Abstract
Background
Basal cell carcinoma (BCC) is the most commonly diagnosed cancer in the United States. In this research, we examine four different feature categories used for diagnostic decisions, including patient personal profile (patient age, gender, etc.), general exam (lesion size and location), common dermoscopic (blue-gray ovoids, leaf-structure dirt trails, etc.), and specific dermoscopic lesion (white/pink areas, semitranslucency, etc.). Specific dermoscopic features are more restricted versions of the common dermoscopic features.
Methods
Combinations of the four feature categories are analyzed over a data set of 700 lesions, with 350 BCCs and 350 benign lesions, for lesion discrimination using neural network-based techniques, including Evolving Artificial Neural Networks and Evolving Artificial Neural Network Ensembles.
Results
Experiment results based on ten-fold cross validation for training and testing the different neural network-based techniques yielded an area under the receiver operating characteristic curve as high as 0.981 when all features were combined. The common dermoscopic lesion features generally yielded higher discrimination results than other individual feature categories.
Conclusions
Experimental results show that combining clinical and image information provides enhanced lesion discrimination capability over either information source separately. This research highlights the potential of data fusion as a model for the diagnostic process.
Keywords: Image Processing, Basal Cell Carcinoma, Skin Lesion, Dermoscopy, Neural Network, Computational Intelligence, Data Fusion
I. INTRODUCTION
Basal cell carcinoma (BCC), characterized as a slow-growing skin malignancy originating within the basal layer of the epidermis, is the most commonly diagnosed cancer with an estimated 3 million new cases annually in the US [1]. In the consultation process when a patient exhibits a skin lesion, physicians gather patient information, determine general information about the skin lesion, and may use devices such as a dermatoscope (3Gen LLC, San Juan Capistrano, CA; Heine Optotechnik, Herrsching, Germany) for determining a preliminary diagnosis. Currently, dermatopathology examination of a biopsy is used as the diagnostic gold standard.
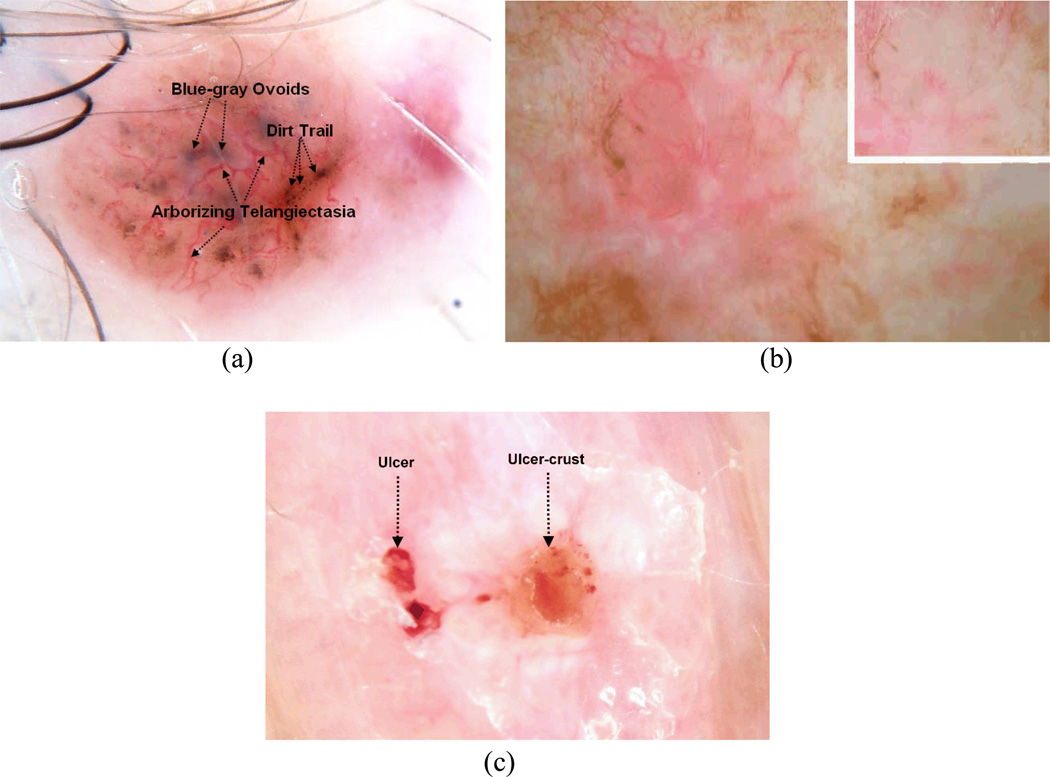
In this research, the diagnostic process, here characterized as BCC versus benign lesion discrimination is examined based on information gathered from the patient, physician, and the dermoscopic image of the lesion. There are several BCC structures identifiable using dermoscopy that strongly suggest a BCC diagnosis. These have been incorporated into the BASAL acronym: Blue-gray ovoids and globules, Arborizing telangiectasia, Semitranslucency / Spoke wheel structures, Atraumatic ulcerations, and Leaf-like structures/dirt-trails [2]. Figure 1(a–c) presents dermoscopic lesion image examples of these structures.
Figure 1.
Examples of BCC BASAL structures visible using dermoscopy, all images are contact dermoscopy images except b. a) Blue-gray ovoids, Arborizing telangiectasia, and dirt trails (rudimentary Leaf-like structures, b) Semitranslucency, non-contact dermoscopy (inset—contact dermoscopy), c) Atraumatic ulceration, with similar ulcer-crust.
While there have been numerous studies based on dermoscopic image feature analysis for pigmented lesion discrimination, few studies have specifically addressed BCC versus benign lesion discrimination by a classifier [3–6]. In those studies, BCC lesion discrimination was focused on the detection and analysis of particular dermoscopic features, including telangiectasia [3,4], leaf-dirt trails [5], and semitranslucency [6]. This research explores the efficacy of fusing clinical and dermoscopic features to enhance skin lesion discrimination capability. Four categories of features were investigated in this research for BCC discrimination: 1) patient personal profile descriptors, 2) general exam descriptors, 3) common dermoscopic skin lesion image features associated with BCC, and 4) specific dermoscopic skin lesion image features used for detecting uncommon BCC presentations. Combinations of these feature categories are examined for skin lesion discrimination using neural network techniques, including standard backpropagation neural networks, Evolving Artificial Neural Networks (EANNs), and Evolving Artificial Neural Network Ensembles (EANNEs).
The remainder of the paper is presented in the following sections: II) Description of Feature Categories, III) Discrimination Algorithms, IV) Experimental Procedure, V) Results and Discussion, and VI) Conclusion.
II. Description of Feature Categories
Table 1 provides an overview of the four overall feature groups. The personal profile descriptors used are patient age, gender, race/ethnicity, and geographic location. Age was given in a binary format for either older than or younger than (and including) 50 years of age. Race/ethnicity is quantized as either non-Hispanic or Hispanic. The geographic location is based on clinic location, either above (Missouri, Connecticut) or below (Florida) the Tropic of Cancer. The final general exam attribute was lesion location. The third category of features are basic and common basal cell carcinoma dermoscopic descriptors BASAL features including blue-gray ovoids, vessels, pink veil (a more general form of semitranslucency), atraumatic ulcerations, and leaf-dirt trails. The fourth category of features comprises more specific dermoscopic features found to be present in this set of 350 BCCs. For both the common and specific dermoscopic features, a dermatologist (W.V.S.) identified the presence or absence of each feature within the lesion. Two of these specific features have been previously described and have high specificity: arborizing telangiectasia and semitranslucency [2,7]. We have determined in this research other features with high specificity found in BCC that have not been previously described. These features are developed here to detect unusual BCC presentations such as white areas, pink areas, purple blotches, pale areas, large (majority of lesion) ulcer/crust plus pink regions, and majority white and pink regions. The common and specific dermoscopic features in each feature group are shown in Table 1.
Table 1.
Overview of feature groups examined for lesion discrimination.
| Patient Personal Profile Descriptors |
General Exam Descriptors |
Common BCC Features |
Specific BCC Features |
|---|---|---|---|
| Age | Size | Blue-gray ovoids | Arborizing telangiectasia |
| Gender | Lesion Location | Pink veil | White/pink areas |
| Race/Ethnicity | |||
| Non-Hispanic | Vessels | Semitranslucency | |
| Hispanic | Leaf-Dirt Trails | Purple Blotches | |
| Atraumatic ulcerations | Pale Areas | ||
| Geographic Location | Majority of Lesion Ulcer/Crust plus Pink Regions | ||
| Above Tropic of Cancer | |||
| Below Tropic of Cancer | Majority Pink/White Regions |
The experimental data examined to evaluate the discrimination capability of the different feature categories includes images acquired from four clinic locations across the United States: The Dermatology Center (Rolla, MO), Drugge & Sheard (Stamford, CT), Skin & Cancer Associates (Plantation, FL) and Columbia Dermatology & Mohs Skin Cancer Surgery. (Columbia, MO). 350 lesions with a BCC diagnosis and 350 benign lesions were used as the test set. Benign lesions consisting of 80 acquired nevi, 71 seborrheic keratoses, 60 actinic keratoses, 51 lentigines, 15 congenital nevi, 10 lichen planus-like keratoses, 9 sebaceous hyperplasia, and 54 lesions of miscellaneous types constituted the competitive set.
Table 2 presents the different feature group combinations examined for BCC versus benign lesion discrimination based on the four feature categories. Column 1 gives the feature combination, Column 2 provides the feature categories included in the feature combination, Column 3 lists the total number of features for the feature combination, and Column 4 gives the multilayer perceptrons (MLP) neural network architecture used for lesion discrimination for the different feature combinations.
Table 2.
Feature combinations for lesion discrimination.
| Feature Combination |
Feature Groups Included | Total Number of Features |
Neural Network Discrimination Algorithm Architecture |
|---|---|---|---|
| 1 | Personal profile descriptors | 4 | 5×5×1 |
| 2 | General exam descriptors | 2 | 3×5×1 |
| 3 | Common lesion features* | 5 | 6×5×1 |
| 4 | Specific lesion features* | 6 | 7×5×1 |
| 5 | Personal profile descriptors, general exam descriptors | 6 | 7×5×1 |
| 6 | Personal profile descriptors, common lesion features | 9 | 10×5×1 |
| 7 | Personal profile descriptors, specific lesion features | 10 | 11×5×1 |
| 8 | General exam descriptors, common lesion features | 7 | 8×5×1 |
| 9 | General exam descriptors, specific lesion features | 8 | 9×5×1 |
| 10 | Common dermoscopic lesion features, specific lesion features | 11 | 12×5×1 |
| 11 | Personal profile descriptors, general exam descriptors, common lesion features | 11 | 12×5×1 |
| 12 | Personal profile descriptors, general exam descriptors, specific lesion features | 12 | 13×5×1 |
| 13 | Personal profile descriptors, common lesion features, specific lesion features | 15 | 16×5×1 |
| 14 | General exam descriptors, common lesion features, specific lesion features | 13 | 14×5×1 |
| 15 | Personal profile descriptors, general exam descriptors, common lesion features, specific lesion features | 17 | 18×5×1 |
Common and specific lesion features determined by dermoscopy
III. Discrimination Algorithms
Using the different combinations of feature categories given in Table 2, Evolving Artificial Neural Networks (EANN’s) and Evolving Artificial Neural Network Ensembles (EANNE’s) are examined for BCC versus benign lesion discrimination. The implementation for each algorithm is given in this section.
A. Evolving Artificial Neural Networks (EANN’s)
EANN’s refer to a class of artificial neural networks (ANN’s) in which both evolution and learning are fundamental forms of adaption. Evolutionary algorithms (EAs) can be used to train the connection weight, design the architecture, and select the input features of the ANN’s. In this research, EAs using particle swarm optimization (PSO) [8] and genetic algorithm (GA) [9] for neural network training are investigated.
PSO is the study of swarms of social organisms such as a flock of birds, in which each particle in the swarm moves toward its previous best location (Pbest) and global best location (Gbest) at each time step [8]. The PSO algorithm utilized in this study is presented in detail in [10] and is overviewed as follows. We initialize M particles, where each particle is a D-dimensional vector () with each element of the vector representing a connection weight and D being the total number of weights. For example, feature combination 1 from Table 2 has an architecture of 5×5×1. The total number of weights (D) is 11 (5+5+1) and each element in this D-dimensional vector is the connection weight. Furthermore, the M particles represent M ANNs.
The connection weights in each ANN are updated when the elements in each particle are trained as follows. The initial value for each element of the vector is randomly set at a value from −0.1 to 0.1. In each training time step, the element’s value of each particle is updated toward Pbest and Gbest. Pbest is the particle of the M particles that gives the least root mean square error (RMSE) between the current training epoch and the previous training epoch. Gbest is the particle among the M particles which generates the minimum RMSE, where the RMSE is calculated based on the difference between the ground truth and the actual ANN’s output. The details for the updating process are given in [8]. The same process is repeated for N epochs. The final Gbest particle is selected for the final ANN weights for the test vector.
Genetic algorithms use the “survival of the fittest” concept where the weak may die and elites are able to progress to the next level [9]. The GA approach investigated here is presented in detail in [10]. We initialize M parents, where each parent is a D-dimensional vector with each element of the vector representing a connection weight, with D being the total number of weights. In a similar fashion defined for the PSO configuration, M parents represent M ANNs. The initial value for each element of the vector is randomly set from −0.1 to 0.1. In each training step, these M parents generate M offspring after implementing random selection, uniform crossover and a mutation operation. Then, the next M parents are selected based on whether the parents or their offspring minimize the RMSE. The same process is repeated for N epochs. From the final parent pool, the parent which minimizes the RMSE is selected for the final ANN weights for the test vectors.
B. Evolving Artificial Neural Network Ensembles (EANNEs)
Neural network ensembles provide an approach for using and combining the outputs from several networks, with each ANN having the same inputs and generating its own outputs [11]. The purpose of EANNs is to make use of the whole population of ANNs rather than a single one. Training the network ensembles and determining the final output from the network ensembles are two main components for EANNEs design.
Negative correlation learning [12,13] is implemented to train neural network ensembles in order to minimize the mutual informal among the networks. We initialize an ensemble with M ANNs with the initial weights in each ANN randomly set to a value from −0.1 to 0.1. In each training time step, each ANN in the ensemble is trained for a certain number of epochs using negative correlation learning firstly. Then, M offspring ANNs will be created by using selection, crossover, and a mutation operation in GA and replacement of the worst ANNs. The same process is repeated for N times.
For the final output determination, there are several criteria such as averaging, winner-taking-all and voting for combining the outputs. Here we simply choose averaging to deliver the final output as shown in Equation 1.
| (6) |
where M is the number of the individual ANNs in the ensemble and Fm is the output for each ANN.
IV. Experimental Procedure
Fifteen different feature combinations were investigated as inputs to the neural network architectures. The feature combinations and the neural network architectures for the EANN approach trained using the GA algorithm, the EANN approach trained using the PSO algorithm, the EANNE approach, and standard backpropagation ANN are given in Table 2. Each neural network architecture includes the total number of features for the feature combination and one bias as the inputs to each classifier algorithm. Each neural network architecture from Table 2 includes five nodes in a hidden layer and one node in the output layer. The input and output layers use linear transfer functions, and the hidden layer utilizes sigmoid transfer functions. A ten-fold cross validation methodology is used for generating training/test sets for each neural network’s architecture [13]. The same training/test sets from the cross-validation process are applied to all feature combinations and classification algorithms presented. The ten-fold cross validation process is repeated five times for each feature combination. Classification results are based on averaging the area under Receiver Operating Characteristic (ROC) curves [14] generated for each of the ten test sets over the five separate ten-fold cross validation sets. The area under the ROC curve was utilized as the classification measure in this research because the area under the ROC curve does not require selecting a decision boundary or threshold to determine detection accuracy.
V. Results and Discussion
Table 3 presents the average area under the ROC curve results over the five sets of ten-fold cross validation test sets for the different feature combinations and discrimination algorithms examined. BP denotes the standard backpropagation ANN algorithm. EANN-GA and EANN-PSO represent the EANN neural network approaches using the genetic algorithm and particle swarm optimization methods for neural network training, respectively.
Table 3.
Average area under the ROC curve discrimination results for different feature and discrimination algorithm combinations.
| Feature Combination | Discrimination Algorithm | |||
|---|---|---|---|---|
| BP | EANNE | EANN-GA | EANN-PSO | |
| 1 | 0.612 | 0.613 | 0.604 | 0.607 |
| 2 | 0.670 | 0.704 | 0.739 | 0.746 |
| 3 | 0.667 | 0.823 | 0.812 | 0.818 |
| 4 | 0.841 | 0.774 | 0.799 | 0.712 |
| 5 | 0.632 | 0.794 | 0.815 | 0.837 |
| 6 | 0.830 | 0.924 | 0.909 | 0.928 |
| 7 | 0.809 | 0.890 | 0.879 | 0.894 |
| 8 | 0.822 | 0.927 | 0.928 | 0.941 |
| 9 | 0.830 | 0.909 | 0.910 | 0.941 |
| 10 | 0.710 | 0.853 | 0.888 | 0.937 |
| 11 | 0.828 | 0.934 | 0.918 | 0.950 |
| 12 | 0.842 | 0.913 | 0.901 | 0.942 |
| 13 | 0.890 | 0.970 | 0.946 | 0.974 |
| 14 | 0.904 | 0.973 | 0.954 | 0.977 |
| 15 | 0.897 | 0.972 | 0.948 | 0.981 |
Several conclusions can be obtained from Table 3.. First, the EANN algorithm using PSO neural network training typically yielded the highest overall area under the ROC curve results from the four discrimination algorithms investigated. Second, using all features (Feature combination 15) gave the highest overall discrimination results, with an area under the ROC curve of 0.981 for the EANN-PSO approach. Feature combinations 13, 14, and 15 yielded comparable results for the EANNE, EANN-GA, and EANN-PSO classification methods, with Feature combination 14 (general exam descriptors, common dermoscopic lesion features, and specific dermoscopic lesion features) providing slightly higher results than using all features (Feature combination 15). This is not a surprising result since the personal profile descriptors (Feature combination 1) yielded the overall lowest BCC discrimination results compared to the other individual feature groups. Third, inspecting the individual feature categories, the common dermoscopic lesion features yielded higher discrimination results than the other feature categories, except for the standard back propagation method. The personal profile descriptors gave consistently lower discrimination results than the other feature categories. Size and location information (general exam descriptors) and specific dermoscopic lesion features gave more discerning discrimination information than personal profile descriptors for all classification algorithms examined.
VI. Conclusion
In this research, BCC versus benign lesion discrimination was explored using four types of clinical and dermoscopic lesion image features: patient personal profile, general exam, common dermoscopic image features associated with BCC, and specific dermoscopic image features with uncommon BCC presentations. Lesion discrimination was performed for different combinations of these features using Evolving Artificial Neural Networks and Evolving Artificial Neural Network Ensembles architectures. Experiment results showed an area under the receiver operating characteristic curve as high as 0.981 when all features were combined. Common dermoscopic lesions features generally gave higher discrimination results than other individual feature categories. Overall, experimental results highlight that combining clinical and image information enhances lesion discrimination capability over either information source separately, demonstrating the potential of data fusion in aiding the lesion diagnostic process.
ACKNOWLEDGEMENTS
This publication was made possible by Grant Number SBIR R43 CA153927-01 of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
REFERENCES
- 1.Rogers HW, Weinstock MA, Harris AR, Hinckley MR, Feldman SR, Fleischer AB, Coldiron BM. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch. Dermatol. 2010 Mar.Vol. 146:283–287. doi: 10.1001/archdermatol.2010.19. [DOI] [PubMed] [Google Scholar]
- 2.Stoecker WV, Stolz W. Dermoscopy and the diagnostic challenge of amelanotic and hypomelanotic melanoma. Arch Dermatol. 2008 Sep.Vol. 144:1120–1127. doi: 10.1001/archderm.144.9.1207. [DOI] [PubMed] [Google Scholar]
- 3.Cheng B, Erdos D, Stanley RJ, Stoecker WV, Calcara D, Gomez D. Automatic detection of basal cell carcinoma using telangiectasia analysis in dermoscopy skin lesion images. Skin Research and Technology. 2011;Vol. 17(No. 3):278–287. doi: 10.1111/j.1600-0846.2010.00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng B, Stanley RJ, Stoecker WV, Hinton K. Automatic telangiectasia analysis in dermoscopy images using adaptive critic design. Skin Research and Technology. 2011 Dec; doi: 10.1111/j.1600-0846.2011.00584.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 5.Cheng B, Stanley RJ, Stoecker WV, Osterwise CTP, Stricklin SM, Hinton KA, Moss RH, Oliviero M, Rabinovitz HS. Automatic dirt trail analysis in dermoscopy images. Skin Research and Technology. 2011 doi: 10.1111/j.1600-0846.2011.00602.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoecker WV, Gupta K, Shrestha B, Wronkiewiecz M, Chowdhury R, Stanley RJ, Xu J, Moss RH, Celebi ME, Rabinovitz HS, Oliviero M, Malters JM, Kolm I. Detection of basal cell carcinoma using color and histogram measures of semitranslucent areas. Skin Research and Technology. 2009;Vol. 15(No. 3):283–287. doi: 10.1111/j.1600-0846.2009.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzies SW, Altamura D, Menzies SW, Argenziano G, Zalaudek I, Soyer HP, Sera F, Avramidis M, DeAmbrosis K, Fargnoli MC, Peris K. Dermatoscopy of basal cell carcinoma: morphologic variability of global and local features and accuracy of diagnosis. J Am Acad Dermatol. 2010 Jan;62(1):67–75. doi: 10.1016/j.jaad.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy J, Eberhart R. Particle swarm optimization. Proceedings of the IEEE International Conference on Neural Networks; Piscataway, NJ. 1995. pp. 1942–1948. [Google Scholar]
- 9.Holland JH. Adaptation in Natural and Artificial Systems. Ann Arbor: University of Michigan Press; 1975. [Google Scholar]
- 10.Cheng B, Stanley RJ, De S, Antani S, Thoma GR. Automatic detection of arrow annotation overlays in biomedical images. International Journal of Healthcare Information Systems and Informatics. 2011;Vol. 6(No. 4):23–41. [Google Scholar]
- 11.Yao X, Islam MM. Evolving artificial neural network ensembles. Computational Intelligence Magazine. 2008;Vol. 3(No. 1):31–42. [Google Scholar]
- 12.Liu L, Yao X. Negatively correlated neural networks can produce best ensembles. Aust. J. Intell. Inf. Proc. Syst. 1998;Vol. 4(No. 3/4):176–185. [Google Scholar]
- 13.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the Fourteenth International Joint Conference on Artificial Intelligence. 1995;Vol.14:1137–1143. [Google Scholar]
- 14.Fogarty J, Baker RS, Hudson SE. Proceedings of Graphics interface, Canadian Human-Computer Communications Society, School of Computer Science. Waterloo, Ontario Victoria, British Columbia: University of Waterloo; 2005. Case studies in the use of ROC curve analysis for sensor-based estimates in human computer interaction; pp. 129–136. [Google Scholar]