Synopsis
Since the discovery of activating mutations in JAK2 in patients with myeloproliferative neoplasms (MPNs) in 2005, gene discovery efforts have identified additional disease alleles which can predate or occur subsequent to acquisition of JAK2/MPL mutations. In 2009, single nucleotide polymorphism (SNP) arrays and comparative genomic hybridization array (aCGH) based profiling led to the identification of somatic copy-number loss and mutations in the genes TET2 and ASXL1 in MPN patients. Biochemical and biological characterization of the TET and ASXL family of proteins have provided valuable insights into new modes of epigenetic regulation of gene transcription. Mutations in TET2 and ASXL1 are also important biomarkers for disease outcome amongst patients with acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS). Despite these important insights, the relevance of these mutations to outcome and to therapeutic response in MPN patients is not yet clear. Genetic analysis of MPN patient cohorts with adequate sample size and clear clinical annotation are needed to understand the importance of these mutations on MPN phenotype, risk of transformation to leukemia, response to therapy, and influence on overall survival.
Keywords: ASXL1, TET2, Myelofibrosis, Myeloproliferative Neoplasms, JAK2
Introduction: Evidence for mutations outside of the JAK-STAT pathway in MPN patients
Myeloproliferative neoplasms (MPNs) are clonal disorders of hematopoiesis characterized by excess production of mature appearing cells within the blood stream. The MPNs were initially grouped together by William Dameshek in 19511. However, in 2005, the first biological basis unifying the pathogenesis of the different MPN was discovered when activating mutations in JAK2 were identified in 95% of patients with polycythemia vera (PV), in 55–60% of patients with essential thrombocytosis (ET), and in 50% of patients with primary myleofibrosis (PMF)2–5. This was quickly followed by the discovery of additional mutations resulting in activation of the JAK-STAT pathway in MPN patients including exon 12 mutations in JAK26, thrombopoietin receptor (MPL) mutations7, and loss-of-function mutations in LNK, a negative regulator of JAK-STAT signaling8, 9 (Table 1).
Table 1.
Frequency of somatic genetic mutations in patients with myeloproliferative neoplasms (MPNs)
| Gene Mutation1 | ||||||
|---|---|---|---|---|---|---|
| MPN | JAK2V617F | JAK2 exon 12 | MPL | LNK | TET2 | ASXL12 |
| Polycythemia vera (PV) | 95% | 3–5% | NR | Present | 9.8 – 16% | 2–5% |
| Essential thrombocytosis (ET) | 55–60% | NR | 3–5% | 3–6% | 4.4 – 5% | 5–10% |
| Primary myelofibrosis (MF) | 50–60% | NR | 8– 10% | 3–6% | 7.7 – 17% | 13–26% |
| Post-PV/ET MF | 50–60% | NR | NR | NR | 14% | 22–38.5% |
NR, not reported.
Several of the manuscripts used to delineate mutational frequency of ASXL1 contain the controversial p.Gly646TrpfsX12 variant which has not definitively proven to be a somatic mutation.
Although the discovery of mutations in JAK2 and MPL provided seminal insight into MPN pathogenesis, several lines of evidence suggest that mutations in genes other than JAK2 and MPL must be present in MPN patients. First was the question of how a single mutation in JAK2, which appeared to be sufficient for MPN pathogenesis from in vivo studies, could result in the development of 3 phenotypically different diseases. One attractive hypothesis to this question was that additional acquired or inherited genetic modifiers outside of JAK2 or MPL could be present and modify the MPN phenotype. Secondly, clonal analysis of patients with JAK2/MPL mutations demonstrated the presence of JAK2 wildtype endogenous erythroid colonies (EECs)- clear evidence that an additional aberration responsible for erythropoietin-independent growth must be present10. Clonality analysis of patients with both a cytogenetic abnormality in conjunction with the JAK2V617F mutation also revealed that some patients had cytogenetically abnormal clones with and without the JAK2V617F mutation11. Finally, a number of reports have reported that leukemic blasts of acute myeloid leukemia (AML) derived from a JAK2V617F MPN are frequently JAK2 wild-type12, 13. This suggests that the MPN and AML clones can arise from 2 different progenitor cells, or that an ancestral clone bearing an abnormality preceding the JAK2V617F mutation can give rise to both the JAK2-positive MPN and the JAK2-negative AML. Thus, additional novel mutations in MPN pathogenesis have been speculated to exist since the discovery of mutations in JAK2.
Discovery of TET2 mutations in MPN patients
Mutations in TET2 (ten eleven translocation two) were the first described recurrent somatic alterations in MPN patients in a gene not directly known to be involved in the JAK-STAT signaling pathway. TET2 mutations were originally described by Delhommeau et al. and Langemeijer et al in 2009 in patients with MPNs and myelodysplastic syndrome (MDS)14, 15. Through careful examination of primary PV patient samples, Delhommeau et al. noticed that majority of JAK2V617F mutant PV patients (~85%) had expansion of CD34+CD38+ committed progenitor cells over CD34+CD38− mutlipotent progenitors in ex vivo liquid cultures. In contrast, a minority (~15%) of JAK2V617F mutant PV patients were characterized by relative expansion of the more immature multipotent progenitor cells (CD34+ CD38−). Hypothesizing that a novel genetic abnormality might be responsible for this immunophenotypic difference in these 2 patient subsets, the authors performed SNP arrays (Affymetrix 500K) and array CGH (Agilent 244K) on a small number of patient samples. They found that 3 of the 5 JAK2V617F mutant PV patients with expansion of CD34+CD38− cells had loss-of-heterozygosity (LOH) at chromosomal locus 4q24. One of these 5 patients had a deletion of a 325 kB region of DNA at 4q24; the only gene present in this region being TET2. This then led to sequencing of TET2 in these patient samples and the identification of somatic TET2 mutations in MPNs. Since then, sequencing of TET2 has led to the identification of TET2 mutations in every myeloid disorder14–20. Mutations in TET2 have been found in all coding regions and can appear as missense, nonsense, or frameshift mutations. Mutations in TET2 are less uncommonly bi-allelic (i.e. involving both copies of TET2) consistent with mutations in TET2 being haploinsufficient loss-of-function mutations in most patients.
Biochemical function of TET2
TET2 is a member of the TET family of genes, the first member of which to be described was TET1. TET1 (ten-eleven translocation 1), located on chromosome 10, was originally identified in cases of adult and pediatric AML as a translocation partner with MLL (located on chromosome 11)21. Although TET1 was the original gene member identified in hematologic malignancies, no sequence alterations in TET1 or TET3 have been identified to date16.
In a landmark publication by Tahiliani and colleagues in 2009, the function of TET1 was first described22. They pursued the identification of human enzymes which modify bases of nucleic acids as a means to understand how catalytic modifications of DNA bases affects the genetic code. As such, they undertook in sllico approach to identify human homologs of the trypanosome proteins JBP1 and JBP2 which are known to oxidize the 5-methyl group of thymine23. Such enzymes were not previously known to exist in higher organisms. Surprisingly, they found that the TET family of genes were human homologues of these trypanosomal enzymes. Further characterization of TET1 revealed that it is an 2-oxoglutarate- and iron(II)-dependent dioxygenase which serves to oxidize the 5-methyl group of cytosine leading to formation of 5-hydroxymethylcytosine (Figure 1).
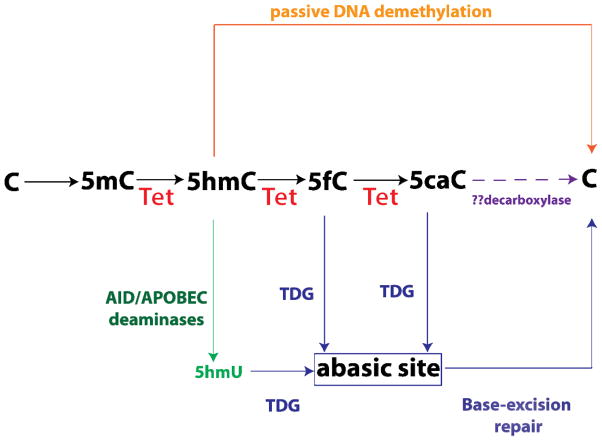
Figure 1. Role of TET2 in DNA hydroxymethylation and DNA demethylation.
The TET family of enzymes (TET1–3) are α-ketoglutarate and Fe(II)-dependent enzymes which hydroxylate the 5-methyl group on methylcytosine (5mC) to create 5-hydroxymethylcytosine (5hmC). TET family of enzymes then further oxidixe 5hmC into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). These activities of the TET family of enzymes may then promote demethylation of DNA in four potential pathways which are being investigated further: (1) because 5hmC is not recognized by maintenance DNA methyltransferases, 5hmC can result in DNA demethylation over time with DNA replication in a passive manner (orange lines and text); (2) 5fC and 5caC can be excised by thymine DNA glycosylase (TDG) into an abasic site which could then be regenerated into an unmodified cytosine by the base-excision repair pathway (BER) (blue lines and text); (3) 5hmC may also be converted by the AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA editing enzyme complex) family of cytosine demethylases into 5-hydroxymethyluracil (5hmU) which may then be repaired through the action of DNA glycosylases and the BER pathway (green text); finally, there is a possibility that 5caC may be further decarboxylated by a yet undiscovered decarboxylase into unmodified cytosine (purple text).
More recent work has identified that all three TET proteins are enzymes which can convert 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC). Moreover, 2 groups have reported that Tet proteins can further convert 5hmC into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in two successive oxidation reactions (Figure 1).
The discovery of these novel enzymatic activities by the Tet proteins has provided insight into potential mechanisms by which 5mC is dynamically regulated as well as demethylated in both active and passive processes. For instance, DNMT1 (DNA methyltransferase 1), the DNA methyltransferase responsible for maintaining DNA methylation, does not recognize 5hmC. Thus, conversion of 5mC into 5hmC may lead to replication-dependent passive demethylation of DNA (Figure 1). Furthermore, oxidized derivatives of 5hmC may serve in a replication-independent, active DNA demethylation process. Proof for this concept comes from the finding that thymine DNA glycosylase (TDG) can excise 5fC or 5caC in the context of CpG sites (TDG has minimal activity towards 5hmC). The resulting abasic site following TDG-excision can then be repaired by the base-excision repair (BER) pathway to generate unmethylated cytosines. There is also evidence that 5hmC can be actively deaminated into 5-hydroxymethyluracil (5hmU) by the AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA editing enzyme complex) complex. The resulting 5hmU could then be removed via the action of DNA glycosylases and the BER pathway. Finally, Wu et al. proposed that 5mC might be converted to cytosine simply by (1) iterative oxidation of 5hmC by Tet enzymes followed by (2) a single decarboxylation of 5caC to regenerate cytosine by a yet unidentified putative decarboxylase. Although no decarboxylase capable of removing the carboxyl group from 5caC has been identified, this latter mechanism of iterative 5mC oxidation followed by decarboxylation is attractive in its simplicity and the fact that no DNA repair mechanism is required to effect DNA demethylation.
Biological role of TET2 in hematopoiesis
In parallel with the biochemical characterization of TET2’s enzymatic function have been extensive functional studies of the biological ramifications of TET2 loss. Initial evidence of the role of TET2 in hematopoiesis came from xenograft studies in the initial reports of TET2 mutations by Delhommeau and colleagues15. They noted that xenograting JAK2V617F-positive CD34+ cells from MPN subjects with (n=2) and those without TET2 mutations (n=3) into NOD-SCID mice revealed a more efficient engraftment of TET2 mutant cells over TET2 wildtype CD34+ cells. Moreover, the hematopoiesis was skewed towards increased frequency of myeloid progenitors over lymphoid progenitors in TET2 mutant patients.
More recently, independent reports of the phenotype of Tet2 knockout mice have been published using at least four different targeting alleles (Figure 2A). Conditional deletion of the first coding exon of Tet2 by Moran-Crusio et al. showed that Tet2 loss leads to a progressive enlargement of the hematopoietic stem cell compartment and eventual myeloproliferation in vivo, including splenomegaly, monocytosis, and extramedullary hematopoiesis24. In addition, Tet2+/− mice displayed increased stem cell self-renewal and extramedullary hematopoiesis, demonstrating Tet2 haploinsufficiency contributes to hematopoietic transformation in vivo (Figure 2B). In a simultaneous publication, Quivoron et al. also found a very similar phenotype in a gene-trap Tet2 knockout model and a conditional Tet2 knockout where the final coding exon of Tet2 was floxed (Figures 2A and 2B).
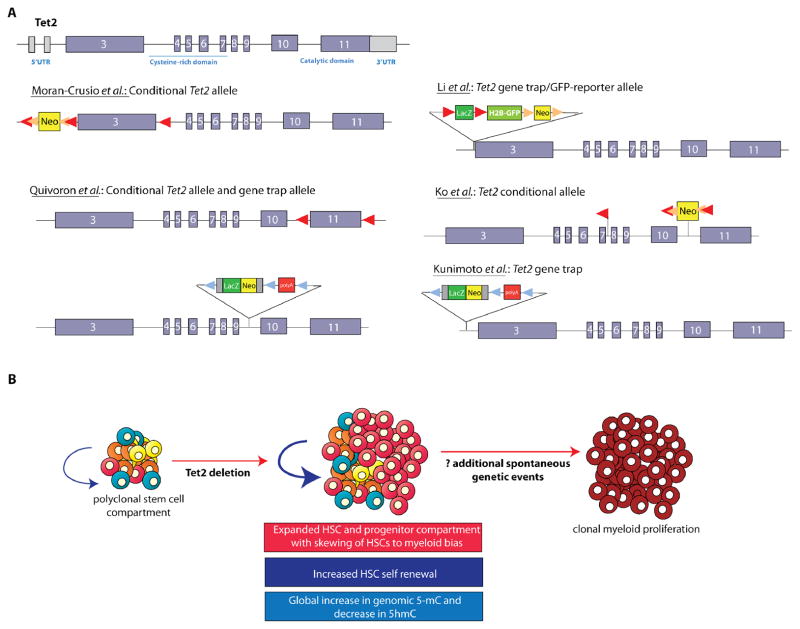
Figure 2. The role of TET2 in hematopoiesis as identified by deletion of Tet2 in murine models.
Targeted disruption of murine Tet2 has now been accomplished using at least six different constructs each targeting Tet2 in a different location and manner (A). This includes conditional deletion of Tet2 by targeting the first coding exon (as done by Moran-Crusio et al.), the last coding exon encoding the enzymatic function of Tet2 (as done by Quivoron et al.), or the exons 8–10 (as done by Ko et al.). Several groups have also studied mice with germline deletion of Tet2 as accomplished by several different gene trap constructs (A). Nearly all of these models have revealed that deletion of Tet2, whether in a conditional, hematopoietic-specific manner, or in the germline, results in expansion of the hematopoietic-stem progenitor compartment and increased hematopoietic stem cell self-renewal shortly after deletion (B). Overt myeloproliferation is also evident following Tet2 deletion in vivo but with a latency of at least 3–6 months, suggesting the acquisition of additional collaborating events are required for disease initiation and progression in vivo.
Just after the initial two Tet2 knockout mouse models were published, two additional reports using different models of Tet2 deletion were published25, 26. All four publications revealed a similar effect of Tet2 loss on increased HSC self-renewal and development of an MPN resembling human chronic myelomonocytic leukemia (CMML). To date, there is no evidence that Tet2 loss in vivo results in development of myelofibrosis in mice24.
Three of the four Tet2 knockout mouse studies have revealed a clear linkage between loss of Tet2 and decreased hmC in vivo25–27. The effect of Tet2 loss on 5mC in patients however has yet to be clarified and the genetic targets of TET2 loss are not yet well understood.
Discovery of ASXL1 mutations
Within the same year as discovery of TET2 mutations in myeloid cancers, mutations were identified in another putative epigenetic modifier in myeloid malignancies, Additional Sex Combs Like 1 (ASXL1). Mutations in ASXL1 were originally identified based on aCGH studies of MDS samples28. Gelsi-Boyer et al. performed Agilent 244K CGH arrays on patients with MDS and noticed deletions in one patient at 20q11. In this particular patient, the 20q deletions involved only 2 possible genes- ASXL1 and DNMT3B. Sequencing efforts of both genes followed and mutations in ASXL1 were found in 4/35 MDS patients (11%). Further sequencing of ASXL1 has delineated the frequency of ASXL1 mutations in MPNs and other myeloid disorders (Table 1)28–30. From these studies, ASXL1 is most commonly mutated amongst MPN patients with PMF and post-PV/ET MF compared with PV or ET. This is in contrast to mutations in TET2, which appear to be somewhat evenly distributed in PV/ET compared with myelofibrosis (Table 1).
Biological role of Asxl1 in hematopoiesis
ASXL1 is the human homologue of Drosophila Additional sex combs (Asx). Asx deletion results in a homeotic phenotype characteristic of both Polycomb (PcG) and Trithorax group (TxG) gene deletions 31 which led to the hypothesis that Asx has dual functions in silencing and activation of homeotic gene expression. In addition, functional studies in Drosophila suggest that Asx encodes a chromatin-associated protein with similarities to PcG proteins 32. The mechanisms by which ASXL1 mutations contribute to myeloid transformation have not been delineated. A series of in vitro studies in non-hematopoietic cells have suggested a variety of activities for ASXL1 including physical cooperativity with HP1a and LSD1 to repress retinoic acid-receptor (RAR) activity and interaction with peroxisome proliferator-activated receptor gamma (PPARg) to suppress lipogenesis 33–35 (Figure 3).
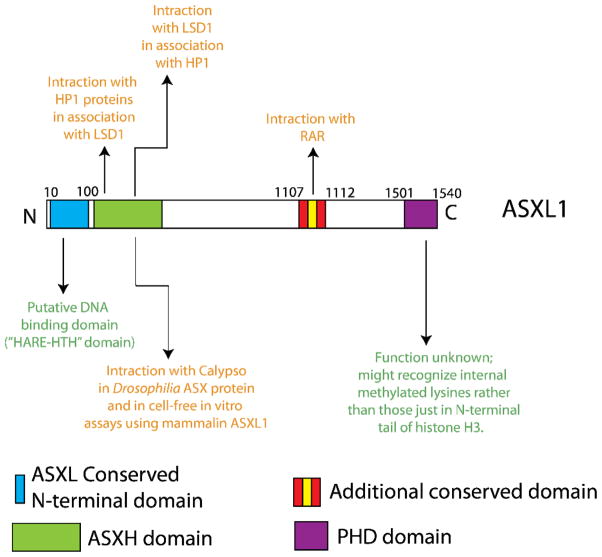
Figure 3. Conserved domains of mammalian Asxl proteins and possible functions of ASXL1.
ASXL1 contains a globular N-terminal domain which is conserved amongst ASX proteins and contains a potential DNA binding motif based on homology with other proteins of known function. This domain has been referred to as a HARE-HTH domain (HB1, ASXL1, restriction endonuclease helix-turn helix domain). Just distal to this domain is a domain which has been shown to bind Calypso (the mammalian homologue of BAP1) and serve as a deubiquitinase for histone H2A lysine 119. This activity has been shown in vivo in Drosophila and in cell-free assays using human ASXL1 purified protein. This same domain has also been suggested to bind to HP1 proteins and LSD1 (this activity has never been studied in a hematopoietic context). Distal to these domains lies a conserved domain which has been suggested to physically interact with the retinoic acid receptor (another activity which has never been verified in hematopoietic cells). Finally, a plant homeofinger domain (PHD domain) lies at the extreme C-terminus of all ASXL proteins. The function of this PHD domain has not yet been identified.
All 3 ASXL family members are characterized by an amino-terminal homology domain and a C-terminal plant homeodomain (PHD domain) (Figure 3)33, 36, 37. Recent bioinformatic analysis of the conserved domains of mammalian ASXL proteins has suggested that the N-terminal domain of ASXL1 (amino acids 10–100) might represent a unique DNA binding motif, termed a HARE-HTH domain (HB1, ASXL1, restriction endonuclease helix-turn helix domain) (Figure 3)38. In addition, based on comparative analysis with other PHD-domain containing proteins, the PHD domain of ASXL proteins appeared unique and were predicted to potentially recognize internal methylated lysines on histone H3 tails as opposed to lysines on the N-terminal tail of histone H3. Further functional investigations of these domains will be needed to understand the role of these domains of ASXL1.
More recently, it was demonstrated that Drosophila Asx forms a complex with the chromatin deubiquitinase Calypso, which constributes the Polycomb-repressive deubiquitinase (PR-DUB) complex. The PR-DUB complex removes monoubiquitin from histone H2A at lysine 119 (Figure 3). The mammalian homologue of Calypso, BAP1, directly associates with ASXL1, and the mammalian BAP1-ASXL1 complex was shown to possess deubiquitinase activity in vitro 39.
The function of ASXL1 in hematopoiesis has not yet been fully delineated. A gene-trap hypomorphic mouse model of Asxl1 has been reported; however this germline allele results in significant perinatal lethality. A small number of surviving mice were analyzed for an overt, short-latency hematopoietic phenotype. This model, created by Fisher et al., placed a PGK promoter-drive neomycin expression cassette into exon 5 of ASXL1 interrupting the reading frame of ASXL1 (this allele is referred to as Asxl1tm1BC), which is predicted to lead to expression of a truncated protein lacking the nuclear interacting domains as well as the PHD domain37. Mice homozygous for this allele (Asxl1tm1BC /tm1BC) however, did not develop any overt hematologic malignancy in the first few months of age, nor did they observe defects in the number of multipotent progenitors. The homozygous Asxl1tm1BC /tm1BC allele in this study was associated with ~75% perinathal lethality and when the mice were backcrossed to a full C57BL/6J background complete embryonic lethality was observed. Thus, further investigation of the function of Asxl1 in hematopoiesis using a conditional knockout model for postnatal and hematopoietic-specific deletion is of importance to the field.
Clinical importance of ASXL1 and TET2 mutations in MPN patients
In contrast to the detailed clinical correlative studies of the effects of the TET2 and ASXL1 mutations on survival and response to therapy amongst patients with AML and MDS, comparatively fewer and smaller studies have been published thus far in MPN patients. Amongst patients with MPNs, ASXL1 mutations are enriched amongst patients with more advanced age as well as in patients with PMF, and post-PV/ET MF compared with PV or ET29, 40, 41 (Table 1). Brecqueville et al. have recently reported that ASXL1 mutant PMF patients have a significantly worsened overall survival compared with their wildtype counterparts42. However, this was a modest study with 9 ASXL1 mutant patients versus 35 wildtype patients and requires detailed study in much larger patient cohorts.
So far few clear clinical associations or correlates have been identified for TET2 mutant MPN patients compared with wildtype counterparts. TET2 mutations cluster roughly equally amongst patients with classic MPNs15 (Table 1). The largest study of the clinical impact of TET2 mutations on outcome amongst MPN patients was performed by Tefferi et al. who did not identify an impact of TET2 mutations on survival or leukemic transformation in a cohort of 89 PV patients and 60 PMF patients15, 43. TET2 mutations were significantly associated with the development of a hemoglobin <10g/dL in patients with PMF.
From the current literature, neither mutations in TET2 nor ASXL1 appear to be increase the risk of developing leukemic transformation in patients with chronic phase MPNs40, 44. However, reports from 4 groups have identified that a substantial subset of MPN patients who transform to AML acquire TET2 mutations in the leukemic state whereas TET2 mutations were not present in the MPN state12, 13, 17, 44. In contrast, analysis of paired samples from the chronic and leukemic state have revealed that ASXL1 mutations are not enriched in the leukemic state compared with the MPN phase of disease40. This data suggests that mutations in ASXL1 may be critical for MPN initiation, and may represent an early event in the clonal evolution of MPNs, in contrast for the more pleiotropic role of TET2 mutations in MPN initiation and progression.
Conclusions
The identification and characterization of mutations in TET2 and ASXL1 have provided important insights into the pathogenesis of MPNs and cancer biology in general. Mutations in TET2 have been recently discovered to be important components in the dynamic regulation of DNA methylation and appear to be valuable biomarkers in prognostication of patients with normal karyotype AML45, 46. Likewise, mutations in ASXL1 predict for worsened outcome in MDS patients47, even in the absence of currently clinically utilized clinical outcome predictors. Despite these important insights, many unresolved questions regarding the biological and clinical importance of these alterations still exist. For example, the prognostic importance of mutations in TET2 and ASXL1 in MPN patients is not clear. Given the relative rarity of these mutations in many chronic phase MPN patients, larger sequencing studies with comprehensive mutational data and pristine clinical annotation are urgently needed. Moreover, further functional studies to understand the effects of these alterations in combination with JAK2 and MPL mutations are needed to better understand the biological contribution of these alterations to MPN phenotype, outcome, and therapeutic response. Lastly, in vitro and in vivo studies in model systems and patient cohorts are needed to address the impact of mutations in epigenetic modifiers on the response to MPN therapies including hydroxyurea, interferon, and JAK2-targeted therapy.
Summary of Important Points and Objectives for Recall.
Mutations in TET2 are loss-of-function mutations present in 9–16% of patients with polycythemia vera (PV), 4–5% of patients with essential thrombocytosis (ET), and 7–17% of patients with primary myelofibrosis (PMF) or myelofibrosis arising from PV or ET.
TET2 is a member of the TET family of α-ketoglutarate and Fe(II)-dependent dioxygenases which oxidize 5-methylcytosine on DNA to produce 5-hydroxymethylcytosine followed by 5-formylcytosine and 5-carboxylcytosine. This enzymatic activity of TET2 is thought to facilitate demethylation of DNA.
ASXL1 is the mammalian homologue of Drosophila Addition of Sex Combs, a protein known to affect both Trithorax group and Polycomb group gene function. Although a number of functions have been ascribed to ASXL1 in non-mammalian and non-hematopoietic cell contexts, the function of ASXL1 in mammalian hematopoietic cells is not yet fully delineated.
Mutations in ASXL1 are most common amongst MPN patients with PMF (13–26%) or post-PV/ET MF (22–38%) as compared with patients with PV (2–5%) or ET (5–10%).
The clinical importance of TET2 and ASXL1 mutations amongst patients with MPNs is not yet clear. ASXL1 mutations may confer worsened overall survival amongst patients with PMF/post-PV/ET MF but this needs to be validated in larger, prospective studies.
Footnotes
Disclosures: The authors have no relationships to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dameshek W. Some speculations on the myeloproliferative syndromes. Blood. 1951;6(4):372–375. [PubMed] [Google Scholar]
- 2.Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 3.James C, Ugo V, Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434(7037):1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 4.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 5.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7(4):387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Scott LM, Tong W, Levine RL, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356(5):459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pikman Y, Lee BH, Mercher T, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardanani A, Lasho T, Finke C, et al. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia. 2010 doi: 10.1038/leu.2010.163. [DOI] [PubMed] [Google Scholar]
- 9.Oh ST, Simonds EF, Jones C, et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116(6):988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kralovics R, Stockton DW, Prchal JT. Clonal hematopoiesis in familial polycythemia vera suggests the involvement of multiple mutational events in the early pathogenesis of the disease. Blood. 2003;102(10):3793–3796. doi: 10.1182/blood-2003-03-0885. [DOI] [PubMed] [Google Scholar]
- 11.Beer PA, Jones AV, Bench AJ, et al. Clonal diversity in the myeloproliferative neoplasms: independent origins of genetically distinct clones. Br J Haematol. 2009;144(6):904–908. doi: 10.1111/j.1365-2141.2008.07560.x. [DOI] [PubMed] [Google Scholar]
- 12.Campbell PJ, Baxter EJ, Beer PA, et al. Mutation of JAK2 in the myeloproliferative disorders: timing, clonality studies, cytogenetic associations, and role in leukemic transformation. Blood. 2006;108(10):3548–3555. doi: 10.1182/blood-2005-12-013748. [DOI] [PubMed] [Google Scholar]
- 13.Theocharides A, Boissinot M, Girodon F, et al. Leukemic blasts in transformed JAK2-V617F-positive myeloproliferative disorders are frequently negative for the JAK2-V617F mutation. Blood. 2007;110(1):375–379. doi: 10.1182/blood-2006-12-062125. [DOI] [PubMed] [Google Scholar]
- 14.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41(7):838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 15.Tefferi A, Pardanani A, Lim KH, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23(5):905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114(1):144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couronne L, Lippert E, Andrieux J, et al. Analyses of TET2 mutations in post-myeloproliferative neoplasm acute myeloid leukemias. Leukemia. 2009 doi: 10.1038/leu.2009.169. [DOI] [PubMed] [Google Scholar]
- 18.Jankowska AM, Szpurka H, Tiu RV, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tefferi A, Levine RL, Lim KH, et al. Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23(5):900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tefferi A, Lim KH, Abdel-Wahab O, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23(7):1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorsbach RB, Moore J, Mathew S, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17(3):637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 22.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer LM, Tahiliani M, Rao A, et al. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8(11):1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Cai X, Cai C, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011 doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko M, Bandukwala HS, An J, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108(35):14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quivoron C, Couronne L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Gelsi-Boyer V, Trouplin V, Adelaide J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 29.Carbuccia N, Murati A, Trouplin V, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23(11):2183–2186. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- 30.Carbuccia N, Trouplin V, Gelsi-Boyer V, et al. Mutual exclusion of ASXL1 and NPM1 mutations in a series of acute myeloid leukemias. Leukemia. 2009 doi: 10.1038/leu.2009.218. [DOI] [PubMed] [Google Scholar]
- 31.Gaebler C, Stanzl-Tschegg S, Heinze G, et al. Fatigue strength of locking screws and prototypes used in small-diameter tibial nails: a biomechanical study. J Trauma. 1999;47(2):379–384. doi: 10.1097/00005373-199908000-00030. [DOI] [PubMed] [Google Scholar]
- 32.Sinclair DA, Milne TA, Hodgson JW, et al. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development. 1998;125(7):1207–1216. doi: 10.1242/dev.125.7.1207. [DOI] [PubMed] [Google Scholar]
- 33.Cho YS, Kim EJ, Park UH, et al. Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J Biol Chem. 2006;281(26):17588–17598. doi: 10.1074/jbc.M512616200. [DOI] [PubMed] [Google Scholar]
- 34.Lee SW, Cho YS, Na JM, et al. ASXL1 represses retinoic acid receptor-mediated transcription through associating with HP1 and LSD1. J Biol Chem. 2010;285(1):18–29. doi: 10.1074/jbc.M109.065862. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Park UH, Yoon SK, Park T, et al. Additional sex comb-like (ASXL) proteins 1 and 2 play opposite roles in adipogenesis via reciprocal regulation of peroxisome proliferator-activated receptor {gamma} J Biol Chem. 2011;286(2):1354–1363. doi: 10.1074/jbc.M110.177816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher CL, Lee I, Bloyer S, et al. Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev Biol. 337(1):9–15. doi: 10.1016/j.ydbio.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher CL, Pineault N, Brookes C, et al. Loss-of-function Additional sex combs-like1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2009 doi: 10.1182/blood-2009-07-230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aravind L, Iyer LM. The HARE-HTH and associated domains: novel modules in the coordination of epigenetic DNA and protein modifications. Cell Cycle. 2012;11(1):119–131. doi: 10.4161/cc.11.1.18475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465(7295):243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdel-Wahab O, Manshouri T, Patel J, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70(2):447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdel-Wahab O, Pardanani A, Patel J, et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25(7):1200–1202. doi: 10.1038/leu.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brecqueville M, Rey J, Bertucci F, et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012 doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- 43.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 44.Beer PA, Delhommeau F, Lecouedic JP, et al. Two routes to leukemic transformation following a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2009 doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 45.Metzeler KH, Becker H, Maharry K, et al. ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood. 2011;118(26):6920–6929. doi: 10.1182/blood-2011-08-368225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]