Abstract
Plasticity is a typical feature of development and can lead to divergent phenotypes. There is increasing evidence that epigenetic mechanisms, such as DNA methylation, are present across species, are modifiable by the environment, and are involved in developmental plasticity. Thus, in the context of the concept of developmental homology, epigenetic mechanisms may serve to create a process homology between species by providing a common molecular pathway through which environmental experiences shape development, ultimately leading to phenotypic diversity. This article will highlight evidence derived from across-species investigations of epigenetics, development, and plasticity which may contribute to our understanding of the homology that exists between species and between ancestors and descendants.
Keywords: DNA methylation, developmental plasticity, homology, inheritance, phenotypic diversity
Development is a dynamic process, involving an elegant interplay between genes and the environment. Within an organism, this interplay leads to increasing cellular complexity and differentiation in genetically homogeneous cells. Though development is certainly dependent on the “presence” of particular genes, the timing of gene activation and selective silencing of genes is equally critical. Thus through cascades of gene expression and recruitment of factors that regulate transcription, tissue specific phenotypes emerge.
Though there are many factors which can alter the transcriptional activity of genes during development, epigenetic mechanisms, defined as factors that alter gene expression without altering underlying gene sequence, may be particularly illustrative of the dynamic interplay between genes and the environment. Epigenetic modifications, such as changes in DNA methylation, are increasingly being explored within the context of developmental studies and may be a highly conserved mechanism for driving phenotype within individuals, across species, and even across taxa (Vasanthi & Mishra, 2008). Moreover, there is increasing evidence that variation in the quality of early life experiences can induce epigenetic variation, thus serving as a mechanism of developmental plasticity.
Emerging evidence for the environmental regulation, stability and potential heritability of epigenetic variation across species raises intriguing questions regarding the role of epigenetics within discussions of developmental homology. Is epigenetic modulation a molecular strategy that confers a homology in the developmental process leading to plasticity in response to the quality of the environment? To illustrate the potential relevance of epigenetics and developmental plasticity to this question, here I will highlight recent advances in our understanding of the basic process of transcriptional regulation by epigenetic factors (with a focus on DNA methylation), evidence for the role of these factors in development across species, the role of epigenetic mechanisms in developmental plasticity, and the heritability of epigenetic effects (Skinner, 2011).
Though there are many ways in which the concept of homology may be applied within the study of epigenetics and developmental plasticity, here I propose that epigenetic mechanisms can be conceptualized as a homology of process – a series of molecular changes involving protein-protein, protein-DNA, and enzymatic reactions that are a highly conserved biological strategy for allowing variation at the level of gene expression and ultimately in the phenotype of the organism in response to environmental experiences. From this perspective, it is the epigenetic mechanisms themselves (and the responsiveness of these mechanisms to environmental modulation) that are homologous across species and taxa. This conceptualization is similar to that proposed for cell signaling pathways involving specific protein-protein interactions that are likewise highly conserved across species and taxa and play a significant role in development (Gilbert & Bolker, 2001). The heritability of epigenetic variation may also have implications for the concept of homology (i.e. as a potential mechanism through which similarities in the character of ancestors and descendants are generated), however at present the role of epigenetics in inheritance and evolution are topics of significant controversy (Haig, 2007; Richards, 2006).
Epigenetics & Development
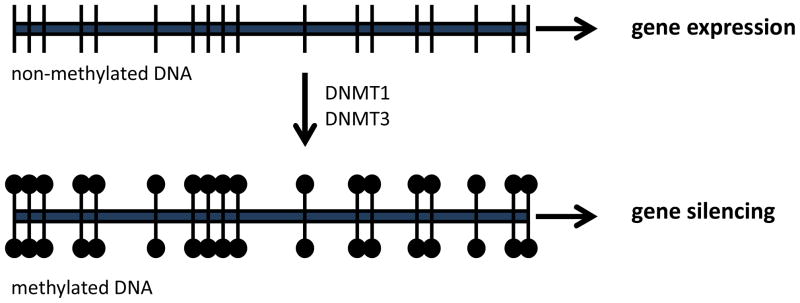
From a homogeneous cluster of progenitor cells, increasing cellular refinement and specialization is achieved through gene silencing – an outcome of epigenetic processes. Historically, the term “epigenetic” has been used to describe the dynamic interplay between genes and the environment which leads to variations in phenotype (Berger, Kouzarides, Shiekhattar, & Shilatifard, 2009; Holliday, 2006; Jablonka & Lamb, 2002). However, more current applications of this term are in reference to the specific molecular mechanisms which can lead to both transient and stable changes in the expression of genes. Gene transcription is dependent on the accessibility of DNA to RNA polymerase and other gene-specific transcription factors. Within the cell nucleus, DNA is wrapped around a core of histone proteins which can undergo multiple post-translational modifications including methylation, acetylation, and ubiquitination (i.e. addition of a methyl or acetyl chemical or ubiquitin protein to the histone) (Peterson & Laniel, 2004; Y. Zhang & Reinberg, 2001). These modifications alter the dynamic interactions between the histones and DNA which either reduce or enhance the accessibility of DNA. DNA methylation is an epigenetic modification through which cytosine nucleotides are converted to 5-methylcytosine. This conversion does not mutate the cytosine (the nucleotide can still form a complimentary base pair with guanine), however, this modification can lead to stable and enduring changes in gene activity. The process of DNA methylation is mediated by methyltransferases such as DNMT1 or DNMT3 (Feng, Fouse, & Fan, 2007; Razin, 1998; Turner, 2001). The conversion of cytosines to 5-methylcytosine typically results in reduced transcriptional activity (see Figure 1); though the location, degree of methylation, and recruitment of methyl binding proteins by methylated DNA will be important predictors of this effect (Jones et al., 1998). During mitosis, patterns of DNA methylation are replicated at the time of DNA synthesis such that daughter cells inherit both genetic and epigenetic information contained within the parental cell (Khavari, Sen, & Rinn, 2010; Wigler, Levy, & Perucho, 1981). The heritability of DNA methylation patterns is thought to be critical for maintaining cell-type specific gene expression patterns and stabilizing the phenotype of differentiated cells. Disruption to the DNA methylation process, through gene deletion of DNA methyltransferase enzymes leads to embryonic lethality (Li, Bestor, & Jaenisch, 1992), highlighting the importance of these epigenetic processes in development.
Figure 1.
DNA methylation and gene expression. When cytosines (vertical lines) in DNA (horizontal line) are not methylated there is increased accessibility to the gene promoter region, leading to increased gene expression. When DNA becomes methylated (black circles) through the enzymatic actions of the DNA methyltransferases (DNMT1 & DNMT3) gene expression is typically reduced or completely silenced.
DNA Methylation and Development Across Species
Though many of the early mechanistic studies on DNA methylation were conducted within in vitro cell culture systems or in laboratory mice, the role of this mechanism in the development of a variety of species and taxa is becoming increasingly evident. In mice, there are very high levels of 5-methylcytosine within the genome and the post-fertilization period is characterized by global demethylation followed by post-implantation tissue-specific increases in 5-methylcytosine levels (Monk, Boubelik, & Lehnert, 1987). In particular, DNA methylation within the male pronucleus undergoes dramatic post-fertilization decreases; an effect observed across many mammalian species including pig, rat, and human (Mayer, Niveleau, Walter, Fundele, & Haaf, 2000; Reik, Dean, & Walter, 2001). However, it is important to note that the dynamics of pronuclear methylation-demethylation are not completely conserved and, for example, paternal pronuclear demethylation may not occur in sheep or rabbits (Beaujean et al., 2004; Dean et al., 2001). Similarly, in frogs (Xenopus), there are high levels of DNA methylation throughout the post-fertilization period and in early embryonic development without the wave of demethylation that occurs in human, rodent, and pig genomes (Veenstra & Wolffe, 2001). Despite the lack of conservation of the temporal dynamics of the DNA methylation system, it is evident that this form of DNA modification is apparent across mammals (Gama-Sosa et al., 1983), in plants (M. Zhang, Kimatu, Xu, & Liu, 2010), and in insects (Wang et al., 2006).
DNA Methylation & Developmental Plasticity
Though it is apparent that epigenetic modifications such as DNA methylation are present within most plants and animals, a critical question in the context of this Special Issue on Developmental Homology is whether epigenetic mechanisms can be conceptualized as an across-species homology in the process through which developmental plasticity is achieved. This question can be further divided into 1) Can DNA methylation patterns be modified during development? and 2) Do developmental modifications in DNA methylation patterns coincide with the emergence of diverse phenotypic outcomes? Both of these issues have been explored across a diverse array of species, providing support for the hypothesis that environmentally-induced changes in epigenetic mechanisms such as DNA methylation are highly conserved across species and that phenotypic variation can be achieved as an outcome of these environmentally-induced changes. Within the context of discussions of process homology, both of these issues may be relevant – though representative of the different levels of biological organization at which the homology can be conceptualized.
Can DNA methylation patterns be modified during development?
Dynamic changes in DNA methylation have been observed across a number of species during early embryonic development. However, a key question here is whether DNA methylation patterns can be altered in response to environmental events. Interestingly, the early studies exploring the across-species conservation of post-fertilization DNA methylation patterns also established that these patterns could undergo modification in response to hormones and cellular cues. Superovulation is a technique used to stimulate increased oocyte production and involves treatment with gonadotrophins. These hormone treatments are routinely used when studying fertilization and early embryonic dynamics in the lab and are also a strategy used in assisted reproduction in humans. Comparison of embryonic DNA methylation patterns derived from superovulated vs. non-superovulated females has indicated that superovulation may induce abnormal DNA methylation patterns (Shi & Haaf, 2002). In vitro fertilization (IVF) was similarly found to induce changes in embryonic DNA methylation dependent on the type of culture medium used for the incubation of sperm and oocytes. Further study of these effects has indicated that DNA methylation patterns in imprinted genes – genes that are expressed or epigenetically silenced in a parent-of-origin specific pattern – are particularly vulnerable to the effects of superovulation/IVF and these abnormalities may account for reports of an increased incidence of imprinting disorders (such as Angelman and Prader-Willi syndrome) in individuals conceived through artificial reproductive technology (Lucifero, Chaillet, & Trasler, 2004).
DNA methylation is clearly subject to environmentally induced change or disruption in response to hormonal and cellular events occurring in early embryogenesis. However, developmental plasticity continues beyond this period. Thus it is important to consider the plasticity of DNA methylation patterns during fetal and postnatal development. There is increasing evidence for this plasticity in response to toxicological, hormonal, nutritional, social, and broad ecological environmental exposures. For example, exposure to the endocrine disrupting chemical bisphenol-A (BPA) during the prenatal period can induce genome-wide changes in brain DNA methylation patterns in mice (Yaoi et al., 2008), and has also been demonstrated to induce variation in DNA methylation in target genes within human cells (Weng et al., 2010). The epigenetic effects of BPA can also be observed following postnatal exposure to this chemical in neonatal rats (Doshi, Mehta, Dighe, Balasinor, & Vanage, 2011). Moreover, epigenetic plasticity continues into adulthood. For example, manipulation of testosterone levels in adult rats is associated with neural changes in DNA methylation within the vasopressin (AVP) and estrogen receptor α (ERα) genes (Auger, Coss, Auger, & Forbes-Lorman, 2011). Exposure to adult social stress is associated with decreased methylation of the corticotrophin releasing factor (CRF) gene in the hypothalamic tissue of mice (Elliott, Ezra-Nevo, Regev, Neufeld-Cohen, & Chen, 2010). Even the process of learning, a phenomenon that demonstrates life-long plasticity in the brain, appears to involve changes in DNA methylation (Levenson & Sweatt, 2005). Thus, although there may certainly be sensitive periods during which plasticity in DNA methylation is heightened (such as during early embryogenesis and fetal development), this epigenetic mechanism appears responsive to environmental experiences occurring across the lifespan.
Do developmental modifications in DNA methylation patterns coincide with the emergence of diverse phenotypic outcomes?
The capacity to induce dynamic changes in gene expression via epigenetic pathways and the role of these pathways in maintaining stable variations in cellular phenotype has led to increasing speculation that divergence in phenotype of the individual (e.g. neurodevelopment, disease risk, behavior) can likewise be achieved through mechanisms such as DNA methylation. In monozygotic twins, there is evidence for discordance in DNA methylation patterns and it would appear that this discordance increases over time (Fraga et al., 2005; Mill et al., 2006; Wong et al., 2010). The critical question raised by these findings is whether these epigenetic modifications serve as a mechanism of developmental plasticity, thus allowing environmental experiences to drive the emergence of phenotypic diversity which is then stably maintained into adulthood. Across species, there is increasing evidence for the influence of early life nutrition, stress, and social experiences on development achieved through environmentally-induced changes in DNA methylation. These studies provide support for the notions of epigenetic plasticity and epigenetic induced developmental plasticity.
Epigenetics & Nutrition
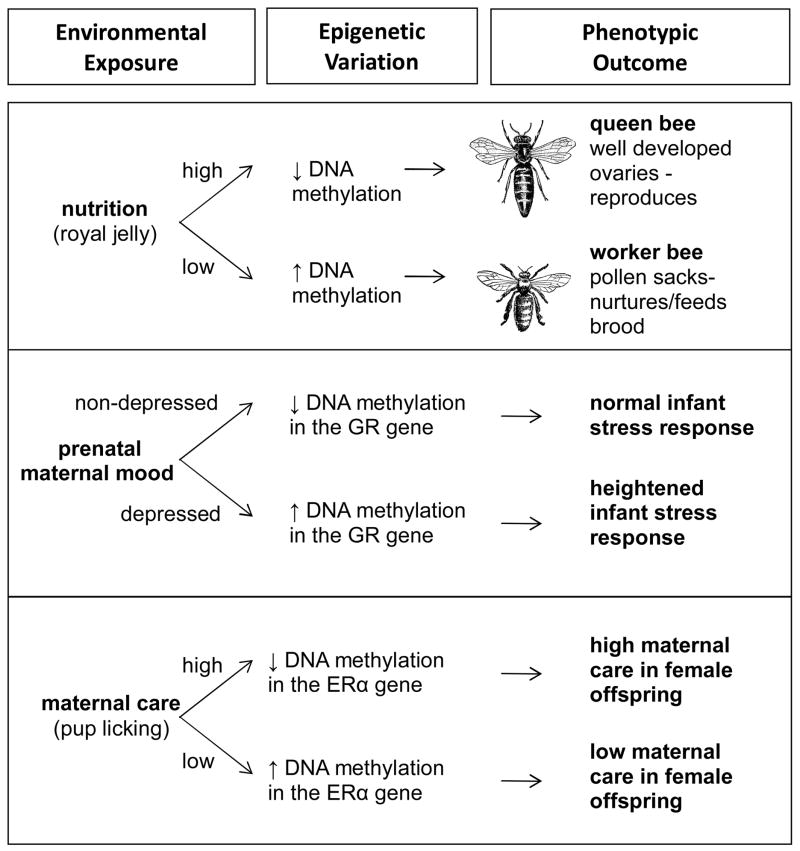
Prenatal and postnatal nutrition is a signal of environmental quality which can predict growth and survival. In human epidemiological studies, analysis of blood samples from famine exposed vs. non-exposed siblings indicates that there is decreased DNA methylation of the insulin-like growth factor 2 (Igf2) gene as a consequence of maternal periconceptual exposure to famine (Heijmans et al., 2008). This epigenetic effect may account for many of the metabolic abnormalities apparent as a function of severe caloric restriction during fetal development (Lumey, Stein, & Susser, 2011). Laboratory studies in rodents have subsequently identified specific nutritional deficits, such as prenatal protein restriction or folic acid/choline deficiency as having similar epigenetic consequences. Offspring of female rats placed on a protein deficient diet throughout gestation were found to have elevated hepatic glucocorticoid receptor (GR) and peroxisomal proliferator-activated receptor (PPAR) gene expression associated with decreased DNA methylation of these genes (Lillycrop, Phillips, Jackson, Hanson, & Burdge, 2005; Lillycrop et al., 2008). Epigenetic modifications in response to the nutritional environment during the early stages of development may also have implications for the morphological changes associated with caste phenotypes in eusocial insects. Honeybees have functional DNA methyltransferases and the degree of methylation of the genome varies during the course of development (Wang, et al., 2006). Amongst female honeybees, social/reproductive caste is determined through early nutritional exposure to royal jelly (with increased royal jelly promoting the development of queen bees and reduced royal jelly promoting the development of worker females – see Figure 2). Manipulation of the activity of the DNA methyltransferase DNMT3 in the honey bee provides evidence that DNA methylation mediates these divergent phenotypes. Under control conditions, 75% of larvae develop as worker bees whereas inhibiting DNMT3 leads to the majority of larvae developing morphologically as queen bees (Kucharski, Maleszka, Foret, & Maleszka, 2008). Taken together, these studies illustrate how epigenetic mechanisms serve a central and developmental role across species, leading to individual variation.
Figure 2.
Phenotypic outcomes associated with environmentally induced epigenetic variation across species. In honeybees (top panel), variation in the amount of royal jelly experienced in early development leads to changes in DNA methylation and phenotypic variation giving rise to caste differences. In humans (middle panel), maternal depression during pregnancy is associated with increased methylation of the glucocorticoid receptor (GR) gene in fetal cord blood leading to increased stress reactivity in infants born to depressed mothers. In rats (bottom panel), high maternal care experienced by female offspring in infancy leads to decreased DNA methylation within the estrogen receptor (ERα) gene, leading to increased ERα gene expression and elevated maternal behavior in adulthood.
Epigenetics & Early Life Stress
The experience of adversity in early development can have a lasting impact on brain, physiology, and behavior. In humans, studies examining the effects of maternal distress during pregnancy are increasingly incorporating an epigenetic perspective. Elevated GR methylation is found in fetal cord blood, associated with increased maternal depression during pregnancy, and this epigenetic modification predicts stress responsivity in infants at 3 months of age (Oberlander et al., 2008) (Figure 2). Among adolescents aged 10–19 years, those born to mothers who experienced intimate partner violence during pregnancy were found to have elevated levels of GR DNA methylation in blood (Radtke et al., 2011). In rats, offspring born to females that undergo bystander stress during pregnancy (through co-housing with a stressed female) have global elevations in DNA methylation levels within the cortex and hippocampus (Mychasiuk et al., 2011). Target gene analyses in male offspring born to female mice that underwent a chronic variable stress regime during pregnancy has revealed decreased DNA methylation of the CRF gene promoter and increased methylation of the GR gene promoter region in hypothalamic tissue. These molecular changes correspond to altered gene expression and increased stress responsivity in stress-exposed offspring.
During the postnatal period, sensitivity to stress and adversity, such as exposure to abuse, neglect, or maternal separation, can have a profound effect in infant development. In human postmortem brain tissue, a history of childhood abuse predicts elevated GR methylation and decreased hippocampal GR gene expression (McGowan et al., 2009). Among rhesus macaques, DNA methylation of the serotonin transporter (5-HTT) gene is increased in peripheral blood mononuclear cells following maternal (and social) separation (Kinnally et al., 2010). This epigenetic change was found to be associated with a decrease in 5-HTT expression and behavioral hyper-reactivity in maternally-deprived infants. In rat pups, exposure to aggressive/abusive encounters with a foster female can induce long-lasting increases in DNA methylation of the brain dervived neurotrophic factor (BDNF) gene in the prefrontal cortex leading to decreased BDNF gene expression and thus increasing the emergence of depressive-like behavior in these offspring (Roth, Lubin, Funk, & Sweatt, 2009). Maternal separation studies in mice indicate effects on DNA methylation of the AVP gene in hypothalamic tissue corresponding to elevations in stress sensitivity (Murgatroyd et al., 2009). Similar to the case of early life nutrition, epigenetic mechanisms appear to play a significant role in linking the experience of adversity during prenatal and postnatal development to long-term variation in offspring phenotype; particularly phenotypes related to stress responsivity.
Epigenetics & Social Experiences
In mammals, the quality of the early life environment is dependent on the pattern and frequency of mother-infant social interactions and there is evidence that variation in these interactions can have persistent epigenetic and neurobehavioral consequences. Postnatal maternal licking/grooming (LG) behavior in rats has been found to induce long-term changes in neuroendocrine function and behavior of offspring, with consequences for stress responsivity and cognition, and cross-fostering studies have confirmed that these effects are mediated by the level of maternal care received during postnatal development (Meaney, 2001). Analysis of the GR gene promoter region suggests that variations in GR expression associated with differential levels of maternal care are maintained though altered levels of DNA methylation (Weaver et al., 2004). Thus, offspring who receive high levels of maternal LG during the early postnatal period have decreased hippocampal GR promoter DNA methylation, increased GR expression and decreased stress responsivity. In contrast, low levels of LG are associated with increased GR DNA methylation, decreased GR expression, and an increased hypothalamic-pituitary-adrenal response to stress. Time course analysis has indicated that these maternally-induced epigenetic profiles emerge during the postnatal period and are sustained into adulthood. Maternal LG also induces increased methylation within the glutamic acid decarboxylase (GAD1) gene in male hippocampal tissue resulting in reduced GAD1 levels and consequences for γ-aminobutyric acid (GABA) circuits and receptor subunit composition (Caldji, Diorio, & Meaney, 2003; T. Y. Zhang et al., 2010). Amongst female offspring, the experience of low levels of LG is associated with increased DNA methylation of the ERα gene in the medial preoptic area of the hypothalamus and consequently these offspring display low levels of LG when caring for their own offspring (Champagne et al., 2006) (Figure 2). These maternally induced epigenetic effects in females may account for the transgenerational continuity in the effects of maternal behavior on neurobiological and behavioral outcomes (Champagne, 2008).
Epigenetics & Inheritance
The transmission of traits across generations can create a homology between the characteristics of descendants and their immediate or distant ancestors. Though traditionally this transmission has been the domain of genetics, there is increasing evidence for the role of epigenetic mechanisms in the inheritance of phenotypes (Daxinger & Whitelaw, 2010; Youngson & Whitelaw, 2008). One route through which this may occur involves the experience-dependent modification of DNA methylation in genes which shape reproductive behavior. For example, variation in maternal LG behavior in rat dams can be sustained across generations through the epigenetic effects of maternal LG on neuroendocrine circuits which regulate LG behavior in offspring (Champagne, 2008). Similarly, the experience of abusive caregiving can have transgenerational effects on DNA methylation of the BDNF gene in female offspring that may account for the emergence of abusive behavior in these offspring. In contrast to this experience-dependent pathway, it may also be possible to inherit environmentally-induced epigenetic effects via the germline. For example, prenatal exposure to the fungicide vinclozolin induces variation in sperm DNA methylation patterns in male rats that persist for multiple generations following exposure with implications for brain gene expression and disease risk (Anway, Cupp, Uzumcu, & Skinner, 2005; Skinner, Anway, Savenkova, Gore, & Crews, 2008). In mice, maternal separation during infancy has similarly been found to affect sperm DNA methylation and offspring phenotypes (Franklin et al., 2010). This type of epigenetic inheritance may be relevant across taxa – being evident in plants (Hauser, Aufsatz, Jonak, & Luschnig, 2011) and mammals (Carone et al., 2010), and may account for the persistence of environmentally-induced phenotypic variation.
Conclusions
Though the investigation of the role of DNA methylation in developmental plasticity has thus far been limited to species in which the genome is well characterized and assays are available to conduct epigenetic analyses, it is evident that epigenetic control of gene expression is an across-species phenomenon. Moreover, the emerging evidence from species ranging from honeybees to humans suggests that environmentally-induced changes in DNA methylation may serve as a mechanism mediating developmental plasticity leading to phenotypic variation. Considered within an evolutionary perspective, it appears likely that these mechanisms are a fundamental feature of the process of adaptation to the environment, leading to adaptive reproductive, behavioral, and metabolic strategies which promote survival. The ability to transmit these developmental effects across generations raises important issues regarding the mechanisms of heritability and the ancestral origins of phenotypic variation (Danchin et al., 2011). Within discussions of developmental homology, evidence for the plasticity of DNA methylation across species may add to our understanding of the process homology that underlies the ability of individuals to adapt and change in response to early life experiences.
Acknowledgments
This research was supported by Grant Number DP2OD001674 from the Office of the Director, National Institutes of Health
References
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–1469. doi: 10.1126/science.1108190. 308/5727/1466 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger CJ, Coss D, Auger AP, Forbes-Lorman RM. Epigenetic control of vasopressin expression is maintained by steroid hormones in the adult male rat brain. Proceedings of the National Academy Sciences of the United States of America. 2011;108(10):4242–4247. doi: 10.1073/pnas.1100314108. 1100314108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, Young L. Non-conservation of mammalian preimplantation methylation dynamics. Current Biology. 2004;14(7):R266–267. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes and Development. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldji C, Diorio J, Meaney MJ. Variations in maternal care alter GABA(A) receptor subunit expression in brain regions associated with fear. Neuropsychopharmacology. 2003;28(11):1950–1959. doi: 10.1038/sj.npp.1300237. [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–1096. doi: 10.1016/j.cell.2010.12.008. S0092-8674(10)01426-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Frontiers in Neuroendocrinology. 2008;29(3):386–397. doi: 10.1016/j.yfrne.2008.03.003. S0091-3022(08)00009-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147(6):2909–2915. doi: 10.1210/en.2005-1119. en.2005-1119 [pii] [DOI] [PubMed] [Google Scholar]
- Danchin E, Charmantier A, Champagne FA, Mesoudi A, Pujol B, Blanchet S. Beyond DNA: Integrating inclusive inheritance into an extended theory of evolution. Nature Reviews Genetics. 2011;12(7):475–486. doi: 10.1038/nrg3028. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: More questions than answers. Genome Research. 2010;20(12):1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proceedings of the National Academy Sciences of the United States of America. 2001;98(24):13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology. 2011;289(2–3):74–82. doi: 10.1016/j.tox.2011.07.011. S0300-483X(11)00271-X [pii] [DOI] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nature Neuroscience. 2010;13(11):1351–1353. doi: 10.1038/nn.2642. nn.2642 [pii] [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatric Research. 2007;61(5 Pt 2):58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy Sciences of the United States of America. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. 0500398102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biological Psychiatry. 2010;68(5):408–415. doi: 10.1016/j.biopsych.2010.05.036. S0006-3223(10)00576-7 [pii] [DOI] [PubMed] [Google Scholar]
- Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, Gehrke CW, Ehrlich M. Tissue-specific differences in DNA methylation in various mammals. Biochimica et Biophysica Acta. 1983;740(2):212–219. doi: 10.1016/0167-4781(83)90079-9. 0167-4781(83)90079-9 [pii] [DOI] [PubMed] [Google Scholar]
- Gilbert SF, Bolker JA. Homologies of process and modular elements of embryonic construction. [Review] Journal of Experimental Zoology. 2001;291(1):1–12. doi: 10.1002/jez.1. [DOI] [PubMed] [Google Scholar]
- Haig D. Weismann Rules! OK? Epigenetics and the Lamarckian temptation. Biology and Philosophy. 2007;22:415–428. 0.1007/s10539-006-9033-y. [Google Scholar]
- Hauser MT, Aufsatz W, Jonak C, Luschnig C. Transgenerational epigenetic inheritance in plants. Biochimica et Biophysica Acta. 2011;1809(8):459–468. doi: 10.1016/j.bbagrm.2011.03.007. S1874-9399(11)00057-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy Sciences of the United States of America. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. 0806560105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Epigenetics: A historical overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ. The changing concept of epigenetics. Annals of the New York Academy of Sciences. 2002;981:82–96. doi: 10.1111/j.1749-6632.2002.tb04913.x. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genetics. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Khavari DA, Sen GL, Rinn JL. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle. 2010;9(19):3880–3883. doi: 10.4161/cc.9.19.13385. [DOI] [PubMed] [Google Scholar]
- Kinnally EL, Capitanio JP, Leibel R, Deng L, LeDuc C, Haghighi F, Mann JJ. Epigenetic regulation of serotonin transporter expression and behavior in infant rhesus macaques. Genes, Brain, and Behavior. 2010;9(6):575–582. doi: 10.1111/j.1601-183X.2010.00588.x. GBB588 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nature Reviews Neuroscience. 2005;6(2):108–118. doi: 10.1038/nrn1604. nrn1604 [pii] [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. 0092-8674(92)90611-F [pii] [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. The Journal of Nutrition. 2005;135(6):1382–1386. doi: 10.1093/jn/135.6.1382. 135/6/1382 [pii] [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. British Journal of Nutrition. 2008;100(2):278–282. doi: 10.1017/S0007114507894438. S0007114507894438 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifero D, Chaillet JR, Trasler JM. Potential significance of genomic imprinting defects for reproduction and assisted reproductive technology. Human Reproduction Update. 2004;10(1):3–18. doi: 10.1093/humupd/dmh002. [DOI] [PubMed] [Google Scholar]
- Lumey LH, Stein AD, Susser E. Prenatal famine and adult health. Annual Reviews Public Health. 2011;32:237–262. doi: 10.1146/annurev-publhealth-031210-101230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000654. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience. 2009;12(3):342–348. doi: 10.1038/nn.2270. nn.2270 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annual Reviews Neuroscience. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- Mill J, Dempster E, Caspi A, Williams B, Moffitt T, Craig I. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2006;141B(4):421–425. doi: 10.1002/ajmg.b.30316. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. nn.2436 [pii] [DOI] [PubMed] [Google Scholar]
- Mychasiuk R, Schmold N, Ilnytskyy S, Kovalchuk O, Kolb B, Gibb R. Prenatal bystander stress alters brain, behavior, and the epigenome of developing rat offspring. Developmental Neuroscience. 2011;33(2):159–169. doi: 10.1159/000330034. 000330034 [pii] [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. 6034 [pii] [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Current Biology. 2004;14(14):R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Elbert T. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Translational Psychiatry. 2011;1:e21. doi: 10.1038/tp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. CpG methylation, chromatin structure and gene silencing--a three-way connection. EMBO Members Journal. 1998;17(17):4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation--revisiting soft inheritance. Nature Reviews Genetics. 2006;7(5):395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. S0006-3223(08)01530-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Haaf T. Aberrant methylation patterns at the two-cell stage as an indicator of early developmental failure. Molecular Reproduction and Development. 2002;63(3):329–334. doi: 10.1002/mrd.90016. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Research Part C: Embryo Today. 2011;93(1):51–55. doi: 10.1002/bdrc.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One. 2008;3(11):e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. Chromatin and Gene Regulation. Oxford: Blackwell Science Ltd; 2001. [Google Scholar]
- Vasanthi D, Mishra RK. Epigenetic regulation of genes during development: A conserved theme from flies to mammals. Journal of Genetics and Genomics. 2008;35(7):413–429. doi: 10.1016/S1673-8527(08)60059-4. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Wolffe AP. Constitutive genomic methylation during embryonic development of Xenopus. Biochimica et Biophysica Acta. 2001;1521(1–3):39–44. doi: 10.1016/s0167-4781(01)00280-9. S0167478101002809 [pii] [DOI] [PubMed] [Google Scholar]
- Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, Robinson GE. Functional CpG methylation system in a social insect. Science. 2006;314(5799):645–647. doi: 10.1126/science.1135213. 314/5799/645 [pii] [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Meaney MJ. Epigenetic programming by maternal behavior. Nature Neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weng YI, Hsu PY, Liyanarachchi S, Liu J, Deatherage DE, Huang YW, Huang TH. Epigenetic influences of low-dose bisphenol A in primary human breast epithelial cells. Toxicology and Applied Pharmacology. 2010;248(2):111–121. doi: 10.1016/j.taap.2010.07.014. S0041-008X(10)00241-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. 0092-8674(81)90498-0 [pii] [DOI] [PubMed] [Google Scholar]
- Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Mill J. A longitudinal study of epigenetic variation in twins. Epigenetics. 2010;5(6):516–526. doi: 10.4161/epi.5.6.12226. 12226 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochemical and Biophysical Research Communications. 2008;376(3):563–567. doi: 10.1016/j.bbrc.2008.09.028. S0006-291X(08)01774-9 [pii] [DOI] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annual Review of Genomics and Human Genetics. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- Zhang M, Kimatu JN, Xu K, Liu B. DNA cytosine methylation in plant development. Journal of Genetics and Genomics. 2010;37(1):1–12. doi: 10.1016/S1673-8527(09)60020-5. S1673-8527(09)60020-5 [pii] [DOI] [PubMed] [Google Scholar]
- Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. The Journal of Neuroscience. 2010;30(39):13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. 30/39/13130 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes and Development. 2001;15(18):2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]