Abstract
BRCA1 and BRCA2 are two major breast and ovarian cancer susceptibility genes. BRCA1 was the first discovered and has been a focus of research for these cancers. BRCA1 mediates tumor suppression in part through pleiotropic interactions with a network of DNA repair proteins on chromatin. BRCA1 mutations cause homologous recombination-mediated DNA repair deficiency, genomic instability, and DNA-damaging agent hypersensitivity. Although BRCA1 and BRCA2 have some shared functions in cancer predisposition and therapy response, there are also key differences indicating divergent roles for each protein. This review summarizes and highlights recent insights into the molecular events responsible for BRCA1 tumor suppression, emphasizing the DNA repair function of BRCA1 as a nexus between its roles in cancer development and therapy.
The breast and ovarian cancer susceptibility genes BRCA1 and BRCA2
The genetic basis underlying common forms of hereditary breast and ovarian cancer was revealed in the early 1990s with the identification of the breast cancer early onset genes, which encode the breast cancer type 1 susceptibility protein (BRCA1) and the breast cancer type 2 susceptibility protein (BRCA2) [1–3]. Although germline BRCA1 and BRCA2 mutations account for only a small percentage of the total breast cancer incidence in the United States and approximately 20% of hereditary breast cancer, intensive basic and clinical efforts have produced copious data regarding the molecular basis of how these tumor suppressors function and how BRCA-mutated cancers can be specifically and efficaciously treated. BRCA1 and BRCA2 are bona fide players in the DNA damage response through enabling the accurate repair of DNA double strand breaks (DSBs) by homologous recombination (HR), which is their most closely associated cellular function to tumor suppression [4,5]. Cells deficient in either gene accumulate characteristic cytogenetic abnormalities indicative of deficiency in HR-mediated DNA repair. Moreover, DNA-damaging agents that necessitate HR for repair have shown promise for treating BRCA1- and BRCA2-mutant cancers [6–8]. Finally, basic investigations into BRCA1 and BRCA2 have led to the identification of other breast and ovarian cancer genes that are required for BRCA DNA repair function. Consequently, observations over the past 18 years following their discovery have solidified the concept of a BRCA-centered tumor suppressor network that is essential for genome integrity and responses to chemotherapy [9]. This review will describe findings related to the roles of BRCA1 in genome integrity, with a particular emphasis on the recent discoveries that have shed light on BRCA1 contributions to DNA repair and tumor suppression.
Tumor phenotypes in BRCA1 and BRCA2 mutation carriers
BRCA1 and BRCA2 are the most frequently mutated genes in hereditary breast and ovarian cancer [10]. Affected individuals inherit one mutated allele and do not display apparent phenotypic abnormalities beyond increased lifetime risk for cancer, which is estimated to be as high as 85% for breast cancer[11]. BRCA1 mutation carriers also have a 30–50% risk of developing ovarian cancer, whereas BRCA2 mutation carrier ovarian cancer risk is roughly half that. Loss of heterozygosity (LOH) at the wildtype BRCA allele and retention of the mutated copy is generally observed in tumors. This feature of a heterozygous patient and a nullizygous tumor presents an opportunity to exploit differences in DNA repair capacity in BRCA mutation-associated cancers, a topic that will be discussed in depth in subsequent passages of this review.
Approximately 80% of BRCA1-mutated breast cancers resemble sporadic basal-like triple-negative breast cancers, which are defined in part as exhibiting a basal histology and while being negative for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2/Neu), [12]. In addition to phenotypic similarities, BRCA1 breast cancers and basal-like breast cancers also demonstrate similar gene expression profiles [12]. Both sporadic and BRCA1 mutant triple-negative breast cancers have high frequencies of p53 and phosphatase and tensin homolog (PTEN) mutations [13]. By contrast, BRCA2-mutated cancers are commonly ER-positive, displaying a luminal phenotype [14].
An unresolved question is why BRCA1 and BRCA2 mutations confer a predilection for breast and ovarian cancer. Arguments have been made that breast epithelial cells are particularly sensitive to DNA damage during developmental windows of estrogen-driven proliferation [15,16]. However, it should not be concluded that BRCA1 and BRCA2 mutations only result in increased breast and ovarian cancer. BRCA2 heterozygotes have increased risk for pancreatic, prostate, and male breast cancers, whereas the predisposition of BRCA1 mutation to these malignancies is less strong [17]. Interestingly, several BRCA1-associated proteins are implicated in Fanconi Anemia (FA), a disease that manifests in bone marrow failure, developmental defects, heightened sensitivity to DNA crosslinking agents, and increased cancer incidence [18]. Notably, biallelic BRCA2 mutations manifest in a severe form of FA [19]. The relationship between the proteins and FA demonstrate the non-exclusivity of the BRCA network for breast and ovarian cancer, and the role of BRCA1 and BRCA2 in interstrand crosslink (ICL) repair.
Structural features of the BRCA1 tumor suppressor protein
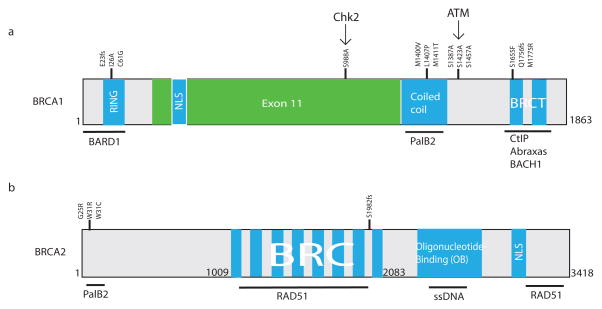
The 1863 amino acids that comprise the BRCA1 polypeptide can be divided into several important domains. It is informative to consider each of these regions in the context of the cancer causing mutations found within them and how these impact interactions with BRCA1-associated proteins (Figure 1). BRCA1 has biochemical interactions with at least 13 different proteins that have been implicated in DNA repair and cancer susceptibility, suggesting that BRCA1 exists within an essential signaling network dedicated to genome integrity [20–24].
Figure 1. The BRCA1 and BRCA2 proteins.
Breast cancer type 1 susceptibility protein (BRCA1) and type 2 susceptibility protein (BRCA2) are structurally highly dissimilar. The protein homology domains are shown in blue, commonly mutated residues are indicated above the protein, and a line underneath the protein indicates protein-protein interaction regions. Chk2 and ATM kinases are shown above the protein with arrows to indicate their substrate residues. “Fs” indicates a frameshift mutation. (a) The first 150 N-terminal amino acids of BRCA1 contain the RING domain that associates with BRCA1-associated RING domain protein 1 (BARD1) and has E3 ubiquitin ligase activity. Adjacent to the RING lies a nuclear localization sequence (NLS). The central region of BRCA1 comprises exon 11, which contains phosphorylation substrate residues for ataxia telangiectasia mutated (ATM) (S1387A, S1423A, and S1524A) and checkpoint kinase 2 (Chk2) (S988A) [79]. The coiled-coil domain associates with partner and localizer of BRCA2 (PalB2). The BRCA1 C-terminal BRCT repeats interact with C-terminal binding protein interacting protein (CtIP), coiled-coil domain containing protein 98 (CCDC98/Abraxas) and BTB and CNC homolog 1 (BACH1). I26A is a synthetic mutation that preserves heterodimerization with BARD1 but loses E3 ligase function [80]. All other residues indicated represent common clinical mutations [11,28,42–44]. (b) The N terminus of BRCA2 mediates PalB2 binding. Its central BRC repeats and C-terminal sequences interact with DNA repair protein Rad51 (Rad51). The oligonucleotide-binding domain associates with single-stranded DNA (ssDNA) [79].
Within the first 150 amino acids of BRCA1 lies an evolutionary conserved RING domain, the most common motif present in E3 ubiquitin ligases. BRCA1 heterodimerizes with its stoichiometric binding partner BARD1, which also contains an N-terminal RING domain [25]. Heterodimerization stabilizes the BRCA1 protein in vivo, and enables the BRCA1 RING domain to interact with E2 ubiquitin ligases and exert maximal E3 ligase activity in vitro [26]. The interaction with BARD1 is important for BRCA1 function in vivo as evidenced by the indistinguishable phenotypes observed for either BRCA1 or BARD1 knockout mice [27]. BARD1 mutations have been found in hereditary breast and ovarian cancers [28] independent of BRCA1 and BRCA2 [29], although the actual relative risk conferred by BARD1 mutation for these cancers is unclear. Interestingly, the BRCA1-BARD1 heterodimer produces non-canonical lysine6-linked ubiquitin (K6-Ub) chains in vitro, in association with the Ubch5c E2 enzyme [30,31]. K6-Ub chains are present at DSBs, their presence has been reported to be BRCA1 dependent [32]. The identification of specific substrates other than autoubiquitinated BRCA1-BARD1 has remained elusive. However, candidates include histone H2A and the BRCA1 interacting partner CtIP [33,34].
Surprisingly, E3 ubiquitin ligase-deficient BRCA1 RING-mutant knock-in mice display different genomic instability and tumor-forming phenotypes, depending on the position of the mutation. Genetically engineered mice that harbor the BRCA1 I26A substitution, a rationally designed synthetic BRCA1 E3 ligase substitution that has not been observed in human patients and disrupts BRCA1 interaction with E2 ubiquitin ligases, were viable and showed no cancer susceptibility or loss of DNA repair function [35,36]. The I26A mice instead displayed slightly decreased body weight and male infertility, a phenotype that has been seen in other BRCA1-deficient models [37]. Conversely, mice expressing a known pathogenic RING substitution, C61G, displayed embryonic lethality, DNA repair deficiency, and cancer predisposition [38]. Prior studies suggested that C61G would impair interaction with BARD1 by leading to structural perturbations in the BRCA1 RING domain [25], whereas BRCA1 I26A maintains interaction with BARD1. Therefore, it is unclear if complete deficiency of BRCA1 E3 ligase activity in the C61G knock-in mouse, or rather the disordered RING domain and diminished interaction with BARD1, is responsible for the more severe phenotypes.
The middle 60% of the BRCA1 protein is derived from a single exon (exon 11) that is present in vertebrates but not lower eukaryote species [39]. Exon 11 encodes a largely unstructured region of the BRCA1 protein that is phosphorylated by the ATM and Chk2 kinases in a DNA damage-inducible manner [40]. Deletion of exon 11 in mice results in genomic instability and impaired HR, but still provides a BRCA1 protein that localizes to DSB sites [41]. Consistent with being partially active, exon 11 mutant mice exhibit perinatal lethality, whereas complete BRCA1-null mice die in utero between embryonic day 5.5–8.5. Moreover, a p53 heterozygote background rescues embryonic lethality in BRCA1 exon 11 mutant mice, but not in a complete BRCA1 null background. Between the exon 11-encoded sequence and the BRCT domain lies a more recently described coiled-coil domain (CCD) that mediates an important interaction with the PalB2 tumor suppressor protein [42,43]. PalB2 was originally identified as a direct BRCA2-interacting partner that is required to target BRCA2 to chromatin and DSBs [44]. PalB2 uses different interaction surfaces to maintain direct interaction with BRCA2 and BRCA1, thereby biochemically connecting the two most important breast and ovarian cancer suppressor proteins. PalB2 is required for the HR function of BRCA2 and appears to facilitate the HR function of BRCA1 as well, given that clinical BRCA1 CCD mutants have HR defects [42–44]. PalB2 mutations have been found in familial breast, ovarian, prostate, and pancreatic cancers [21,22,28,45], and biallelic germline PalB2 mutations result in a severe form of FA that mimics biallelic BRCA2 mutations [45,46]
The BRCA1 C terminus harbors the tandem BRCA1 carboxy terminal (BRCT) repeats. These motifs recognize phospho-serine residues on the direct binding partners Abraxas, BACH1, and CtIP [47,48] and are required for DSB localization. The BRCT repeats recognize a S(p)XF motif present in each of these direct binding partners. Numerous clinical missense mutations disrupt BRCA1 interaction with these proteins and localization to ionizing radiation induced foci (IRIF), which occur at the genomic locations of DSBs. BRCA1 BRCT-mutant knockin mice have been genetically engineered using alleles known to be pathogenic in humans. BRCA1-S1598F BRCT mutant mice (analogous to the S1655F substitution in human patients) are HR deficient, sensitive to interstrand crosslinking agents, and tumor prone to a similar extent as BRCA1-null mice [36]. Interestingly, BRCA1 BRCT-mutant mice are viable through adulthood, demonstrating that partial BRCA1 function is present in BRCT missense mutant alleles, but that this hypomorphic activity does not necessarily rescue the cancer susceptibility phenotype of BRCA1-null mice.
The various domains of BRCA1 allow it to integrate signals from upstream DNA damage sensing proteins such as ATM and Chk2, and to recruit phosphoproteins BACH1, CtIP, PalB2, as well as heterodimerize with BARD1 to mediate E3 ligase activity. By contrast, BRCA2 does not appear to have the same damage-sensing and recruitment capabilities, suggesting that BRCA2 is an effector whereas BRCA1 is more of a coordinator that mediates various responses to DSBs.
Multifactorial recognition of DNA damage by BRCA1 protein complexes
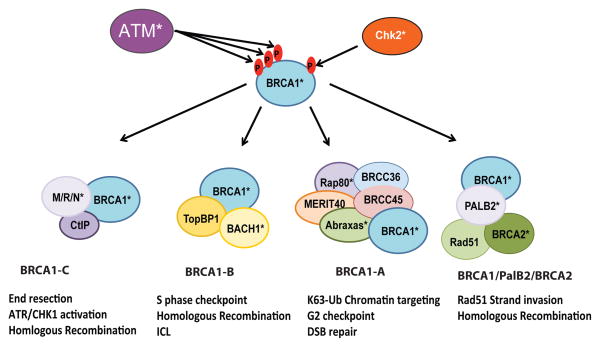
BRCA1 forms multiple distinct protein complexes that all localize at IRIF but perform different aspects of DSB repair (Figure 2) [9,49,50]. Although not appreciated at the time of discovery, BRCA1 IRIF were the earliest indication that chromatin responses are central to BRCA1 DSB localization. BRCA1 localization to IRIF hinges upon ATM, ATR, and DNA-PKcs-mediated phosphorylation of Serine 139 on the histone H2A variant H2AX. Phosphorylated H2AX is designated γH2AX because the phosphorylation occurs at the γ amino acid residue from the H2AX carboxy terminus. γH2AX is crucial for the sustained recruitment of BRCA1 and various protein complexes to the lesion site and for ensuring optimal repair. One of the key proteins recruited by γH2AX is mediator of DNA-damage checkpoint 1 (MDC1), which gets phosphorylated in a series of TQXF repeats [51]. Phospho-MDC1 provides a scaffold for the phospho-binding ubiquitin E3 ligase ring finger protein 8 (RNF8), which ubiquitylates histones H2A, H2B, and H2AX [52–54]. It is thought that this initial ubiquitylation of the chromatin flanking DSBs is able to recruit RNF168, a second E3 ubiquitin ligase, through the RNF168 ubiquitin-binding domains, allowing amplification of the ubiquitin signal along chromatin in cis to DSBs [55,56]. BRCA1 accesses ubiquitin chains at DSBs through its interaction with a 5-member ubiquitin recognition complex that is coordinated by RAP80, a protein that utilizes tandem ubiquitin interaction motifs to specifically recognize lysine63-linked ubiquitin (K63-Ub) chains [57–59]. Interestingly, the RAP80 complex also contains BRCA/BRCA2-containing complex subunit 36 (BRCC36), a deubiquitylating enzyme that specifically hydrolyzes K63-Ub chains, thus limiting the accumulation of DSB-associated K63-Ub [60,61]. Although BRCA1 forms DSB foci through chromatin-associated K63-Ub chains, this does not account for all, or even probably most, of the function of BRCA1 in DSB repair [57–59]. BRCA1 is a central component of at least 3 different protein complexes that function independently of the RAP80-containing BRCA1-A complex (Figure 2) to interact with PalB2 and BRCA2 (BRCA1/PalB2/BRCA2 supercomplex), BACH1 (BRCA1-B), and CtIP (BRCA1-C).
Figure 2. BRCA1 protein complexes.
During the DNA damage response ataxia telangiectasia mutated (ATM) and checkpoint kinase 2 (Chk2) become activated and phosphorylate many DDR mediators, including BRCA1. Upon ATM and Chk2 phosphorylation, BRCA1 becomes activated and associates with at least four different protein complexes at sites of damage [50], BRCA1-A, BRCA1-B, BRCA1-C, and BRCA1/PalB2/BRCA2 supercomplex. The known functions of these complexes are indicated, and proteins that have clinical mutations in breast and ovarian cancer are marked with an asterisk (*)[20–24,28].
New insights and therapeutic implications of the DNA repair function of BRCA1
A crucial determinant of whether HR or non-homologous end joining (NHEJ) DNA repair is utilized to repair a DSB is the process of end resection. This is executed in a 5′ to 3′ dependent fashion by several different nucleases, which include CtIP in association with Mre11, Rad50, and Nbs1 (the MRN complex); exodeoxyribonuclease 1 (Exo1); and DNA replication ATP-dependent helicase (Dna2) [62,63]. The single strand binding protein replication protein A (RPA) then loads onto the 3′ overhanging single-strand DNA (ssDNA) for further structural processing, eventually to be replaced by Rad51, which conducts a homology search and facilitates strand invasion into the template sister chromatid (Figure 3). BRCA2- and PalB2-mediated displacement of RPA with Rad51 has been shown in vitro [64,65], and mutation of either gene leads to severely impaired Rad51 foci formation and HR in cells. Although BRCA1 directly interacts with PalB2 and utilizes this interaction to maintain association with BRCA2 and Rad51, the BRCA1 contribution to Rad51 strand invasion has yet to be directly tested in vitro. Circumstantial evidence suggests that BRCA1 assists BRCA2 in Rad51 strand invasion reactions; namely, clinical missense mutations in the BRCA1 coiled-coil domain (CCD) that disrupt interaction with PalB2 can reduce HR efficiency. Moreover, BRCA1 knockdown reduces PalB2, BRCA2, and Rad51 foci formation [42,43], suggesting that BRCA1 provides a DSB recognition element for this complex.
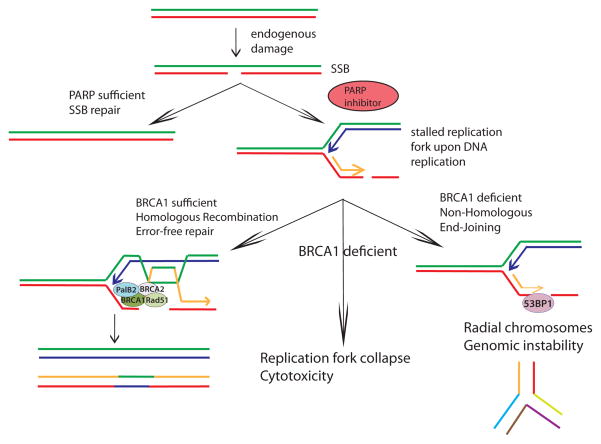
Figure 3. Synthetic lethality of PARP inhibition in BRCA1-mutated cancers.
Poly(ADP-ribose) polymerase (PARP) inhibition causes persistent single strand DNA breaks that must be repaired by DNA replication-dependent homologous recombination (HR). During DNA replication, these single strand breaks turn into stalled replication forks that must be resolved by double strand break repair mechanisms. In cells expressing at least one wildtype BRCA1 allele, BRCA1 localizes to sites of damage, promoting end resection, enabling these forks to be resolved by BRCA2-Rad51-mediated HR with high fidelity and the cell to continue replication (left side of the figure). In BRCA1-deficient cells (such as in a BRCA1-mutated tumor), 53BP1 occupies sites of damage, preventing adequate end resection and promoting inappropriate NHEJ and radial chromosome formation (right side of the figure)[61,62].
Cells that accumulate stalled or collapsed replication forks are particularly vulnerable to deficiency in either BRCA1 or BRCA2 because this form of DNA damage results in DSBs and necessitates HR for accurate repair. A failure of HR-mediated repair leads to inappropriate utilization of NHEJ DNA repair mechanisms and the formation of radial chromosome structures and commensurate cellular toxicity. Poly(ADP-ribose) polymerase (PARP) is an NAD-dependent enzyme that is required for single strand break (SSB) repair. PARP inhibition (PARPi), results in unresolved SSBs that become one-sided DSBs when replication forks encounter them in S phase (Figure 3). HR effectively deals with replication induced breaks, resulting from PARPi, in a BRCA1- and BRCA2-dependent manner. The requirement of PARP-deficient cells for HR genes has been exploited as a synthetic lethal strategy to preferentially kill BRCA1 and BRCA2-null cancer cells, while producing minimal toxicity in BRCA heterozygous cells [6,7]. Indeed BRCA1−/− and BRCA2−/− cells display more than two orders of magnitude increased sensitivity to PARPi compared to BRCA1+/− and BRCA2+/− cells. There are currently numerous PARP inhibitors in various stages of clinical trials, with AZD2281 (olaparib) being the best characterized. Olaparib had shown early efficacious and selective response in BRCA1- and BRCA2-mutated cancer patients refractory to other therapies [8] However, the clinical BRCA1 C61G RING mutation caused mammary cancers in mice that became resistant to therapy more rapidly than BRCA1 null tumors [38], suggesting that the specific location of a BRCA1 mutation will influence response to PARPi. Furthermore, studies revealed that patients often developed resistance due to reversal mutations that rescued HR [66,67]. In clinical trials the PARPi olaparib did not show efficacy in broader populations of triple-negative breast cancer patients (most of whom did not have BRCA mutations) [68,69].
The sensitivity of many BRCA1-mutant cells to PARPi seemingly places BRCA1 as a requisite member of the BRCA2-Rad51 complex. In an apparent contradiction to this model, much of the HR deficiency in BRCA1-null backgrounds, including restoration of Rad51 post-irradiation foci formation by immunofluorescence (IRIF), can be rescued by concomitant deficiency of p53-binding protein 1 (53BP1) [70,71]. 53BP1 is a chromatin-associated protein that recognizes dimethylated histone H4 lysine 20 residues (H4K20Me2) and is required for NHEJ that is utilized in class switch recombination at immunoglobulin loci and in end-to-end fusions of deprotected telomeres [72–74]. Deficiency in 53BP1 results in excessive end resection and increased HR utilization. The prevailing thought is that increased single stranded DNA in BRCA1−/− 53BP1−/− cells enables BRCA2-Rad51 dependent strand invasion and HR even in the absence of BRCA1. In support of this model, 53BP1 nullizygosity rescues the embryonic lethality, HR activity, and PARPi sensitivity of BRCA1 knockout mice [70,71,75]. Conversely, 53BP1 nullizygosity does not rescue these phenotypes in the BRCA2 knockouts, highlighting the differences in BRCA1 and BRCA2 requirement for Rad51-dependent strand invasion. In simplistic terms, one might conclude that the BRCA1 DNA repair function primarily serves to prevent 53BP1 from blocking end resection, rather than playing an essential role in the delivery of Rad51 to DSBs as part of the BRCA1-PalB2-BRCA2 complex. This is likely not the entire story, however, because BRCA1−/− 53BP1−/− mice are still sensitive to interstrand crosslinking (ICL) agents (another class of cancer therapeutics) such as cisplatin and mitomycin C (MMC), and males display infertility due to defective spermatogenesis [76]. The sensitivity to DNA interstrand crosslinking agents like cisplatin is partially alleviated by deletion of NHEJ proteins and has been suggested to be the result of reduced FA protein localization to DNA crosslinks in the absence of BRCA1 [76]. These data suggest that the mechanism for cisplatin sensitivity is distinct from the DSB repair defect involved in PARPi treatment. In addition, BRCA1 has been implicated in ICL repair after UV damage, although it is not known what impact combined 53BP1 deficiency would have on this process [77]. Collectively, these findings suggest that BRCA1 impacts tumor suppression and response to therapy beyond limiting excessive 53BP1 at breaks. Moreover, resistance to PARPi might not necessarily produce cross-resistance to cisplatin, suggesting crosslinking agents may be a valuable option to treat PARPi-resistant cancers.
New concepts in BRCA1 tumor suppression: BRCA1 E3 ubiquitin ligase-dependent silencing of heterochromatin
The BRCA1-BARD1 heterodimer can mono-ubiquitylate histone H2A in vitro [26]. This chromatin modification has an established role in transcriptional repression of genes that are developmentally regulated, within heterochromatin, or located in cis to DSBs. An exciting recent report indicates that BRCA1 E3 ligase activity contributes to genome integrity by ubiquitylation of H2A at pericentromeric heterochromatin, thus repressing expression of repetitive α-satellite DNA [78]. BRCA1-mutant cells demonstrated elevated α-satellite RNA levels and reduced H2A-Ub at pericentromeric heterochromatin, whereas expression of an ectopic H2A-Ub fusion protein rescued silencing of satellite repeats. The authors proposed that this function of BRCA1 in transcriptional silencing of heterochromatin is a unifying mechanism to explain the pleiotropic roles of BRCA1 in maintaining genomic stability. Indeed, overexpression of α-satellite RNA led to increased γH2AX foci formation, reduced HR, centrosome duplication, and checkpoint deficiency, which are all characteristics of BRCA1 deficiency. Moreover, ectopic expression of an H2A-ub fusion protein restored the DNA repair capacity in BRCA1 mutant cell lines [78]. Satellite RNA transcription has been reported to associate with a variety of cancers, but it is not clear whether satellite derepression plays a causative role in tumorigenesis or is simply a correlative observation. Regardless, these findings present an opportunity to explore new avenues related to BRCA1 function in genome integrity. For example, it will be interesting to determine if ectopic expression of the H2A-Ub fusion protein can restore DNA damage responses and viability in the C61G knockin mice. Another outstanding question is whether 53BP1 deficiency restores transcriptional silencing of satellite DNA in BRCA1 null cells, given that HR and genome integrity are largely rescued in BRCA1−/− 53BP1−/− cells.
Concluding remarks
In the 18 years since BRCA1 was discovered, considerable insight into its roles in genome integrity and cancer have led to the discovery of new cancer susceptibility genes, surgical interventions to reduce cancer incidence in mutation carriers, and selective therapies to treat BRCA mutant tumors. Many of these findings stem from a biochemical understanding of BRCA1 interactions with other DNA repair proteins, as well as a detailed knowledge of its functions in DNA repair. Given the central importance of the BRCA1 network in all proliferating cells, insights into the underlying mechanisms of BRCA1 function on chromatin might extend beyond hereditary cancers and lead to an improved understanding of genome and epigenome maintenance as well as therapies that target HR deficiency in a broader range of malignancies.
Acknowledgments
R.A.G. acknowledges support by 1R01CA138835–01 from the NCI, a Research Scholar Grant from the American Cancer Society, a pilot grant from the Fox Chase Cancer Center/University of Pennsylvania Ovarian SPORE, and funds from the Abramson Family Cancer Research Institute. M.L.L. is supported by NRSA PHS grant 5T32GM7170–37 and the Medical Scientist Training Program at the Pereleman School of Medicine, University of Pennsylvania.
Key terms
- Homologous recombination (HR)
a DNA double strand break (DSB) repair mechanism that utilizes strand invasion into a homologous template (usually a sister chromatid) followed by replication of the template. It has higher fidelity than non-homologous end joining. It is preferred during S phase
- Non-homologous end joining (NHEJ)
a DSB repair mechanism that joins the ends of broken double-strand DNA (dsDNA) segments without using homology search. It has lower fidelity and can lead to the formation of radial chromosomes (non-linear chromosomes that contain multiple segments). It is utilized throughout the cell cycle
- Synthetic lethality
a situation in which the deficiency of two factors together causes cell death, but when only one is deficient the cell is viable. In the setting of using Poly(ADP-ribose) polymerase inhibitors (PARPi) in BRCA1 or BRCA2 null tumors, the use of PARPi in BRCA sufficient cells or mock treatment in BRCA deficient cells do not lead to lethality, but using PARPi in BRCA deficient cells does
- PARP inhibitors
Poly(ADP-ribose) polymerase inhibitors. PARP is a protein that mediates single strand break (SSB) repair. When PARP is inhibited, the SSBs lead to stalled replication forks that require BRCA-mediated HR for repair. Without BRCA function the stalled replication forks become persistent DSBs that utilize NHEJ instead, ultimately leading to genomic instability and cell death
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 3.Futreal PA, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, et al. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 5.Moynahan ME, et al. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 6.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 7.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 8.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117:305–317. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 11.King M-C, et al. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 12.Foulkes WD, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 13.Saal LH, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavaddat N, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21:134–147. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elledge SJ, Amon A. The BRCA1 suppressor hypothesis: an explanation for the tissue-specific tumor development in BRCA1 patients. Cancer Cell. 2002;1:129–132. doi: 10.1016/s1535-6108(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 17.Stratton MR. Recent advances in understanding of genetic susceptibility to breast cancer. Hum Mol Genet. 1996;5:1515–1519. doi: 10.1093/hmg/5.supplement_1.1515. [DOI] [PubMed] [Google Scholar]
- 18.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 19.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 20.Solyom S, et al. Breast cancer-associated Abraxas mutation disrupts nuclear localization and DNA damage response functions. Sci Transl Med. 2012;4:122ra23. doi: 10.1126/scitranslmed.3003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 22.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 24.Nikkilä J, et al. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009;28:1843–1852. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu LC, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 26.Xia Y, et al. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy EE, et al. Loss of Bard1, the heterodimeric partner of the Brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol Cell Biol. 2003;23:5056–5063. doi: 10.1128/MCB.23.14.5056-5063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh T, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratajska M, et al. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res Treat. 2012;131:89–97. doi: 10.1007/s10549-011-1403-8. [DOI] [PubMed] [Google Scholar]
- 30.Wu-Baer F, et al. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem. 2003;278:34743–34746. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa H, et al. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:3916–3924. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- 32.Morris JR, Solomon E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13:807–817. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, et al. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes & Development. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thakar A, et al. BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. J Biomol Struct Dyn. 2010;27:399–406. doi: 10.1080/07391102.2010.10507326. [DOI] [PubMed] [Google Scholar]
- 35.Reid LJ, et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc Natl Acad Sci U S A. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shakya R, et al. BRCA1 Tumor Suppression Depends on BRCT Phosphoprotein Binding, But Not Its E3 Ligase Activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 2003;130:2001–2012. doi: 10.1242/dev.00410. [DOI] [PubMed] [Google Scholar]
- 38.Drost R, et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Boulton SJ, et al. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol. 2004;14:33–39. doi: 10.1016/j.cub.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 40.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 41.Huber LJ, et al. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol Cell Biol. 2001;21:4005–4015. doi: 10.1128/MCB.21.12.4005-4015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sy SM-H, et al. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 47.Manke IA, et al. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, et al. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 49.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & Development. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 54.Huen MS-Y, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 56.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 57.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H, et al. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 59.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao G, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes & Development. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao G, et al. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mazón G, et al. SnapShot: Homologous recombination in DNA double-strand break repair. Cell. 2010;142:646, 646.e1. doi: 10.1016/j.cell.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 64.Yang H, et al. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 65.Jensen RB, et al. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhillon KK, et al. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102:663–669. doi: 10.1111/j.1349-7006.2010.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Norquist B, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008–3015. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ledford H. Drug candidates derailed in case of mistaken identity. Nature. 2012 Mar 29;483:519. doi: 10.1038/483519a. [DOI] [PubMed] [Google Scholar]
- 69.Powell K. Molecular oncology: The positive in the negative. Nature. 2012;485:S52–3. doi: 10.1038/485S52a. [DOI] [PubMed] [Google Scholar]
- 70.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimitrova N, et al. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bunting SF, et al. BRCA1 Functions Independently of Homologous Recombination in DNA Interstrand Crosslink Repair. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pathania S, et al. BRCA1 Is Required for Postreplication Repair after UV-Induced DNA Damage. Mol Cell. 2011;44:235–251. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roy R, et al. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horwitz AA, et al. A mechanism for transcriptional repression dependent on the BRCA1 E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2007;104:6614–6619. doi: 10.1073/pnas.0610481104. [DOI] [PMC free article] [PubMed] [Google Scholar]