 cells harboring YCpTYS1-myc have no detectable defects in nuclear export (Figure 7, A–D). Even though tys1-1 cells harboring YCpTYS1-nls1-myc or YCpTYS1-nls2-myc have equivalent levels of tyrosyl-tRNA synthetase activity as do cells with Tys1p-myc, they had substantial nuclear accumulation of tRNATyr, tRNAMet, and tRNAIle but not poly(A) RNA when incubated at the nonpermissive temperature (Figure 7, E–H and I–L). Thus, the mutations that reduce the nuclear pool of Tys1p without substantially reducing catalytic activity cause tRNA to accumulate in the nucleus. The data support the model proposed by Lund and Dahlberg (1998) that nuclear tRNA aminoacylation plays a role in the tRNA nuclear export process.
Figure 7.
In situ hybridization for cells containing Tys1p nls mutations. ts2 cells containing YCpTYS1-myc (A–D and A′–D′), YCpTYS1-nls1-myc (E–H and E′–H′), or YCpTYS1-nls2-myc (I–L and I′–L′) were studied. Cells in A, E, and I were probed with an oligonucleotide specific for tRNATyr; B, F, and J were probed with an oligonucleotide specific to tRNAMet; C, G, and K were probed with an oligonucleotide specific for tRNAIle; and D, H, and L were probed with oligo(d)T, specific for poly(A) RNA. A′ to L′ are the same cells stained with DAPI to located nuclei. Bar, 5 μm.
To determine the consequences of redistributing Tys1p such that there is reduced or no detectable nuclear pool upon the growth of yeast, we monitored the ability of the TYS1-nls mutations to complement the ts growth defect of tys1-1 mutant cells. Mutant tys1-1 cells harboring vector alone were unable to grow at 37°C. When the cells were provided with YCpTYS1-myc, YCpTYS1-nls1-myc, or YCpTYS1-nls2-myc they were able to grow at 37°C, and there were little apparent differences in growth rate among the various transformed yeast (Figure 2). Thus, Tys1p redistribution appears to have little consequence on cell growth, indicating that the nuclear Tys1p pool may be unessential.
Lack of Genetic Interactions between Los1p and Aminoacyl Synthetases
Our results indicate that nuclear tRNA aminoacylation may be unessential. Interestingly, Los1p, the putative tRNA nuclear exportin is also unessential. An interpretation of these genetic results is that Los1p and tRNA nuclear aminoacylation each define redundant tRNA export pathways so that elimination of one can be compensated by the other (Sarkar et al., 1999; Grosshans et al., 2000b). If Los1p and tRNA nuclear aminoacylation provide the two major alternative paths for tRNA nuclear export, then cells containing mutations of both should be severely defective in tRNA nuclear export and consequently unable to grow.
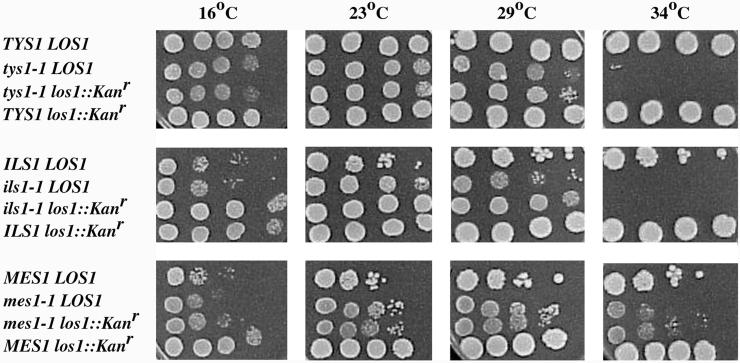
We determined the physiological consequences of mutations in both putative pathways. Haploid cells containing ts mutations of methionyl-tRNA or isoleucyl-tRNA synthetases, mes1-1 or ils1-1, respectively, were mated with haploid cells containing los1-1. For controls, we also mated the parents of these strains to each other and generated all combinations of diploids with single mutations. We performed tetrad analyses to test the viability and growth of the double mutants. We found approximately the same viabilities of progeny from each of the genetic crosses. Moreover, progeny with mes1-1 los1-1 or ils1-1 los1-1 genotypes did not have more severe growth defects at intermediate temperatures than cells with mes1-1 or ils1-1 alone. Therefore, by the genetic crosses we found no evidence for genetic interactions between genes encoding aminoacyl-tRNA synthetases and Los1p. To confirm these observations and to extend them to Tys1p, we disrupted LOS1 in mes1-1, ils1-1, and tys1-1 mutants. The los1::Kanr mes1-1, los1::Kanr ils1-1, and los1::Kanr tys1-1 double mutants were viable and had no more severe growth defects than any of the single synthetase mutations (Figure 8). Thus, no synthetic interactions between Los1p and aminoacyl-tRNA synthetases could be detected, arguing against Los1p and tRNA nuclear aminoacylation acting in parallel to provide the two major alternative paths for tRNA nuclear export.
Figure 8.
Growth of cells containing los1 and aminoacyl-tRNA synthetase mutations. Cells were grown and serially diluted, and aliquots were spotted onto solid rich media and were incubated at 16, 23, 29, or 34°C, as indicated for 3 days. Strains utilized: TYS1 LOS1, SS328; tys1-1 LOS1, ts2; tys1-1 los1:: Kanr, SSS708; TYS1 los1:: Kanr, SSS706, derived from SS328; ILS1 LOS1, A364a; ils1-1 LOS1, ts341, derived from A364a; ils1-1 los1:: Kanr, SSS705, derived from ts341; ILS1 los1:: Kanr, SSS707, derived from A364a; MES1 LOS1, A364a; mes1-1 LOS1, ts19:3:4, related to A364a; mes1-1 los1:: Kanr, SSS703, derived from ts19:3:4; MES1 los1:: Kanr, SSS707.
DISCUSSION
Evidence for Nuclear Tys1p Pools in Budding Yeast
Previous studies indicating that tRNAs can be aminoacylated before their export to the cytosol (Lund and Dahlberg, 1998; Sarkar et al., 1999; Grosshans et al., 2000a) led to the prediction that there should be functional pools of the aminoacyl-tRNA synthetases in the nucleus. Here we provide several lines of data showing that this prediction has been met. First, by cellular fractionation studies, a small portion (∼1.5%) of the S. cerevisiae Tys1p pool cofractionates with nuclei under conditions in which a cytosolic protein does not. Second, yeast Tys1p possesses an ADEPT (Stanford et al., 2000), resembling the classic basic NLS motif (Dingwall and Laskey, 1991), that efficiently delivers a reporter protein to the nucleoplasm and delivery is dependent on consensus basic amino acids. Other eukaryotic Tys1 proteins also possess carboxyl-terminal extensions that could function in delivery to the nucleus. The putative fungal ADEPTs resemble classical NLS motifs, but the analogous region in the metazoan Tys1p counterparts do not. If it is assumed that there is a Tys1p nuclear pool in higher eukaryotes, either the NLSs reside in the same carboxyl-terminal region and define new motifs or the information for nucleus location resides elsewhere. Some plants lack the C-terminal addition but contain instead extra sequences located at the amino terminus; however, these additions also do not resemble known NLS motifs. Third, mutations of the basic amino acids of the putative NLS to acidic amino acids in endogenous Tys1p substantially decrease the amount of altered Tys1p that cofractionates with nuclei. In our previous studies to assess the aminoacylation status of nucleus-located tRNAs we were unable to detect any uncharged tRNAs in the nucleus (Sarkar et al., 1999), leading to the prediction that all 20 aminoacyl-tRNA synthetases may, like Tys1p, be located in the nucleus as well as in the cytosol. In fact, a very recent report demonstrated that 13 aminoacyl-tRNA synthetases cofractionate with nuclei of rodent cells (Nathanson and Deutscher, 2000).
Cytosolic pools of aminoacyl-tRNA synthetases are required for protein synthesis. Therefore, the subcellular distribution of aminoacyl-tRNA synthetases must be regulated such that the majority of the enzyme resides in the cytosol, available for protein synthesis. The putative NLS we mapped in the C-terminal region of Tys1p delivers nearly all β-galactosidase to the nucleus, even though only a small fraction of endogenous Tys1p appears to be nuclear. The data indicate that there is information in Tys1p that counteracts the NLS and functions in maintaining a cytosolic pool of this enzyme. Three ways in which this can happen are: 1) a Tys1p nuclear export sequence (NES) returns nucleoplasmic Tysp1 to the cytosol; 2) a Tys1p region tethers the major fraction of Tys1p in the cytosol; 3) the Tys1p NLS has limited availability to the nuclear import machinery. We transferred the Tys1p-GFP construct to yeast strains with mutations of the various exportins to determine whether a known exportin functions in Tys1p nucleus/cytosol shuttling, but none of the exportins appeared to affect Tys1p subcellular distribution (Azad, Stanford, Sarkar, and Hopper, unpublished results). Provided that Tys1p is not served by more than one exportin, an interpretation of the data is that it is more likely that Tys1p cytosolic location is due to cytosolic retention or to NLS masking than to Tys1p shuttling. Protein modification often functions in NLS masking and cytosolic retention (for review, see Jans and Hüber, 1996), and there is some indication that there may be multiple Tys1p forms (Figure 3). If confirmed, it will be interesting to learn whether protein modification affects Tys1p subcellular distribution. It also will be very interesting to identify the cis-acting determinants that assure predominant cytosolic pools.
Because cytosolic pools of aminoacyl-tRNA synthetases are required for continuous protein synthesis, one cannot use mutations or drugs that affect catalytic activity of these enzymes to distinguish between roles of the aminoacyl-tRNA synthetases in the nucleus from roles in the cytosol. Here, we generated new TYS1 alleles that alter information for Tys1p nuclear distribution without significantly altering tyrosyl-tRNA synthetase catalytic activity. That these new alleles cause defects in tRNA nuclear export supports the hypothesis that nucleus-located Tys1p functions in tRNA nuclear export. Although we have not completely ruled out the caveat that small decreases in catalytic activity of the nls mutant synthetases, undetectable by the in vitro charging assays, cause the defects in tRNA nuclear export, we do not favor this interpretation because cells with Tys1p-nls-myc have equivalent growth characteristics and tyrosyl-tRNA synthetase activities as do cells with Tys1p-myc that do not accumulate nuclear tRNA.
Previously, we showed that the tys1-1 mutation that affects Tys1p enzymatic activity caused defects in noncognate as well as cognate tRNATyr nuclear export (Sarkar et al., 1999). It is interesting that the new TYS1 alleles that affect Tys1p nuclear distribution, rather than catalytic activity, also affect export of noncognate tRNAs. Because these new alleles encode sufficient activity to support protein synthesis, the data support previous suggestions that defects in tRNA nuclear export that occur upon inhibition of tRNA charging are unlikely due to inhibition of protein synthesis (Lund and Dahlberg, 1998; Sarkar et al., 1999; Grosshans et al., 2000a).
tRNA Nuclear Export Pathways
Even though tys1-nls-myc mutations cause nuclear tRNA accumulation, cells with these alleles have no detectable growth disadvantage compared with cells with wild-type TYS1-myc. The data indicate that tRNA charging in the nucleus is not absolutely required for nuclear export. The data are consistent with the demonstration in Xenopus oocytes that a mutant tRNA, unable to be aminoacylated but able to interact with exportin-t, can be exported to the cytosol, albeit less well than normal tRNAs (Arts et al., 1998b). Likewise, LOS1, encoding the yeast tRNA nuclear exportin, is not an essential gene (Hurt et al., 1987). An interpretation of the data is that Los1p and tRNA aminoacylation do not function in all of the tRNA nuclear export pathways.
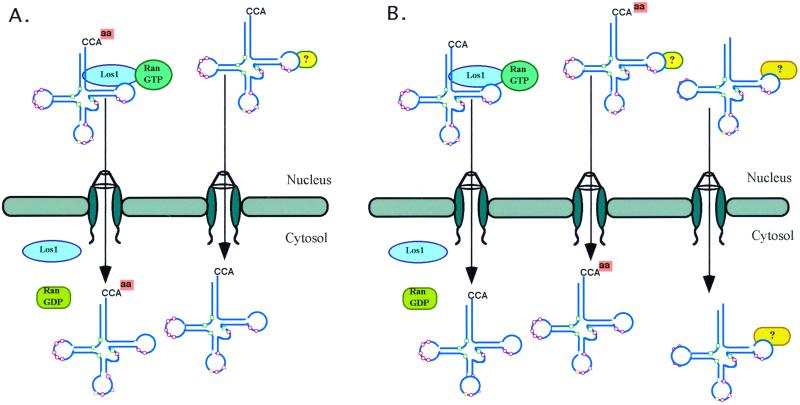
We interpret the lack of genetic interactions of mutations in any of three genes, mes1-1, ils1-1, and tys1-1, encoding ts aminoacyl synthetases and los1, to mean that the synthetases and Los1p function in the same path (Figure 9A). However, there are alternative explanations. First, because MES1, ILS1, and TYS1 are essential genes, it is possible that the mutant alleles have sufficient enzyme activity at intermediate temperatures such that it is not possible to assess synthetic lethality with los1. We do not favor this interpretation because in our studies we used ts mutations of three different genes encoding defective aminoacyl-tRNA synthetases. Second, it is possible that tRNA aminoacylation and Los1p function in two of many parallel paths so that alterations of just two have no serious physiological consequences (Figure 9B). The gene products that function in the alternative paths can be identified by continued searches for ts mutants defective in tRNA nuclear export and by searches for mutations that cause lethality when cells contain both los1 and aminoacyl-tRNA synthetase NLS mutations. We intend to conduct both types of studies.
Figure 9.
Two possible models for the different tRNA nuclear export pathways. (A) tRNA aminoacylation in the nucleus and Los1p act in series in one tRNA nuclear export pathway, and alternative pathway(s) remain unidentified. (B) tRNA aminoacylation in the nucleus and Los1p act in parallel in two tRNA nuclear export pathways, and there are other important export pathways yet to be identified.
If aminoacylation functions in the Los1p tRNA nuclear export pathway, it will be important to learn whether Los1p discriminates between charged versus uncharged tRNAs. Exportin-t/Los1p can bind to uncharged tRNAs, but preferentially associates with tRNAs that have mature 5′- and 3′-termini (Arts et al., 1998b; Lipowsky et al., 1999). However, it appears that there has been no comparison of the relative affinities of exprotin-t/Los1p to aminoacylated versus nonaminoacylated tRNAs. If exportin-t/Los1p has higher affinity for aminoacylated tRNAs, it would provide a simple explanation for how aminoacylation and Los1p might function in series to proofread and export mature functional tRNAs to the cytosol.
ACKNOWLEDGMENTS
We thank G. Peng, W. Feng, and D. Eisaman for valuable scientific insights and comments on the manuscript. This work was supported by a Public Health Service grant from the National Institutes of Health to A.K.H.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998a;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998b;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- Denhardt DT. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966;23:641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences: a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dove JE, Brockenbrough JS, Aris JP. Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 1998;53:33–46. doi: 10.1016/s0091-679x(08)60873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Benko AL, Lee J-H, Stanford DR, Hopper AK. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Hurt EC, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000a;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Simos G, Hurt EC. Review: transport of tRNA out of the nucleus-direct channeling to the ribosome? Struct Biol. 2000b;129:288–294. doi: 10.1006/jsbi.2000.4226. [DOI] [PubMed] [Google Scholar]
- Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, McLaughlin CS. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc Natl Acad Sci USA. 1968;62:468–474. doi: 10.1073/pnas.62.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt EC, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1998;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt DJ, Wang SS, Lin YH, Hopper AK. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4323–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans DA, Hüber S. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- Jones EW. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Zhao Y, Tartakoff AM. A conditional yeast mutant deficient in mRNA transport from the nucleus to cytoplasm. Proc Natl Acad Sci USA. 1992;89:2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schäfer S, Gross HJ, Beier H, Görlich D. Coordination of tRNA nuclear export with processing of tRNA. RNA. 1999;5:539–549. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- McLaughlin CS, Hartwell LH. A mutant of yeast with a defective methionyl-tRNA synthetase. Genetics. 1969;61:557–566. doi: 10.1093/genetics/61.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland RB, Langevin GL, Singer RH, Garcea RL, Hereford LM. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987;7:4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson L, Deutscher MP. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J Biol Chem. 2000;275:31559–31562. doi: 10.1074/jbc.C000385200. [DOI] [PubMed] [Google Scholar]
- Natsoulis G, Hilger F, Fink GR. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW. GeneDoc: Analysis and Visualization of Genetic. Variation EMBNEW NEWS. 1997;4:14. [Google Scholar]
- Peng G, Hopper JE. Evidence for Gal3p's cytoplasmic location and Gal80p's dual cytoplasmic-nuclear location implicates new mechanisms for controlling Gal4p activity in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5140–5148. doi: 10.1128/mcb.20.14.5140-5148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenko VI, Ivanova JL, Cherny NE, Filonenko VV, Beresten SF, Wolfson AD, Kisselev LL. Compartmentalization of certain components of the protein synthesis apparatus in mammalian cells. Eur J Cell Biol. 1994;65:60–69. [PubMed] [Google Scholar]
- Sarkar S, Azad AK, Hopper AK. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P, Ribas De Pouplana L. Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem Sci. 2000;25:207–209. doi: 10.1016/s0968-0004(00)01553-x. [DOI] [PubMed] [Google Scholar]
- Schimmel P, Wang CC. Getting tRNA synthetases into the nucleus. Trends Biochem Sci. 1999;24:127–128. doi: 10.1016/s0968-0004(99)01369-9. [DOI] [PubMed] [Google Scholar]
- Stanford DR, Martin NC, Hopper AK. ADEPTs: information necessary for subcellular distribution of eukaryotic sorting isozymes resides in domains missing from eubacterial and archaeal counterparts. Nucleic Acids Res. 2000;28:383–392. doi: 10.1093/nar/28.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Plewniak TJ, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151:57–75. doi: 10.1093/genetics/151.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]