Abstract
The huntingtin exon 1 proteins with a polyglutamine repeat in the pathological range (51 or 83 glutamines), but not with a polyglutamine tract in the normal range (20 glutamines), form aggresome-like perinuclear inclusions in human 293 Tet-Off cells. These structures contain aggregated, ubiquitinated huntingtin exon 1 protein with a characteristic fibrillar morphology. Inclusion bodies with truncated huntingtin protein are formed at centrosomes and are surrounded by vimentin filaments. Inhibition of proteasome activity resulted in a twofold increase in the amount of ubiquitinated, SDS-resistant aggregates, indicating that inclusion bodies accumulate when the capacity of the ubiquitin–proteasome system to degrade aggregation-prone huntingtin protein is exhausted. Immunofluorescence and electron microscopy with immunogold labeling revealed that the 20S, 19S, and 11S subunits of the 26S proteasome, the molecular chaperones BiP/GRP78, Hsp70, and Hsp40, as well as the RNA-binding protein TIA-1, the potential chaperone 14–3-3, and α-synuclein colocalize with the perinuclear inclusions. In 293 Tet-Off cells, inclusion body formation also resulted in cell toxicity and dramatic ultrastructural changes such as indentations and disruption of the nuclear envelope. Concentration of mitochondria around the inclusions and cytoplasmic vacuolation were also observed. Together these findings support the hypothesis that the ATP-dependent ubiquitin–proteasome system is a potential target for therapeutic interventions in glutamine repeat disorders.
INTRODUCTION
Huntington's disease (HD) is an inherited neurodegenerative disorder characterized by personality changes, motor impairment, and subcortical dementia (Harper, 1991). The disease is associated with selective neuronal cell death occurring mainly in the cerebral cortex and the striatum (Vonsattel et al., 1985). The mutation causing HD is a CAG repeat expansion located within exon 1 of the IT-15 gene encoding huntingtin, a ∼350 kDa protein of unknown function. The CAG repeat is translated into a polyglutamine (polyQ) sequence. In HD patients huntingtin is expressed with 38–182 glutamine residues, whereas in healthy individuals the same protein is synthesized with only 8–37 glutamine residues (Rubinsztein et al., 1996; Sathasivam et al., 1997). Thus, HD manifests itself only when a critical length of ∼37 glutamine residues (pathological threshold) is exceeded.
Perutz et al. (1994) proposed that polyQ chains that exceed a critical length of 41 residues form anti-parallel β-strands held together by hydrogen bonds (polar zippers). In vitro evidence in support of the polar zipper hypothesis has been presented. Scherzinger et al. (1999) demonstrated that HD exon 1 proteins with a polyQ tract in the pathological range (≥37 glutamines), but not in the normal range (20–32 glutamines), form SDS-insoluble protein aggregates with a fibrillar morphology. Other laboratories arrived at similar results (Cooper et al., 1998; Lunkes and Mandel, 1998; Martindale et al., 1998; Kazantsev et al., 1999). Thus, similar to the in vivo observations, a polyQ repeat in the pathological range (≥37 glutamines) is critical for the formation of huntingtin protein aggregates in vitro and in cell culture model systems.
The accumulation of insoluble polyQ-containing protein aggregates in intranuclear and perinuclear inclusions has also been detected in brains of HD transgenic mice (Davies et al., 1997) and HD patients (DiFiglia et al., 1997). These findings have led to the hypothesis that HD as well as the related glutamine repeat disorders spinal bulbar muscular atrophy, dentatorubral pallidoluysian atrophy, and the spinocerebellar ataxia types 1, 2, 3, 6, and 7 (for review, see Paulson, 1999) are caused by the accumulation of insoluble protein aggregates in neuronal inclusions; however, to this day it is still unclear whether the formation of inclusion bodies causes dysfunction and neurodegeneration, or whether it is merely a defense mechanism to protect neuronal cells from the toxicity of misfolded proteins. In support of the second possibility, Saudou et al. (1998) and Klement et al. (1998) presented evidence that the formation of inclusion bodies with aggregated polyQ-containing protein is nontoxic or even beneficial for neuronal cells. In strong contrast to these findings, other investigators have demonstrated that formation of protein aggregates correlates with disease progression and the development of neuronal symptoms (Davies et al., 1997; Ona et al., 1999; Yamamoto et al., 2000). Very recently, using a conditional mouse model of HD, Yamamoto et al. (2000) showed that expression of mutant HD exon 1 protein results in inclusion body formation and progressive motor dysfunction. Blockage of HD exon 1 expression in symptomatic mice led to disappearance of the inclusions and the behavioral phenotype. Thus, inclusion body formation and disease progression appear to be clearly linked. Furthermore, the development of an HD-like pathology is dependent on the continuous expression of a truncated huntingtin protein with a polyQ repeat in the pathological range.
Immunohistochemical and ultrastructural studies have shown that the aggregated huntingtin protein in neuronal inclusions of HD transgenic mice and patients is ubiquitinated (Davies et al., 1997; DiFiglia et al., 1997). These findings suggest that the mutant huntingtin protein has been marked for degradation by the ubiquitination machinery but that it is apparently resistant to degradation. Degradation of most proteins by the proteasome requires the conjugation of multiple ubiquitin molecules (Bonifacino and Weissman, 1998). Ubiquitinated proteins are then recognized and hydrolyzed by the 26S proteasome (Voges et al., 1999). The 26S proteasome is composed of two major subcomplexes: the 20S proteasome, a barrel-shaped multicatalytic protease, and the 19S (PA700) regulatory complex, which associates with the 20S proteasome. The 19S regulatory complex is required for the recognition of ubiquitinated proteins. In addition to the 19S complex, a second regulator of the 20S proteasome has been described. This ring-shaped structure was termed 11S or PA28 and binds to the 20S proteasome in an orientation similar to that of 19S. The 11S subcomplex is mainly required for the degradation of short peptides rather than large ubiquitinated proteins.
Cummings et al. (1998) showed that ubiquitin-positive nuclear inclusions in neurons of spinocerebellar ataxia type 1 patients and transgenic mice stain positively for the 20S proteasome and the molecular chaperone HDJ-2/HSDJ, indicating that subcomplexes of the 26S proteasome as well as heat shock proteins are redistributed to the sites of ataxin-1 protein aggregation. These results were confirmed in cell culture, transgenic mouse as well as fly model systems, using different polyQ-containing proteins (Chai et al., 1999; Stenoien et al., 1999; Warrick et al., 1999). Together these findings suggest that the redistribution of the proteasomal machinery and molecular chaperones to polyQ-containing protein aggregates is a natural response of cells to remove misfolded aggregation-prone proteins.
A general relevance of the ubiquitin–proteasome pathway with regard to the degradation of misfolded proteins has been proposed. Ward et al. (1995) showed that degradation of wild-type and mutant cystic fibrosis transmembrane conductance regulator (CFTR) is blocked by specific proteasome inhibitors resulting in the accumulation of polyubiquitinated forms of CFTR. Immunofluorescence and electron microscopy also revealed that CFTR molecules aggregate in distinct perinuclear inclusions (Johnston et al., 1998). The formation of these structures, termed “aggresomes,” could be a general response of cells that occurs whenever the degradative capacity of the proteasome is exceeded. Recently, Wigley et al. (1999) found that under normal conditions the components of the 26S proteasome, as well as ubiquitin and heat shock proteins, are concentrated at the centrosome. Interestingly, this structure enlarges in response to inhibitors of proteasome activity, and additional pools of chaperones, ubiquitin, and proteasomal subcomplexes are recruited into the inclusion body. Together these findings suggest that mammalian cells contain a specific organelle located at the centrosome that is specialized in the degradation of misfolded proteins. Disease proteins containing a polyglutamine tract in the pathological range may resist the natural degradation in this organelle or disrupt the ubiquitin–proteasome system, or both.
In this study we have examined the cellular response to the formation of polyQ-containing huntingtin aggregates in 293 Tet-Off cells. We found that besides the polyQ repeat-length, aggregation of huntingtin protein in mammalian cells is critically dependent on the proteasomal activity. Inhibition of the activity of the 26S proteasome significantly enhanced the accumulation of mutant HD exon 1 protein aggregates. Our data also show that huntingtin aggregation leads to distinct perinuclear inclusion bodies that are structurally very similar to CFTR aggresomes (Johnston et al., 1998). Accumulation of insoluble huntingtin aggregates is toxic for mammalian cells and results in the redistribution of several cellular factors, such as stress proteins and components of the proteasome system to the inclusion bodies. Taken together, our data support the hypothesis that inclusion bodies with aggregated huntingtin protein accumulate in mammalian cells mainly because the natural proteasome system is unable to degrade the expressed mutant huntingtin protein.
MATERIALS AND METHODS
Antibodies
The following antibodies were used for immunofluorescence analysis: rabbit polyclonal CAG53b IgG (diluted 1:250; Davies et al., 1997), rabbit polyclonal HD1 antibody (diluted 1:2000; Scherzinger et al., 1997), rabbit polyclonal 14–3-3 antibody (diluted 1:300; this study), rabbit polyclonal α-synuclein antibody (diluted 1:150; this study), mouse monoclonal γ-tubulin antibody (diluted 1:100; Sigma, St. Louis, MO), mouse monoclonal vimentin antibody (clone V9, diluted 1:20; Sigma), mouse monoclonal Flag antibody (diluted 1:2000; Sigma), rabbit polyclonal antibody to 11S regulator subunit α (PA28α, diluted 1:250; Affiniti Research Products, Exeter, United Kingdom), rabbit polyclonal antibody to 19S regulator (nonATPase S10a, diluted 1:80; Affiniti Research Products), rabbit polyclonal antibody to 20S proteasome core (diluted 1:300; Affiniti Research Products), mouse IgG2a BiP/GRP78 antibody (diluted 1:500; Transduction Laboratories, Lexington, KY), goat polyclonal antibody to Hsp40 (C-20, diluted 1:300; Santa Cruz Biotechnology, Santa Cruz, CA), goat polyclonal antibody to Hsp70 (K-20, diluted 1:300; Santa Cruz Biotechnology), mouse monoclonal antibody to TIA-1 (ML-29, IgG1, 1:200; kindly supplied by Dr. N. Kedersha, Division of Rheumatology and Immunology, Brigham and Women's Hospital, Boston), rabbit polyclonal antibody to α-mannosidase II (diluted 1:100; kindly supplied by Kelley Moremen, University of Georgia, and Marilyn G. Farquhar, University of California, San Diego), secondary donkey anti-mouse or anti-rabbit IgG conjugated to either CY3 or FITC (diluted 1:200; Jackson ImmunoResearch, West Grove, PA), and secondary donkey anti-goat IgG conjugated to Alexa 546 (diluted 1:200; Molecular Probes, Eugene, OR).
Preparation of Antibodies
A His6-tagged 14–3-3 fusion protein was generated by inserting the cDNA coding for amino acids 94–255 of the epsilon isoform of 14–3-3 (Accession No. U54778) into the bacterial pQE-32 expression vector (Qiagen, Hilden, Germany). Similarly, cDNA coding for amino acids 1–142 (Accession No. AI739317) of α-synuclein was cloned into pQE-32. The respective fusion proteins were expressed in Escherichia coli, affinity purified under denaturing conditions on Ni-NTA agarose, and injected into rabbits. The resulting immune sera were used in immunofluorescence studies.
Plasmid Construction
The pTet-CMV-Hyg plasmid used for subcloning of HD exon 1 fragments was generated from the pTetCMV-F° vector (Wu and Chiang, 1996) as follows. First, a new multiple cloning site with recognition sites for NdeI, BamHI, SphI, SalI, EcoRV, NotI, and PstI was ligated 3′-terminal to the Flag-tag of the pTetCMV-F° vector. Second, the hygromycin gene together with the HSV TK promotor was PCR amplified from pTK-Hyg (Clontech, Palo Alto, CA) and inserted counterclockwise into the XhoI site of modified pTetCMV-F°. HD exon 1 fragments with 20, 51, and 83 CAG repeats were excised from pCAG20, pCAG51, and pCAG83 with BamHI and SalI (Scherzinger et al., 1997) and subcloned into the BamHI and SalI sites of pTet-CMV-Hyg yielding pTetCMV-Hyg-CAG20, -CAG51, and -CAG83, respectively.
Generation of Stable, Inducible Cell Lines
A premade 293 Tet-Off cell line purchased from Clontech was transfected with 10 μg of pTetCMV-Hyg-CAG20, -CAG51, and -CAG83 using the calcium phosphate method. The vector pTetCMV-Hyg was used as a control. Selection of stable cell lines was initiated 4 d after transfection using 150 μg/ml hygromycin. Fourteen days after transfection, several colonies were isolated and cultured in 24-well plates and then transferred into 12-well and 6-well plates and finally into 25 cm2-T flasks. Transgene expression of induced and noninduced cells was verified by Western blot analysis and immunofluorescence using antibodies directed against huntingtin (HD1 and CAG53b) and the Flag-tag.
Culturing of Cell Lines
Stable transfected 293 Tet-Off cells were grown in l-glutamine–free MEM with Earle's Salts (Life Technologies, Gaithersburg, MD) and supplemented with 10% FBS, 2 mM l-glutamine, 100 IU/ml penicillin plus 100 μg/ml streptomycin, 100 μg/ml G418, 150 μg/ml hygromycin B, and 10 ng/ml doxycycline in poly-l-lysine–coated cell culture flasks. Expression was induced by thoroughly washing the cells with PBS and adding fresh medium lacking doxycycline. The medium was changed on the following day to remove the doxycycline completely. Expression was induced for different times as indicated in each individual experiment.
For inhibition of proteasome activity 24 h after induction of protein expression, cell cultures were adjusted to 10 μM clasto-lactacystin β-lactone (Affiniti Research Products) and incubated for an additional 48 h. Control cells were incubated with an equivalent amount of the solvent DMSO. The cell culture medium and the proteasome inhibitor were replaced every day.
Preparation of Protein Extracts
Cells were washed, scraped in ice-cold PBS, and pelleted. Cell lysis was performed on ice for 30 min in 50 mM Tris-HCl (pH 8.8), 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, and protease inhibitors (2 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 1 μg/ml aprotinin, and 50 μg/ml antipain). Insoluble material was pelleted by centrifugation at 14,000 rpm for 10 min at 4°C. The pellet was resuspended in 100 μl 20 mM Tris-HCl (pH 8.0)/15 mM MgCl2 containing 0.5 mg/ml DNAseI and incubated for 1 h at 37°C. Protein of the supernatant and pellet fraction was determined using the Bradford protein assay (Bio-Rad, Munich, Germany).
For preparation of whole-cell extracts, cells were washed, scraped in ice-cold PBS, and pelleted. Lysis was performed on ice for 30 min in a buffer containing 50 mM Tris-HCl (pH 8.8), 100 mM NaCl, 5 mM MgCl2, 0.5% NP-40, protease inhibitors (as described above), and 250 U/ml benzonase. Protein concentration was determined using the Bradford protein assay.
Western Blot Analysis and Filter Retardation Assay
Proteins were denatured, reduced, separated by SDS-PAGE (12.5%), and transferred to nitrocellulose according to standard procedures. Membranes were blocked with 3% nonfat dry milk in PBS containing 0.05% Triton X-100 and incubated with the anti-huntingtin antibody HD1 (diluted 1:1000) or an anti-ubiquitin antibody (diluted 1:500), respectively. The secondary antibody was a peroxidase-conjugated anti-rabbit antibody. Immunoreactive protein was detected by using enhanced chemoluminescence (ECL, Boehringer Mannheim, Mannheim, Germany). For the filter retardation assay (Scherzinger et al., 1997; Wanker et al., 1999), protein extracts were heated at 98°C for 3 min in 2% SDS and 50 mM DTT and filtered through a 0.2-μm cellulose acetate membrane (Schleicher & Schuell, Dassel, Germany) using a BRL dot-blot filtration unit. Filters were processed for immunodetection as described above, using an alkaline–phosphatase-conjugated anti-rabbit secondary antibody and the fluorescent substrate AttoPhos (JBL Scientific, San Luis Obispo, CA). The relative amount of captured aggregate was evaluated with the Fuji-Imager LAS 2000 (Tokyo, Japan).
Immunofluorescence and Electron Microscopy
Induced cells were grown in leighton tubes (Costar, Cambridge, MA) for the indicated times. After cells were washed with PBS, they were fixed in 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100 for 15 min, and blocked with 3% BSA for 30 min. Cells were then incubated for 1 h at room temperature with the appropriate primary antibodies diluted in PBS, washed three times for 10 min with 0.1% Triton X-100, and incubated for 1 h at room temperature with the secondary antibodies diluted in PBS containing 3% goat serum. After the leightons were washed in 0.1% Triton X-100 and in PBS, cell nuclei were counterstained with Hoechst (Sigma). Samples were viewed with the fluorescence microscope Axioplan-2 (Zeiss, Thornwood, NY).
For electron microscopic analysis, cells were grown on poly-l-lysine–coated Thermanox coverslips (13 mm diameter; Nunc, Naperville, IL), and transgene expression of the HD exon1 proteins was induced for 3 d. For embedding in LR Gold Resin (London Resin Company, Berkshire, United Kingdom), coverslips with cells were washed in PBS and fixed for 1 h in a mixture of 1% formaldehyde and 0.2% glutaraldehyde. After dehydration in an ethanol series, cells were infiltrated with LR Gold as described by suppliers. Polymerization was performed under visible light for 3 d in the presence of benzil. For embedding in Spurr's resin, cells were fixed in 2% glutaraldehyde, treated for 1 h with 2% osmium tetroxide, and dehydrated in an acetone series. After gradual transfer from 100% acetone to pure resin, polymerization was performed at 60°C overnight. For polymerization, coverslips were placed upside down on top of resin-filled gelatin capsules in both cases.
For immunolabeling, 60-nm sections on nickel grids were incubated for 10 min in buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl), followed by another 10 min of incubation in buffer B (buffer A containing 6 mg/ml Aurion BSA-c, Wageningen, The Netherlands). Antibodies against HD1 (1:400), ubiquitin (1:30), or 14–3-3 (1:400) were added to buffer B and incubated for 2 h at room temperature. After four 10 min washes in buffer B, secondary antibody conjugated with 10 nm gold (1:100; British Bio Cell, Cardiff, United Kingdom) was applied in buffer B for 2 h at room temperature. After extensive washing in buffer A, sections were post-stained for 1 min with 0.5% uranyl acetate and for 45 s with lead citrate and then viewed in a Philips CM100 EM (Philips Electron Optics, Eindhoven, The Netherlands).
Cell Viability Assay
Cell viability was measured by cleavage of the yellow tetrazolium salt XTT to form an orange formazan dye by metabolic active cells (Roche Molecular Biochemicals, Mannheim, Germany). One day before measuring XTT cleavage, cells were plated in 96-well plates at a density of ∼7000 cells per well. XTT was added 4 d after induction of HD exon 1 expression following the manufacturer's protocol. After 5 h of incubation at 37°C, the optical density was measured at 450 nm with a reference wave length set at 690 nm. For each cell line examined, nine independent values were measured and evaluated statistically.
RESULTS
Expression of Mutant HD Exon 1 Proteins in 293 Tet-Off Cells Results in Aggregate Formation
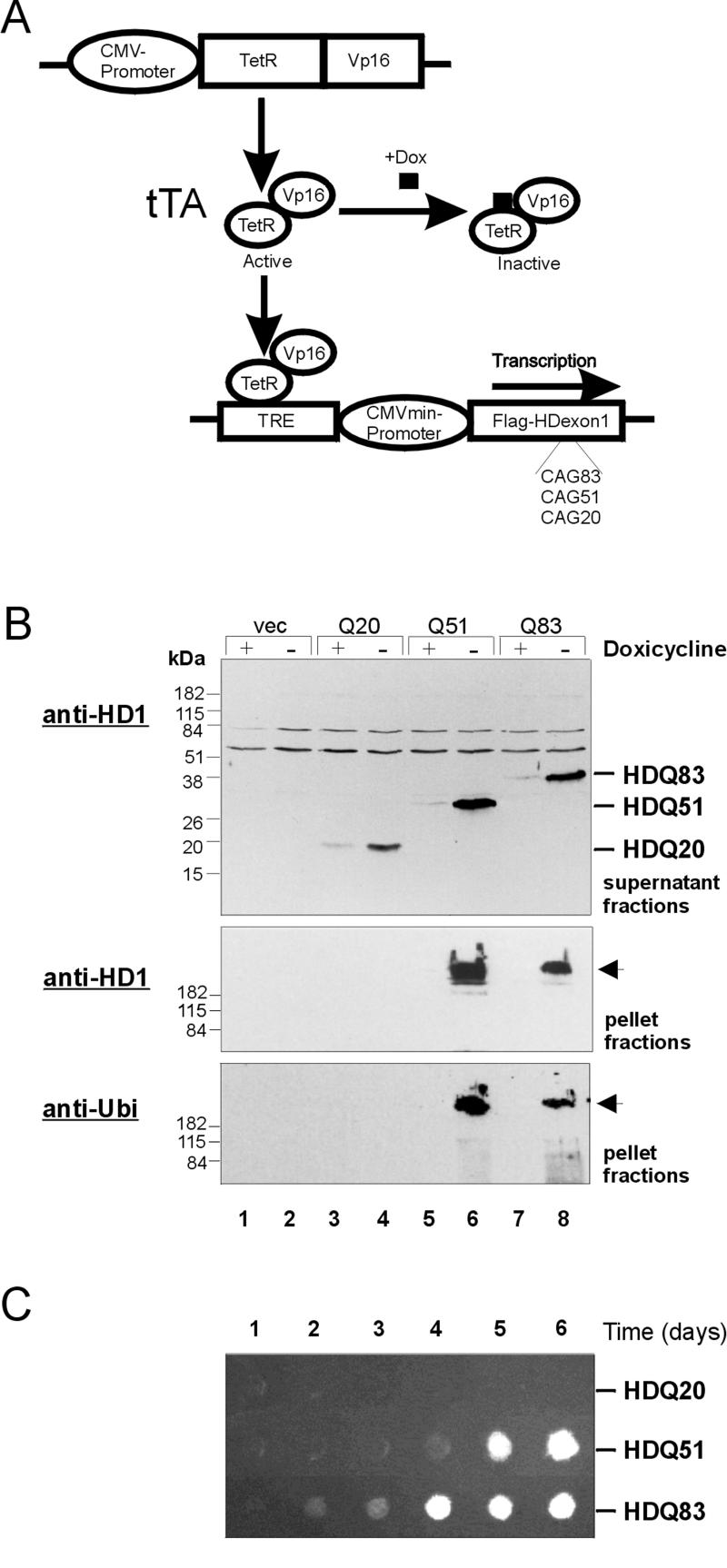
The tetracycline (tet)-regulated system (Gossen and Bujard, 1992) has been used for the expression of Flag-tagged HD exon 1 proteins with 20 (HDQ20), 51 (HDQ51), and 83 (HDQ83) glutamines in 293 Tet-Off cell lines (Figure 1A). Regulation of this system is achieved through a tet-regulated transactivator, a fusion protein consisting of the tet-repressor, and a VP16 activation domain. This hybrid protein binds specifically to a tetracycline-responsive DNA element TRE and promotes transcription from the adjacent CMV promoter. Tetracycline and its analogues such as doxycycline can bind to the transactivator and thereby prevent the hybrid protein from binding to the TRE element. Thus, if doxycycline is present in the cell culture medium, transcription of recombinant HD exon 1 constructs in 293 Tet-Off cells is inhibited, whereas in its absence HD exon 1 expression is induced.
Figure 1.
Cell model design and analysis of HD exon 1 protein aggregation in 293 Tet-Off cells. (A) Activation of the transactivator tTA, a fusion protein consisting of the tet-repressor and the VP16 activation domain, by removal of doxycycline (Dox) from the culture medium results in the binding of tTA to the tet-responsive element TRE and transcription of Flag-tagged HD exon 1 fragments with 20, 51, and 83 CAG repeats in 293 Tet-Off cells. (B) Immunoblot analysis of soluble (supernatant) and insoluble (pellet) protein fractions prepared from induced and uninduced 293 Tet-Off cells. For induction of HD exon 1 expression, cells were grown for 3 d in the absence of doxycycline. Proteins were resolved by SDS-PAGE, blot ted onto nitrocellulose membranes, and probed with the anti-huntingtin antibody HD1 (top and middle panel) or with anti-ubiquitin antibody (bottom panel). Lanes 1 and 2: protein fractions prepared from induced and uninduced control cells lacking the HD exon 1 construct; lanes 3, 5, and 7: protein fractions prepared from uninduced cells; lanes 4, 6, and 8: protein fractions prepared from induced cells expressing the proteins HDQ20, HDQ51, and HDQ83, respectively. Arrows mark the origin of electrophoresis. (C) Time course of HD exon 1 aggregation in 293 Tet-Off cells. Expression of the recombinant proteins was induced by removal of doxycycline from the culture medium. Protein extracts were prepared at the indicated times after induction and analyzed by a filter retardation assay. Captured HD exon 1 protein aggregates were detected with the HD1 antibody.
293 Tet-Off cell lines were cultivated for 3 d in either the absence or presence of doxycycline. Whole-cell extracts were prepared, and after centrifugation the soluble (supernatant) and insoluble (pellet) fractions were analyzed by SDS-PAGE and Western blotting (Figure 1B). In the supernatant fractions from cells that have been grown in the absence of doxycycline, prominent bands migrating at 20, 35, and 45 kDa corresponding to the proteins HDQ20, HDQ51 and HDQ83, respectively, were detected by the anti-huntingtin antibody HD1 (lanes 4, 6, and 8). These bands were barely detected in the corresponding fractions of uninduced cells (lanes 3, 5, and 7). In supernatant fractions prepared from control cells lacking the HD exon 1 construct, recombinant HD exon 1 protein was not observed by immunoblotting using the HD1 antibody (lanes 1 and 2) in either the absence or presence of doxycycline. Insoluble protein aggregates that did not enter the stacking gel were detected in the pellet fractions prepared from cells expressing the proteins HDQ51 and HDQ83 (lanes 6 and 8), but not in the corresponding fraction of HDQ20-expressing cells (lane 4). These results confirm earlier results (Scherzinger et al., 1999) showing that a polyQ repeat in the pathological range is critical for the formation of aggregates in 293 Tet-Off cells. Gel-excluded HD exon 1 aggregates were also detected by an anti-Flag antibody and an antibody specifically directed against ubiquitin, indicating that they are ubiquitinated (Figure 1B).
The time course of HD exon 1 aggregation in the stable transfected 293 Tet-Off cell lines is shown in Figure 1C. Expression of HD exon 1 proteins was induced by removal of doxycycline from the culture medium. After incubation for various times (1–6 d), cell extracts were prepared and analyzed for the presence of insoluble HD exon 1 aggregates by a filter retardation assay. SDS-resistant HDQ83 aggregates were detected after a lag phase of 2–3 d, whereas under the same conditions the HDQ51 protein required 4–5 d to develop detectable high molecular weight aggregates. In induced cells expressing HDQ20, no SDS-insoluble protein aggregates were detected, even after an incubation of 6 d. These findings confirm our previous results using COS cells that HD exon 1 aggregation is polyglutamine-length dependent, and that a polyglutamine tract in the pathological range (≥37 glutamines) is required for the formation of high molecular weight protein aggregates in vitro (Scherzinger et al., 1999).
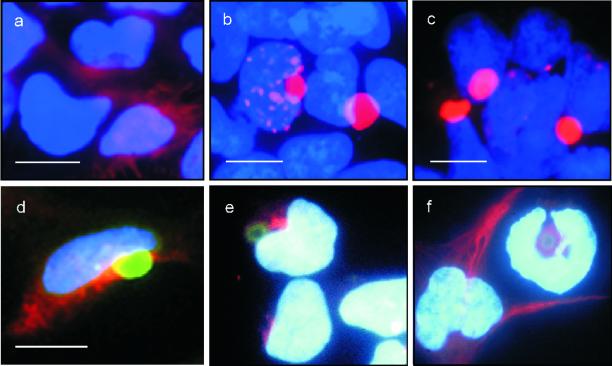
Aggregate formation in 293 Tet-Off cells was also assessed by indirect immunofluorescence microscopy (Figure 2). After induction of HD exon 1 expression for 3 d by removal of doxycycline from the culture medium, cells were immunolabeled with the anti-Flag antibody; nuclei were counterstained with Hoechst. Homogenous cytoplasmic staining was observed in cells expressing HDQ20 protein (Figure 2a). In sharp contrast, distinct cytoplasmic and nuclear inclusions containing aggregated huntingtin protein were detected in cells expressing the proteins HDQ51 or HDQ83 (Figure 2, b and c). In the vast majority (>80%) of cells, the HD exon 1 protein aggregates accumulated in the perinuclear region and appeared as spherical structures with a diameter of 1–3 μm. Interestingly, inclusion formation resulted in a change of the shape of the adjacent nuclei, suggesting that the accumulation of insoluble HD exon 1 protein aggregates demolishes the structure of the nuclei. Both nuclear and cytoplasmic inclusions reacted with the anti-Flag monoclonal antibody and also with the anti-ubiquitin antibody (our unpublished results).
Figure 2.
HD exon 1 aggregates form at the centrosome and result in the redistribution of the intermediate filament vimentin. (a–c) Expression of the proteins HDQ20, HDQ51, and HDQ83 was induced for 3 d by removal of doxycycline from the culture medium. Cells were processed for immunofluorescence using the anti-Flag antibody followed by the CY3-coupled secondary antibody. Nuclei were counterstained with Hoechst. Cells expressing the HDQ20 protein showed homogenous cytoplasmic staining (a), whereas in cells expressing the proteins HDQ51 (b) or HDQ83 (c), distinct inclusion bodies were observed. Immunofluorescence double labeling of cells reveals that FITC-labeled HDQ83 aggregates (green) partially colocalize with CY3-labeled γ-tubulin (red), a centrosomal marker (d), but not with CY3-labeled α-mannosidase II, a known Golgi marker (e). Induction of HDQ83 expression for 3 d also results in a redistribution of CY3-labeled vimentin to the perinuclear inclusion bodies (f). Note the ring-like appearance of vimentin fluorescence in cells containing HDQ83 protein aggregates. In comparison, in control cells lacking HDQ83 protein, the vimentin filaments extend to the cell periphery. Bar, 10 μm.
Cytoplasmic HD Exon 1 Protein Aggregates Colocalize with the Centrosome/Microtubule-organizing Center and Lead to Intracellular Redistribution of Vimentin
Johnston et al. (1998) showed that misfolded, ubiquitinated CFTR protein accumulates in characteristic perinuclear inclusions, which they termed aggresomes. Aggresomes form specifically at the centrosome and are surrounded by the intermediate filament vimentin. To examine whether the perinuclear HD exon 1 protein aggregates are related in structure to CFTR aggresomes, colocalization studies were performed using immunofluorescence microscopy. Comparison of the fluorescence of HDQ83 protein aggregates with the immunostaining of γ-tubulin, a known marker for the centrosome (Dictenberg et al., 1998), revealed that both structures colocalize at least partially (Figure 2d). On the other hand, the fluorescence of the HD exon 1 protein aggregates did not overlap with the immunostaining of α-mannosidase II, a marker for the Golgi apparatus (Figure 2e), indicating that the Golgi apparatus, which is also located in the perinuclear region (Velasco et al., 1993), is distinct from the HDQ83 protein aggregates. Accumulation of mutant HDQ83 protein in 293 Tet-Off cells also appeared to cause the reorganization of the intermediate filament vimentin. As shown in Figure 2f, vimentin filaments form a ring-like structure around the HDQ83 protein aggregates. In comparison, a branching cytoskeletal network that extends to the cell periphery was detected in control cells. Together, these findings indicate that the perinuclear inclusion bodies containing aggregated HDQ51 or HDQ83 protein are structurally similar to the aggresomes that have been described previously for CFTR (Johnston et al., 1998).
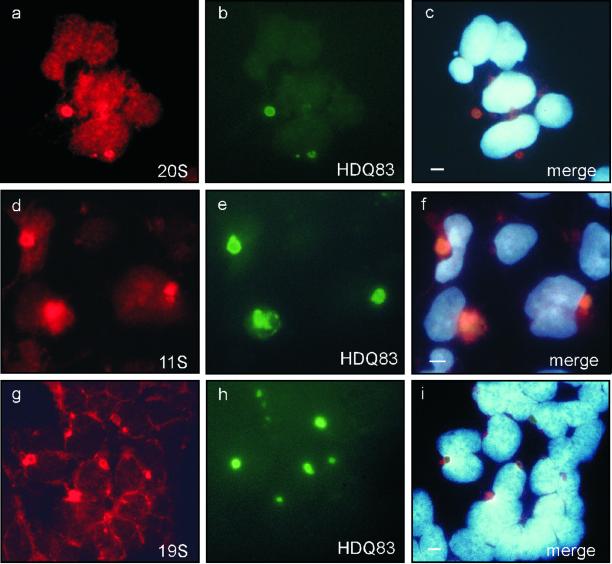
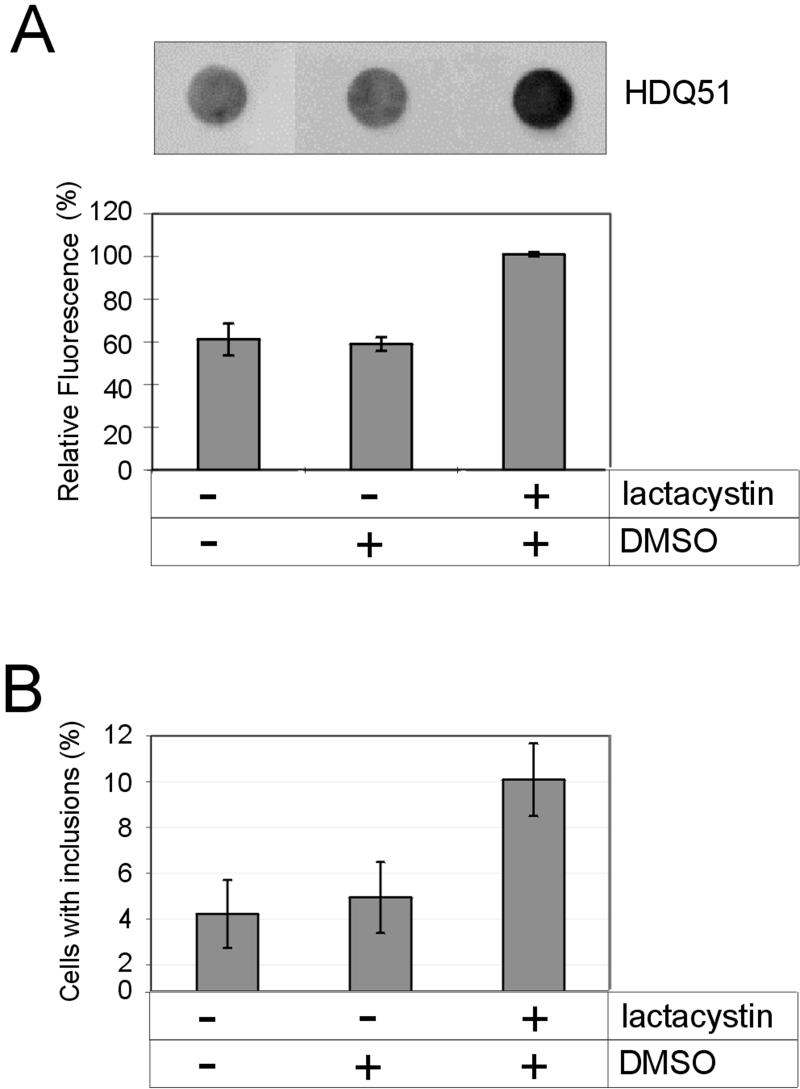
Colocalization of Proteasomal Components with HD Exon 1 Aggregates and Enhancement of Aggregate Formation in Response to Proteasome Inhibition
The proteasome has been generally implicated in the degradation of misfolded proteins (Kopito, 1997). To determine whether the key components of the 26S proteasome are redistributed to perinuclear inclusion bodies, induced 293 Tet-Off cells expressing HDQ83 protein were analyzed by immunofluorescence microscopy. Figure 3 shows that the proteasomal subcomplexes 20S, 19S, and 11S colocalize with the insoluble HDQ83 protein aggregates, suggesting that the 26S proteasome is directly involved in the degradation of aggregated HD exon 1 protein. To further test this hypothesis, cells expressing the HDQ51 protein were treated with the proteasome inhibitor clasto-lactacystin β-lactone, and the formation of SDS-insoluble HD exon 1 protein aggregates was analyzed using the filter retardation assay. The amount of HDQ51 aggregates in clasto-lactacystin β-lactone–treated cells was found to be roughly twofold higher than that of untreated cells or cells that had been treated with the solvent DMSO, indicating that proteasomal activity of the cell is critical for the accumulation of perinuclear polyQ-containing protein aggregates (Figure 4A). Essentially the same result was obtained when the number of cells with perinuclear inclusion bodies was determined by indirect immunofluorescence microscopy (Figure 4B).
Figure 3.
Colocalization of HDQ83 protein aggregates with the proteasomal subcomplexes 20S, 11S, and 19S. After induction of HDQ83 expression for 3 d, cells were double immunolabeled with antibodies directed against 20S/HDQ83 (a–c), 11S/HDQ83 (d–f), and 19S/HDQ83 (g–i). HDQ83 protein aggregates were immunolabeled with anti-Flag antibody coupled to a FITC-conjugated antibody (green); proteasomal subcomplexes were labeled with anti-20S, anti-11S, and anti-19S antibodies coupled to CY3 antibody (red). Cells were counterstained with Hoechst, allowing the detection of nuclei. Bar, 5 μm.
Figure 4.
The amount of HDQ51 aggregates in 293 Tet-Off cells increases in response to proteasome inhibition. (A) After induction of HDQ51 expression for 1 d, cells were treated for 48 h with lactacystin. Protein extracts were prepared as described in MATERIALS AND METHODS and analyzed by the filter retardation assay. Captured aggregates were detected by incubation with HD1 antibody, followed by incubation with alkaline phosphatase-conjugated anti-rabbit secondary antibody and the fluorescent substrate AttoPhos. The relative amount of aggregates for each sample was quantified on a Fuji-Imager (LAS2000). The dot with the highest signal intensity was arbitrarily set as 100. The data shown are representative of three independent experiments. (B) After induction of HDQ51 expression for 8 h, cells were treated for 24 h with lactacystin (10 μM). Formation of inclusion bodies was followed by indirect immunofluorescence microscopy using the CAG53b antibody. A total of ∼6000 lactacystin-treated and untreated cells were examined in each experiment.
The Endoplasmic Reticulum (ER) Resident Chaperone BiP/GRP78 Is Recruited into Perinuclear Inclusion Bodies
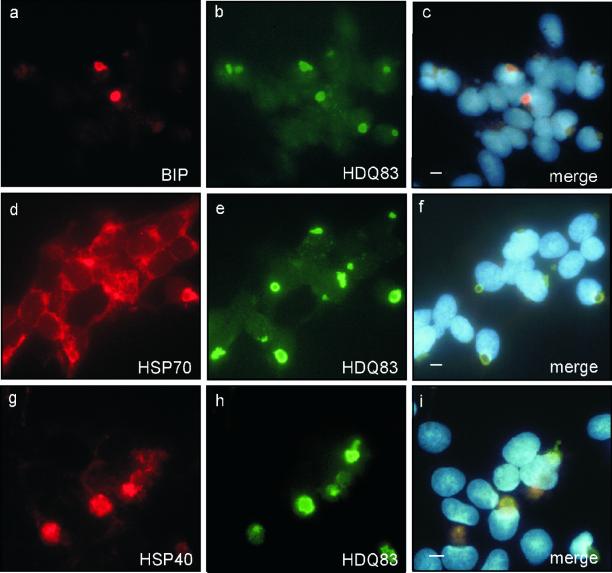
Chaperones are expected to play a protective role during polyQ aggregation in mammalian cells (Cummings et al., 1998; Warrick et al., 1999). Recently, we have shown in vitro that both Hsp40 and Hsp70 are effective in preventing the formation of SDS-insoluble polyQ-containing fibrillar structures (Muchowski et al., 2000). Here, we used indirect immunofluorescence to examine whether distinct classes of chaperones associate with insoluble HDQ83 aggregates in 293 Tet-Off cells. First, the colocalization of GRP78, an ER resident chaperone, with the perinuclear inclusion bodies was analyzed. In the ER, GRP78 associates with misfolded or incompletely assembled proteins, which are destined for proteolytic degradation (Voges et al., 1999). An association of this chaperone with cytoplasmic polyglutamine-containing protein aggregates has not been described previously. Unexpectedly we found that GRP78 colocalizes with the core of the HDQ83 inclusion bodies (Figure 5, a–c). In a typical experiment, 40–50% of the inclusion bodies examined contained GRP78. Colocalization between GRP78 and inclusion bodies was detected at an early stage of the HDQ83 aggregation process. Thus, 2 d after induction of HDQ83 expression, when the first perinuclear inclusion bodies could just be detected by immunofluorescence microscopy, colocalization between GRP78 and HDQ83 inclusion bodies was apparent. This suggests that recruitment of GRP78 into HD exon 1 aggregates belongs to the earliest responses of cells in their fight to prevent the accumulation of misfolded protein. In comparison, at this stage colocalization between HDQ83 aggregates and the cytoplasmic chaperones Hsp70 and Hsp40 was barely detectable. A significant enrichment of Hsp70 and Hsp40 at the inclusion bodies was not detected before 5–7 d after induction of HDQ83 expression, indicating that the recruitment of these cytoplasmic chaperones is a late event in the process of inclusion formation (Figure 5, d–i).
Figure 5.
Chaperones are recruited into HD exon 1 aggregates. HDQ83 expression was induced by removal of doxycycline from the culture medium. Three days after induction of HDQ83, expression cells were double immunolabeled with antibodies directed against BiP and HDQ83 (a–c). Double immunolabeling of cells with antibodies directed against Hsp70/HDQ83 (d–f) and Hsp40/HDQ83 (g–i) was performed on day 6 after induction of HDQ83 expression. HDQ83 protein aggregates were immunolabeled with anti-CAG53b coupled to FITC-conjugated secondary antibody (green); chaperones were labeled with antibodies directed against BiP, Hsp70, and Hsp40 coupled to a CY3-conjugated secondary antibody (red). Nuclei were counterstained with Hoechst. Colocalization of HDQ83 aggregates with BiP, Hsp70, and Hsp40 is shown in c, f, and i, respectively. Bar, 5 μm.
Colocalization of TIA-1, 14–3-3ε, and α-Synuclein with Perinuclear Inclusions
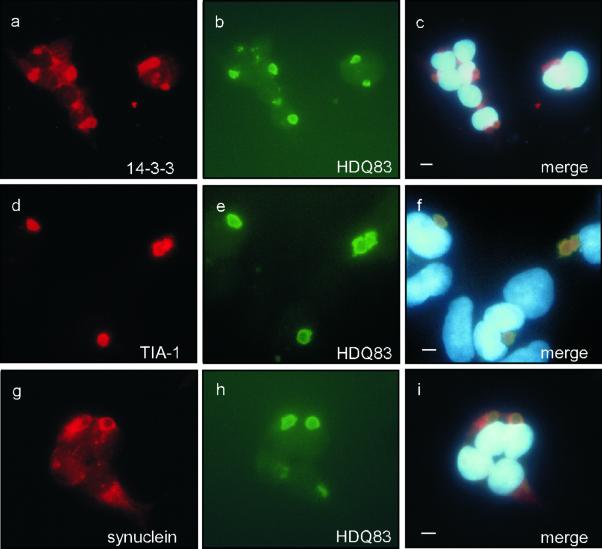
To assess whether in addition to chaperones and components of the ubiquitin–proteasome system other cellular factors are recruited into inclusion bodies, a number of different antibodies were tested by indirect fluorescence microscopy. Two to 3 d after induction of HDQ83 expression, the RNA binding protein TIA-1 (Kedersha et al., 1999), the potential chaperone 14–3-3ε (Shaw, 2000), and α-synuclein (Souza et al., 2000) were found to be present in perinuclear inclusion bodies (Figure 6), suggesting that recruitment of these proteins into HD exon 1 aggregates belongs to an early response of a cell to remove the misfolded protein. Five to 7 d after induction of HDQ83 expression, colocalization of amyloid-β (Masters et al., 1985) and presenilin 1 (Sherrington et al., 1995) with inclusion bodies could also be detected, indicating that the accumulation of amyloid-β and presenilin-1 are relatively late events in the process of inclusion body formation (our unpublished results).
Figure 6.
Colocalization of endogenous 14–3-3, TIA-1, and α-synuclein with HD exon 1 aggregates. Expression of HDQ83 was induced for 3 d by removal of doxycycline from the culture medium. Cells were double immunolabeled with antibodies recognizing 14–3-3/HDQ83 (a–c), TIA-1/HDQ83 (d–f), and α-synuclein/HDQ83 (g–i). HDQ83 aggregates were immunolabeled with anti-Flag antibody coupled to FITC-conjugated antibody (a–c, g–i; green) or CAG53b antibody coupled to FITC-conjugated antibody (d–f). The proteins 14–3-3, TIA-1, and α-synuclein were detected with antibodies coupled to a CY3-conjugated antibody (red). Nuclei were stained with Hoechst. Bar, 5 μm.
Ultrastructure of Perinuclear Inclusions
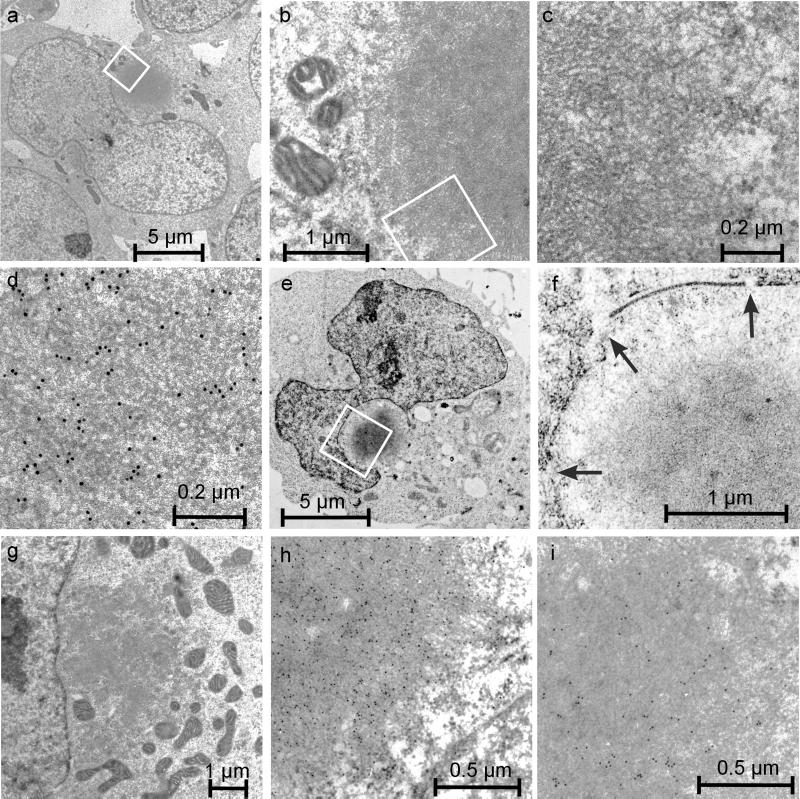
Cells of transgenic 293 Tet-Off lines were incubated for 2–5 d in the absence of doxycycline and then analyzed by transmission electron microscopy. Although cells expressing HDQ20 appeared normal, in cells expressing the mutant HD exon 1 proteins, characteristic morphological changes were observed. At the ultrastructural level, cells expressing HDQ83 contained relatively large membrane-free, spherical structures (diameter ∼1–5 μm) composed of electron-dense filamentous material (Figure 7, a and b). At higher magnification it was possible to determine that the fibrous region consists of individual filaments with a diameter of ∼10 nm (Figure 7, c and d). These filaments are reminiscent of HD exon 1 fibrils that have been produced previously in vitro (Scherzinger et al., 1997) as well as in HD transgenic mouse models (Davies et al., 1997). The identity of the HDQ83 filaments was confirmed by immunoelectron microscopy using the anti-huntingtin antibody HD1 and a gold colloid secondary antibody. At a higher magnification, decoration of the 10 nm fibrils with 10 nm gold particles is evident (Figure 7d).
Figure 7.
Electron micrographs of 293 Tet-Off cells containing HDQ83 aggregates. After HDQ83 expression for 3–5 d, cells were fixed as described in MATERIALS AND METHODS and viewed by electron microscopy. (a–c) Different magnifications of a cell containing a typical perinuclear inclusion body. At higher magnifications, HDQ83 fibrils with a diameter of ∼10 nm can be observed. Immunogold labeling of cells with the HD1 antibody confirmed the identity of the HDQ83 fibrils (d). Immunogold labeling of cells also reveals that the proteins ubiquitin (h) and 14–3-3 (i) are present in the interior of inclusion bodies. In cells with perinuclear inclusion bodies, ultrastructural changes such as large indentations of the nuclear envelope (e), disruption of the nuclear membrane (f), and concentration of mitochondria in the vicinity of inclusion bodies (g) are observed. In f, alterations of the nuclear envelope are indicated by arrows.
To further characterize the inclusion bodies, cells expressing HDQ83 were immunogold labeled with antibodies directed against ubiquitin and 14–3-3ε. As shown in Figure 7, h and i, the inclusions were selectively labeled with each antibody, confirming the presence of ubiquitin and 14–3-3ε in the core region of the inclusion body. These findings support the hypothesis that aggregated HDQ83 protein is ubiquitinated and that certain cellular proteins are recruited into these structures. Frequently, the nuclei from cells containing inclusion bodies were seen to contain large indentations in their envelope (Figure 7, a and e). Similar changes of nuclei have also been described for neuronal cells of HD transgenic mice and patients (Roizin et al., 1979; Davies et al., 1997). At higher magnification, a clear disruption of the nuclear membrane was obvious (Figure 7f), suggesting that inclusion body formation not only alters the shape of nuclei but also disrupts, at least partially, the nuclear envelope. Frequently, disintegration of organelles such as mitochondria, vacuoles, and vesicular structures was observed in cells containing large inclusion bodies, and mitochondria, especially, appeared to be trapped at the periphery of the filamentous inclusion bodies (Figure 7g). In the periphery, but not in the interior of inclusions, bundles of the intermediate filament vimentin were also detected, confirming the results obtained by immunofluorescence microscopy (Figure 2f). None of these alterations were found in cells expressing HDQ20 protein (our unpublished results).
Expression of Mutant HD Exon 1 Protein Is Toxic for 293 Tet-Off Cells
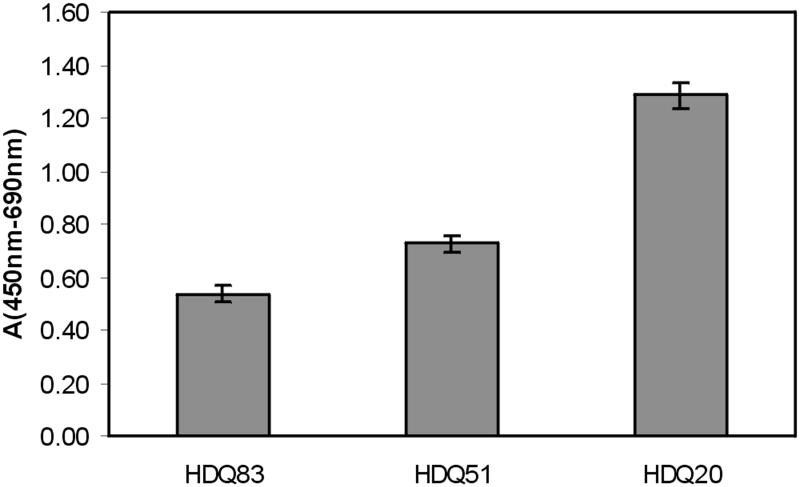
Expression of HDQ83, HDQ51, and HDQ20 in stable transfected cell lines was induced for 4 d, and cell viability was determined using the XTT assay. As shown in Figure 8, the viability of 293 Tet-Off cells decreased in a glutamine repeat length–dependent manner. For example, viability of cells expressing the HDQ83 protein was reduced approximately twofold compared with cells expressing the HDQ20 protein. At the time when cell viability was examined, ∼30–50% of all cells expressing the proteins HDQ51 or HDQ83 contained large perinuclear inclusions; no such structures were observed in cells expressing the HDQ20 protein. Thus, cells containing inclusion bodies appear to be significantly less viable than cells lacking aggregated protein. These results suggest that the process of aggregate formation causes toxicity in the 293 Tet-Off cell lines.
Figure 8.
Expression of mutant HD exon 1 protein is toxic for 293 Tet-Off cells. Expression of HD exon 1 proteins with 20, 51, and 83 glutamines was induced for 4 d, and cell viability was determined using the XTT assay as described in MATERIALS AND METHODS. Values shown are the average of nine determinations ± SE.
DISCUSSION
In this study we have generated inducible cell lines for the detailed analysis of huntingtin protein aggregation in mammalian cells. Our data demonstrate that overexpression of HD exon 1 protein fragments with a polyQ repeat in the pathological range (51 and 83 glutamines), but not with a polyQ repeat in the normal range (20 glutamines), results in the formation of massive cytoplasmic inclusions that are toxic for 293 Tet-Off cells. These inclusions contain aggregated, ubiquitinated HD exon 1 protein with a fibrillar morphology and are structurally very similar to the neuronal inclusions that have been detected previously in brains of HD patients and transgenic animals (Davies et al., 1997; DiFiglia et al., 1997). Recently, Wigley et al. (1999) presented evidence that the centrosome or a closely associated structure may play a functional role in the degradation of misfolded proteins in mammalian cells. Using immunofluorescence microscopy, they identified a specific structure in the centrosomal region in which components of the 26S proteasome, as well as ubiquitin and heat shock proteins, are concentrated under basal conditions. Inhibition of the proteasome activity as well as overexpression of mutant CFTR (Johnston et al., 1998) resulted in an enlargement of this centrosome-associated structure and the formation of large perinuclear inclusion bodies with undegraded protein. These results suggest that mammalian cells contain a specific organelle that is specialized in the degradation of misfolded aggregation-prone proteins and peptides. Our results obtained with mutant and wild-type HD exon 1 proteins are in agreement with this prediction, and we propose to term this structure “degrasome.” We found that inclusion bodies with aggregated huntingtin protein accumulate in the perinuclear region at or near the centrosome. Furthermore, the amount of ubiquitinated huntingtin protein aggregates in inclusions was found to increase twofold when the activity of the proteasome was inhibited by lactacystin. These results indicate that the amount of huntingtin protein that accumulates in inclusion bodies is highly dependent on the activity of the proteasome.
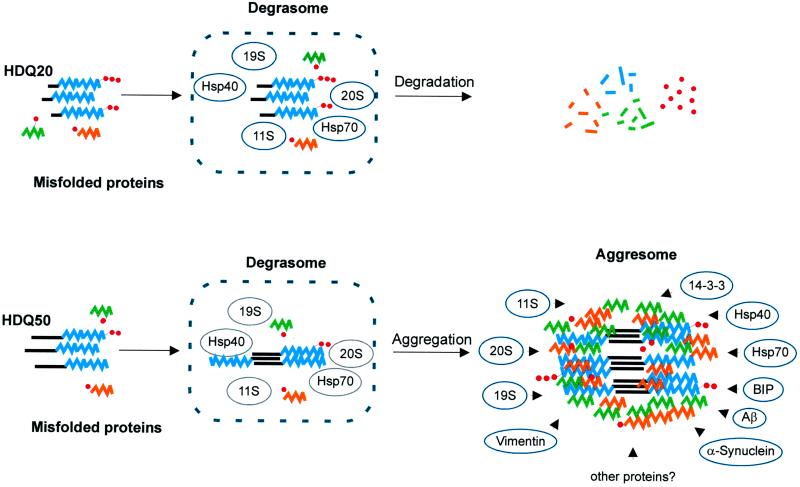
A model for the accumulation of polyQ-containing protein aggregates in 293 Tet-Off cells is outlined in Figure 9. We propose that under normal conditions, misfolded cellular proteins as well as wild-type and mutant HD exon 1 protein fragments are transported to the degrasome for proteolytic digestion; however, although the HD exon 1 protein with 20 glutamines can be successfully degraded by the ubiquitin–proteasome system, which is concentrated in the degrasome, the HD exon 1 protein with a long polyQ repeat (51 and 83 glutamines) polymerizes into insoluble fibrils that resist proteolytic degradation. Once the first nuclei and fibrillar structures have formed, the cell responds to the accumulation of mutant protein with the recruitment of additional proteins such as ubiquitin, heat shock proteins, and proteasomal subcomplexes that coassemble with aggregated huntingtin protein. Thus, we have shown that the resulting inclusion bodies, termed aggresomes by Johnston et al. (1998), contain a number of different proteins such as GRP78, 14–3-3, α-synuclein, 19S, 20S, and 11S in addition to the aggregated huntingtin protein (Figures 3, 5, and 6). We suggest that the accumulating huntingtin fibrils inhibit the proteasomal activity and thereby obstruct the cellular stress response pathway for misfolded protein. Thus, after a certain time, in addition to the mutant huntingtin fragments, other misfolded cellular proteins, which are normally digested in the degrasome, are also accumulating. Immunofluorescence microscopy revealed that 5–7 d after induction of HDQ83 expression, proteins and peptides such as amyloid-β or presenilin-1 also accumulate in the perinuclear inclusion bodies (our unpublished results). The redistribution of the cytoplasmic heat shock proteins Hsp40 and Hsp70 and the intermediate filament vimentin is another response to the accumulation of insoluble huntingtin fibrils. We speculate that after a certain time the cell recognizes that it will lose the battle against the accumulating protein aggregates and begins to isolate them from their natural environment. The encapsulation of aggregated huntingtin protein with the intermediate filament vimentin may be one such strategy.
Figure 9.
Proposed mechanism for the formation of perinuclear inclusion bodies. Top, misfolded proteins, including N-terminal huntingtin fragments with 20 glutamines (HDQ20; blue) are ubiquitinated (small red circles) and degraded in a hypothetical organelle, the degrasome, containing the proteasomal subcomplexes 11S, 19S, and 20S and the heat shock proteins Hsp40 and Hsp70. Bottom, huntingtin fragments with 51 glutamines (HDQ51; blue) form insoluble aggregates that resist proteolysis and accumulate in the degrasome. As a response, additional proteins like the potential chaperones BiP and 14–3-3, α-synuclein, Aβ, and presumably many others are recruited and coated with vimentin, forming the aggresome.
Our data, however, indicate that this strategy is not very successful. Ultrastructural studies showed that the process of aggregate formation dramatically alters the cellular environment (Figure 7). Inclusion body formation not only changes the shape of the adjacent nuclei, it also destroys the nuclear envelope. Furthermore, mitochondria are dislocated to the area surrounding the inclusion bodies; it seems as if they are caught by the fibrillar structures. The reason for the concentration of mitochondria in the vicinity of the inclusion bodies is not immediately obvious. It is well known that mitochondria play a crucial role in the activation of specific apoptotic pathways: under certain conditions they are concentrated in the perinuclear region (Desagher and Martinou, 2000). Furthermore, there is accumulating evidence that mitochondrial dysfunction is linked to the pathogenesis of neurodegenerative disorders such as HD (Beal, 2000). We suggest that mitochondria concentrate at inclusion bodies because the ubiquitin–proteasome system, which tries to degrade the amyloid-like huntingtin protein, requires additional ATP; however, the energetics of cells containing inclusion bodies with aggregated huntingtin protein has to be examined in more detail to understand this phenomenon.
Our data suggest that the redistribution of the proteins GRP78, 14–3-3, α-synuclein, and TIA-1 to inclusion bodies belongs to an early stress response of cells in their struggle for survival. There is experimental evidence to indicate that GRP78, 14–3-3, and α-synuclein function as chaperones (Bonifacino and Weissman, 1998; Shaw, 2000; Souza et al., 2000), whereas TIA-1 has been characterized as a stress-induced RNA-binding protein (Kedersha et al., 1999). Chaperones are known to bind to misfolded aggregation-prone polypeptides, thereby diminishing the likelihood of aggregate formation (Hartl, 1996). Thus, we suggest that chaperones accumulate at inclusion bodies either to refold misfolded protein or to totally unfold polypeptide chains for their efficient degradation. In particular, the recruitment of the ER-resident chaperone GRP78 into the cytoplasmic inclusion bodies was unexpected. GRP78 is known to associate with misfolded aggregation-prone proteins in the ER. Retrograde transport of this protein from the ER into the cytoplasm, however, has not been described. It is possible that GRP78 is transported into the cytoplasm via the Sec61 protein complex (Plemper et al., 1997) or, alternatively, is present in the cytoplasm because the huntingtin fibrils have disrupted the ER membrane and therefore released the protein. Additional experiments will be necessary to address this question in more detail.
Recently, Ostrerova et al. (1999) showed that α-synuclein is homologous to the group of 14–3-3 proteins and that it binds to 14–3-3 as well as to several 14–3-3 interacting proteins such as protein kinase C and BAD. Evidence also has been presented that both α-synuclein and 14–3-3 proteins function as chaperones (Souza et al., 2000); however, the involvement of these proteins in signaling pathways as well as their participation in the pathogenesis of neurodegenerative disorders such as Parkinson's, Alzheimer's, and Huntington's disease has also been described (Layfield et al., 1996; Polymeropoulos et al., 1997; Charles et al., 2000;). For example, α-synuclein immunoreactivity has been detected in inclusion bodies with aggregated, ubiquitinated huntingtin protein of HD patients and transgenic mice, suggesting that the redistribution of this protein to inclusions might influence disease progression (Charles et al., 2000). Our results strongly support the importance of these findings. Wild-type and mutated α-synuclein have also been identified in Lewy bodies of Parkinson's disease patients. Furthermore, genetic studies have identified point mutations in the α-synuclein protein that cause familial Parkinson's disease and enhance the aggregation of this protein (Polymeropoulos et al., 1997; Conway et al., 1998). With regard to 14–3-3, association of this class of proteins with neurofibrillary tangles in Alzheimer's disease brains has been reported (Layfield et al., 1996). Neurofibrillary tangles are formed from the hyperphosphorylated microtubule-associated tau protein, and there is evidence that the mitogen-activated protein kinase pathway is responsible, at least in part, for the phosphorylation of tau (Drewes et al., 1992). 14–3-3 proteins are highly abundant in brain, and they have been shown to play a central role in the mitogen-activated protein kinase signaling cascade (Xing et al., 2000). Binding of 14–3-3 proteins to target proteins is dependent on their phosphorylation of Ser/Thr residues (Aitken, 1996). Recently, Tzivion et al. (2000) showed that 14–3-3 proteins interact with phosphorylated vimentin. Moreover, an association of these proteins with the centrosome has been described (Pietromonaco et al., 1996). These results suggest that 14–3-3 proteins link intracellular signaling cascades to the protein degradation machinery at the centrosome; however, further studies will be required to assess the biochemical and biophysical properties of this class of proteins. Furthermore, it will be interesting to see whether 14–3-3 proteins are also present in cytoplasmic or nuclear inclusion bodies, or both, of HD patients.
Our data demonstrate that the protein TIA-1 is another constituent of the perinuclear inclusion bodies. TIA-1 is a poly(A)+ RNA binding protein that continuously shuttles between the nucleus and the cytoplasm, suggesting that it participates in the nucleocytoplasmic transport of selected mRNAs. Recently, evidence has been presented that TIA-1 and its related homologue TIAR regulate the general translational arrest that accompanies environmental stress (Kedersha et al., 1999). After a stress stimulus, these proteins recruit most cytoplasmic mRNAs to discrete cytoplasmic foci known as stress granules. Concentration of mRNAs in these granules prevents their translational initiation. Thus, TIA-1 and TIAR might function as translational silencers that influence the duration of a stress-induced translational arrest. Very recently, experimental evidence in support of this hypothesis was presented. Piecyk et al. (2000) showed that TIA-1 represses the expression of tumor necrosis factor alpha in mouse cells. The reason for the accumulation of TIA-1 in HD exon 1 protein aggregates is presently unknown; however, in light of these observations, it will be interesting to examine whether in addition to TIA-1, mRNAs are also concentrated in the perinuclar inclusions. A stress-induced accumulation of mRNAs could significantly alter the expression of proteins in mammalian cells. In this regard it is interesting to note that polyQ repeat length–dependent changes in the expression levels of neurotransmitter receptors and proteins involved in calcium signaling have recently been described (Luthi-Carter et al., 2000).
During the last years, several independent in vivo and in vitro studies have presented evidence that the formation of intracellular inclusion bodies with aggregated polyQ-containing proteins is nontoxic or even beneficial for mammalian cells (Klement et al., 1998; Saudou et al., 1998). Our present studies do not support these results. On the contrary, we find that the process of inclusion body formation correlates with toxicity in 293 Tet-Off cells. Viability of cells expressing HD exon 1 protein with 83 and 51 glutamines was reduced by ∼60 and 40%, respectively, compared with cells expressing an HD exon 1 protein with 20 glutamines (Figure 8). The molecular mechanism by which the process of aggregate formation causes toxicity to cells is unclear; however, our data suggest that formation of insoluble fibrillar structures interferes with proteasomal activity. It is possible that the 19S and 11S regulatory subcomplexes of the 26S proteasome recognize and bind the misfolded HD exon 1 protein; however, because of aggregate formation, the ubiquitinated polyQ-containing proteins cannot be degraded efficiently, and therefore the release of free ubiquitin molecules from the huntingtin fibrils by the cellular deubiquitinating enzymes is blocked. This could lead to the depletion of free ubiquitin in the cell, particularly when the amount of insoluble HD exon 1 protein increases. Because it has been shown that the primary function of the proteasome is the rapid degradation of proteins with abnormal folding (Voges et al., 1999), the formation of HD exon 1 protein aggregates could lead indirectly to the accumulation of regulatory proteins such as transcription factors, oncogene products, tumor suppressors, and rate-limiting enzymes that might be highly toxic for mammalian cells.
Our findings have several implications for the development of a treatment against HD and the related glutamine disorders (Heiser et al., 2000). We propose that either inhibition of huntingtin aggregation or stimulation of the natural clearance of accumulated disease protein by small molecules is an effective therapeutic strategy against these neurodegenerative diseases. Also, reducing the cellular concentration of the mutant huntingtin protein by antisense oligonucleotides or ribozymes could significantly slow down disease progression. Previously, we have shown that overexpression of the heat shock proteins Hsp70 and Hsp40 inhibits the formation of HD exon 1 aggregates in both yeast and mammalian cells (Muchowski et al., 2000; E. Wanker, unpublished results), and similar results were obtained when transiently transfected COS cells expressing a mutated HD exon 1 protein were treated with certain chemical compounds (Heiser et al., 2000). Together, these findings suggest that the inhibition of aggregate formation and the stimulation of the heat shock response by small molecules might be a feasible strategy toward a therapy for HD and related disorders. To our knowledge, chemical compounds that specifically stimulate the proteolytic digestion of a mutant disease protein have not yet been described. Therefore, a future challenge will be to identify chemical compounds that selectively activate the ubiquitin–ATP-dependent pathway and thereby inhibit the accumulation of aggregation-prone huntingtin protein in mammalian cells.
ACKNOWLEDGMENTS
We thank S. Schnögl for critical reading of this article and A. Dröge for providing Figure 9. This work has been supported by grants from the Deutsche Forschungsgemeinschaft (WA1151/1-2 and WA1151/2-1), the Huntington's Disease Society of America, the Human Frontier Science Program Organization, and the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BioFuture project: 0311853).
REFERENCES
- Aitken A. 14–3-3 and its possible role in coordinating multiple signaling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- Beal MF. Energetics in the pathogenesis of neurodegenerative diseases. Trends Neurosci. 2000;23:298–304. doi: 10.1016/s0166-2236(00)01584-8. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- Charles V, Mezey E, Reddy PH, Dehejia A, Young TA, Polymeropoulos MH, Brownstein MJ, Tagle DA. Alpha-synuclein immunoreactivity of huntingtin polyglutamine aggregates in striatum and cortex of Huntington's disease patients and transgenic mouse models. Neurosci Lett. 2000;289:29–32. doi: 10.1016/s0304-3940(00)01247-7. [DOI] [PubMed] [Google Scholar]
- Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- Cooper JK, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Drewes G, Lichtenberg-Kraag B, Doring F, Mandelkow EM, Biernat J, Goris J, Doree M, Mandelkow E. Mitogen activated protein (MAP) kinase transforms tau protein into an Alzheimer-like state. EMBO J. 1992;11:2131–2138. doi: 10.1002/j.1460-2075.1992.tb05272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper PS. Huntington's Disease. London: W. B. Saunders; 1991. [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, Lehrach H, Wanker EE. Inhibition of huntingtin fibrillogenesis by specific antibodies and small molecules: implications for Huntington's disease therapy. Proc Natl Acad Sci USA. 2000;97:6739–6744. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine- induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Layfield R, Fergusson J, Aitken A, Lowe J, Landon M, Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14–3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Mandel JL. A cellular model that recapitulates major pathogenic steps of Huntington's disease. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Martindale D, et al. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet. 1998;18:150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ona VO, et al. Inhibition of caspase-1 slows disease progression in a mouse model of Huntington's disease. Nature. 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- Ostrerova N, Petrucelli L, Farrer M, Mehta N, Choi P, Hardy J, Wolozin B. α-Synuclein shares physical and functional homology with 14–3-3 proteins. J Neurosci. 1999;19:5782–5791. doi: 10.1523/JNEUROSCI.19-14-05782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Am J Hum Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietromonaco SF, Seluja GA, Aitken A, Elias L. Association of 14–3-3 proteins with centrosomes. Blood Cells Mol Dis. 1996;22:225–237. doi: 10.1006/bcmd.1996.0103. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Roizin L, Stellar S, Liu JC. Neuronal nuclear-cytoplasmic changes in Huntington's chorea: electron microscope investigations. Adv Neurol. 1979;23:95–122. [Google Scholar]
- Rubinsztein DC, et al. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am J Hum Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K, Amaechi I, Mangiarini L, Bates G. Identification of an HD patient with a (CAG)180 repeat expansion and the propagation of highly expanded CAG repeats in lambda phage. Hum Genet. 1997;99:692–695. doi: 10.1007/s004390050432. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates GP, Lehrach H, Wanker EE. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. The 14–3-3 proteins. Curr Biol. 2000;10:R400. doi: 10.1016/s0960-9822(00)00519-4. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Souza JM, Giasson BI, Lee VM, Ischiropoulos H. Chaperone-like activity of synucleins. FEBS Lett. 2000;474:116–119. doi: 10.1016/s0014-5793(00)01563-5. [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN, Marcelli M, Weigel NL, Mancini MA. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Luo ZJ, Avruch J. Calyculin A induced vimentin phosphorylation sequesters 14–3-3 and displaces other 14–3-3 partners in vivo. 2000. J Biol Chem. 2000;275:29772–29778. doi: 10.1074/jbc.M001207200. [DOI] [PubMed] [Google Scholar]
- Velasco A, Hendricks L, Moremen KW, Tulsiani DR, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase I and II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. Establishment of stable cell lines expressing potentially toxic proteins by tetracycline-regulated and epitope-tagging methods. Biotechniques. 1996;21:718–722. doi: 10.2144/96214rr05. , 724–725. [DOI] [PubMed] [Google Scholar]
- Xing H, Zhang S, Weinheimer C, Kovacs A, Muslin A. 14–3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 2000;19:349–358. doi: 10.1093/emboj/19.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]