Abstract
The lack of reliable noninvasive diagnostic biomarkers of biliary atresia (BA) results in delayed diagnosis and worsened patient outcome. Circulating microRNAs (miRNAs) are a new class of noninvasive biomarkers with encouraging diagnostic utility. In this study we examined the ability of serum miRNAs to distinguish BA from other forms of neonatal hyperbilirubinemia. BA-specific serum miRNAs were identified using a microfluidic array platform and validated in a larger, independent sample set. The miR-200b/429 cluster was significantly increased in the sera of BA patients and has promising diagnostic clinical performance.
INTRODUCTION
Biliary atresia (BA) is an idiopathic neonatal liver disease characterized by inflammatory and fibrotic obliteration of the extrahepatic bile ducts, leading to severe cholestasis and biliary cirrhosis. The only therapies are Kasai portoenterostomy (KPE) and liver transplantation. The success rate of KPE decreases significantly with increased age at the time of surgery (1, 2). A definitive diagnosis of BA is often delayed, and currently requires invasive procedures including liver biopsy and intra-operative cholangiography. New, noninvasive diagnostic indicators may speed diagnosis, thereby improving patient outcome following KPE.
MicroRNAs (miRNAs) are short, noncoding RNAs that regulate target genes via transcript degradation or translational repression. Cell- and tissue-specific miRNA expression are essential for vertebrate organ development (3–5) and miRNAs are disregulated in numerous pathologies, including experimental BA (6). Recent studies have identified stable populations of miRNAs present in cell-free plasma and serum preparations (7–10). These circulating miRNAs can reflect altered expression or release from injured and diseased tissues and can distinguish similar disorders (11–13).
We hypothesize that tissue injury associated with BA results in a unique circulating miRNA profile that can be used as a diagnostic biomarker. Here we describe a cluster of miRNAs elevated in BA sera compared to cholestatic control samples that possess promising diagnostic characteristics.
MATERIALS AND METHODS
Sample collection and RNA isolation
Human serum samples were obtained from the Childhood Liver Disease Research and Education Network's (ChiLDREN) prospective longitudinal study of cholestasis in infancy (Prospective Database of Infants With Cholestasis (PROBE), clinicaltrials.gov NCT00061828). All BA diagnoses were confirmed by cholangiography, operative exploration, and/or histology. All mice were housed, handled, and euthanized in accordance with federal and institutional guidelines under the supervision of the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee. Following euthanasia, liver and extrahepatic bile duct (EHBD) tissue were excised, flash frozen and pulverized. Total RNA was isolated from 60 µL of human serum and from mouse tissue samples using the mirVana miRNA Isolation Kit (Ambion, Austin, TX). The C.elegans miRNAs cel-miR-54 and cel-miR-238 were added to serum samples as exogenous normalizing controls immediately following serum denaturation. RNA was eluted in 100 µL elution solution (95°C) and stored at −80°C.
miRNA analysis by low density array (LDA
TaqMan® Array Human MicroRNA A Cards (Applied Biosytems Inc., Foster City, CA) were used to quantify serum miRNA content according to manufacturer’s instructions. Briefly, 3 µL of total RNA from each sample was reverse transcribed. RT products were pre-amplified and diluted in 0.1X TE buffer, pH 8.0. Arrays were processed and analyzed by the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems). Detector threshold cycle (Ct) values were normalized to the mean Ct of each array.
qRT-PCR
For serum samples, total RNA volumes of 1.33 µL were reverse transcribed using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems Inc.) and miRNA-specific RT primers from the appropriate TaqMan® miRNA Assay Kit (Applied Biosystems). For tissue samples, 10 ng total RNA was reverse transcribed as above. Real-time quantitative polymerase chain reaction (qRT-PCR) was performed on a Stratagene MX3000P using TaqMan® miRNA assay kits and TaqMan® Universal PCR Master Mix (Applied Biosystems). Serum miRNA levels were normalized to two exogenous C.elegans miRNAs (cel-miR-54 and cel-miR-238) and tissue miRNA levels were normalized to endogenous sno-202 and sno-234 levels.
Statistical analysis
Statistical calculations were performed with Stata 11.0 (StataCorp, College Station, TX). Differences in miRNA levels between sample populations were assessed by the Wilcoxon rank-sum test. Correlation was determined using Spearman’s rank correlation test. Null hypotheses were rejected at p < 0.05. Hierarchical cluster analysis using complete-linkage clustering with Euclidean (L2) distance was performed using the 104 miRNAs detected in all 10 LDA samples.
RESULTS
Identification of BA-associated circulating miRNA
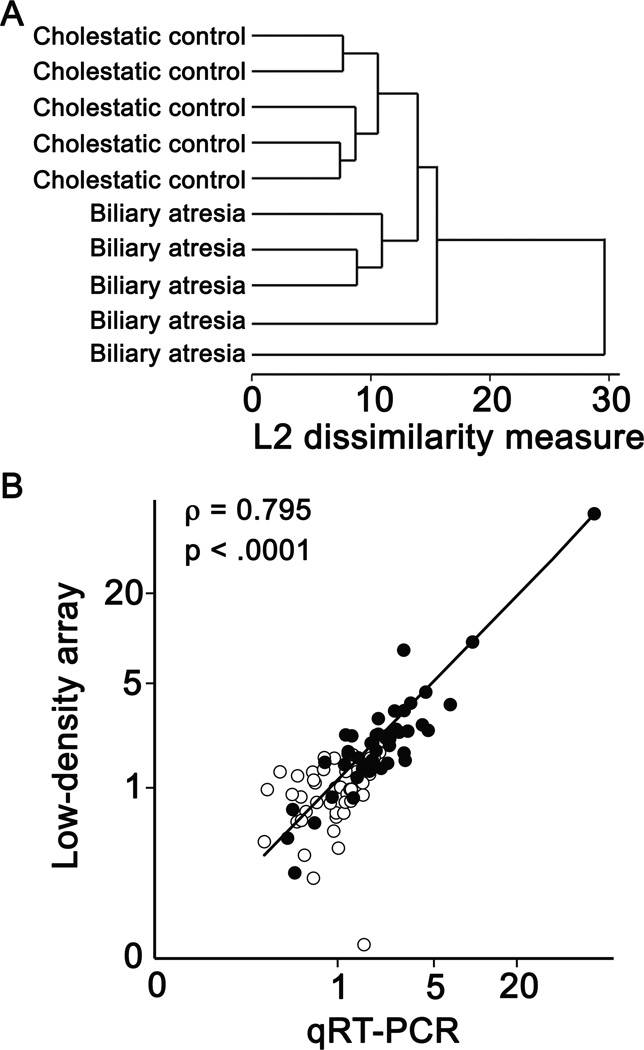
Serum miRNA profiles of BA patients and age- and sex-matched controls with indeterminate cholestasis (n=5 each) were determined using the TaqMan® Array Human MicroRNA A Card platform (Applied Biosystems). Hierarchical cluster analysis showed BA samples posses an altered serum miRNA profile compared to cholestatic controls (Fig. 1A). 134 of 375 miRNAs were detected in at least 3 out of 5 samples in the case or control group. A total of 11 miRNAs were selected for further analysis by applying the following criteria: a fold change >1.5, mean Ct <30 in at least one group, and p-value <0.05. Individual qRT-PCR reactions for these miRNAs were performed on the same RNA samples used in the LDA experiment. Of the 11 candidate miRNAs, 9 were confirmed by qRT-PCR as significantly altered in BA sera (online-only supplemental Table 1, http://links.lww.com/MPG/A153). MiRNA levels measured by qRT-PCR were significantly correlated to those by LDA (Fig. 1B).
FIGURE 1.
Low-density array (LDA) analysis of serum miRNA in human biliary atresia (BA). A, Dendrogram of unsupervised hierarchical analysis using 104 miRNAs detected in all samples by LDA. B, Scatterplot of relative serum miRNA levels of 9 candidate miRNAs as determined by LDA and individual qRT-PCR. Control sample means are scaled to 1. Open circles, cholestatic control samples; filled circles, BA samples; ρ, Spearman rank correlation coefficient.
Validation of BA-associated circulating miRNA
To validate the candidate BA-specific circulating miRNAs, qRT-PCR was performed on serum RNA from an independent set of BA and cholestatic controls (n=24 each) (online-only supplemental Table 2, http://links.lww.com/MPG/A154). The two patient groups displayed similar demographic and clinical characteristics, including total and direct serum bilirubin levels (Table 1). Of the nine candidate miRNAs, only miR-200b was significantly altered in BA sera compared to controls. To confirm the elevation of miR-200b, we measured levels of miR-200a and miR-429, which are co-transcribed with miR-200b from the miR-200b/429 locus. Both miR-200a and miR-429 were significantly elevated in BA sera (Fig. 2A). Serum levels of each miR-200b/429 member were significantly correlated with the other cluster members at p < 0.0001 (data not shown). Members of the paralagous miR-200c/141 cluster, miR-200c and miR-141, were unaltered in BA sera (Fig. 2B).
Table 1.
Validation set patient characteristics
| Cholestatic controls (n=24) | Biliary atresia (n=24) | |
|---|---|---|
| Gender, male/female | 16/8 | 12/12 |
| Age at collection, days | 51.00 ± 4.12 | 55.75 ± 3.33 |
| Total bilirubin, mg/dL | 8.51 ± 0.56 | 8.28 ± 0.46 |
| Direct bilirubin, mg/dL | 5.32 ± 0.38 | 5.08 ± 0.35 |
| AST, IU/L | 165.54 ± 21.47 | 180.55 ± 16.52 |
| ALT, IU/L | 118.13 ± 15.26 | 122.79 ± 13.34 |
| Albumin, g/dL | 3.40 ± 0.09 | 3.62 ± 0.10 |
| Total protein, g/dL | 5.60 ± 0.11 | 6.07 ± 0.13 * |
| GGTP, U/L | 243.24 ± 70.38 | 571.33 ± 72.97 * |
| INR | 0.98 ± 0.02 | 1.32 ± 0.34 |
| PTT, seconds | 32.11 ± 0.96 | 39.24 ± 5.91 |
| Hemoglobin, g/dL | 10.73 ± 0.46 | 10.82 ± 0.30 |
| WBCs, 1000/mcl | 10.93 ± 0.93 | 12.86 ± 0.67 |
| Birth weight, kg | 3.85 ± 0.18 | 4.25 ± 0.14 |
AST=aspartate aminotransferase; ALT=alanine aminotransferase; GGTP=γ-glutamyl transpeptidase; INR=international normalized ratio; PTT=partial thromboplastin time; WBCs=white blood cells.
P<0.05.
FIGURE 2.
Validation of circulating miRNAs in biliary atresia (BA). A,B Box-whisker plots of serum levels of miR-200b/429 cluster members (A) and miR-200c/141 and miR-122 (B) in an independent set of cholestatic controls and BA cases (n=24 each). Box, 25% to 75%; whisker, upper and lower adjacent values; line, median; points, outside values. Cholestatic control sample means are scaled to 1. *p < 0.05 vs. cholestatic control. C, Receiver-operating characteristic curve of miR-200a in cholestatic control and BA sera (n=24 each). AUC = area under the curve. D, Box-whisker plot of relative miRNA expression in liver and extrahepatic bile duct (EHBD) tissue of 8-week-old Balb/c mice (n=6). Box, 25% to 75%; whisker, upper and lower adjacent values; line, median; points, outside values. *p < 0.05 vs. liver.
Diagnostic utility and source of BA-specific circulating miRNA
The diagnostic utility of the miR-200b/429 cluster was examined using receiver operating characteristic (ROC) analysis. All three miRNAs of the cluster displayed good diagnostic properties, with area under the ROC curve (AUC) values greater than 0.80 (Table 2 and Fig. 2C). At optimal thresholds, the miR-200b/429 cluster correctly classified up to 85% of patient samples, with sensitivity and specificity values ranging from 71% to 92% (Table 2). As shown in Table 1the total protein and γ-glutamyl transpeptidase (GGTP) levels were elevated in the sera of BA patients compared to controls, so we also performed ROC analysis on these parameters. The potential diagnostic utility of the total protein was low (AUC 0.706), while the serum GGTP displayed a comparable AUC value (0.893) to those of the miR-200b/429 cluster.
Table 2.
Diagnostic properties of BA-specific serum miRNAs
| miRNA | Sensitivity, % | Specificity, % | Correctly classified, % |
AUC | 95% CI |
|---|---|---|---|---|---|
| miR-200b | 79.17 | 79.17 | 79.17 | 0.807 | 0.675–0.939 |
| miR-200a | 83.33 | 83.33 | 85.42 | 0.862 | 0.750–0.974 |
| miR-429 | 70.83 | 91.67 | 81.25 | 0.806 | 0.677–0.934 |
AUC=area under the curve; CI=confidence interval
To identify the tissue source of circulating miR-200b/429 cluster miRNAs, we isolated RNA from liver and extrahepatic bile duct (EHBD) tissue of 8-week-old Balb/c mice and measured miRNA levels by qRT-PCR. Interestingly, miR-200b/429 levels were up to 29-fold higher in EHBD tissue compared to liver (Fig. 2D), suggesting that cholangiocytes are the source of elevated miR-200b/429 cluster levels in BA.
DISCUSSION
A timely diagnosis of BA is critical for optimal patient outcome following KPE, yet current non-invasive tests are unable to reliably distinguish BA from other forms of neonatal hyperbilirubinemia. Earlier reports had suggested GGTP as a biomarker of BA (14, 15), and this is supported by our results. However, subsequent studies have found that serum GGTP levels are frequently elevated in other forms of neonatal cholestasis (16, 17). Wang et al. used a proteomic approach to identify 11 serum proteins significantly altered in BA compared to non-BA cholestasis (18). A classifier derived from the proteins correctly identified nine of ten BA and non-BA cholestatic control samples. Other proposed serum biomarkers, including interleukins, hyaluronic acid, insulin-like growth factor-1, and α-enolase have not been validated as clinically useful indicators of BA (19–22). In this report, we have identified a BA-specific circulating miRNA signature that may serve as a novel biomarker. A screen of serum miRNA by TaqMan® array identified nine miRNAs that are significantly altered in BA versus cholestatic controls. Subsequent validation in a larger, independent sample set confirmed a single miRNA cluster, miR-200b/429, as significantly increased in BA sera.
The miR-200b/429 cluster performed well diagnostically, correctly classifying up to 85% of patients. Although not all samples were properly distinguished, elevated circulating miR-200b/429 levels may prove a reliable indicator for an accelerated diagnostic workup in the jaundiced infant or in population-based screening tests. Future studies with large sample sets are needed to confirm our findings, establish reliable diagnostic thresholds for BA, and to compare or combine miRNA with other markers.
The high miR-200b/429 expression we observed in EHBD implicates this tissue as a source of the circulating fraction in BA. We propose that the elevation of miR-200b/429 in BA sera results from the destruction of EHBD, a feature unique to BA compared to the control patients. If serum miR-200b/429 levels correlate with EHBD damage, the cluster may also have prognostic value by predicting native liver survival in patients undergoing KPE. High expression may also occur in the intrahepatic bile ducts (IHBD); this would likely be masked in our liver RNA samples, as IHBD tissue represents a small fraction of total liver mass. Serum levels of the hepatocyte-specific miR-122 were not altered between groups (Fig. 2B), suggesting that hepatocyte injury was similar between BA and cholestatic controls.
Circulating miRNAs with prognostic potential have previously been reported in a variety of pathological contexts (23–25), and this study suggests that circulating miRNAs are of diagnostic value in BA. Future work will include the development and testing of diagnostic and prognostic logistical regression models using serum miR-200b/429 levels in larger populations and in combination with other biochemical parameters in an effort to facilitate early detection and improved patient outcome.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the Childhood Liver Research and Education Network (ChiLDREN) investigators, coordinators, and families who participated and made this work possible. The authors are also grateful to all members of the Fred and Suzanne Biesecker Pediatric Liver Center for support and helpful discussions. This work was supported by the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and its core facilities.
This study was supported by the Fred and Suzanne Biesecker Pediatric Liver Center, NIH grant R01DK079881 (JRF), and NIH grant U01DK062456 (ChiLDREN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
The authors report no conflicts of interest.
REFERENCES
- 1.Ohi R. Surgery for biliary atresia. Liver. 2001;21:175–182. doi: 10.1034/j.1600-0676.2001.021003175.x. [DOI] [PubMed] [Google Scholar]
- 2.Mieli-Vergani G, Vergani D. Biliary atresia. Semin Immunopathol. 2009;31:371–381. doi: 10.1007/s00281-009-0171-6. [DOI] [PubMed] [Google Scholar]
- 3.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 4.Lynn FC, Skewes-Cox P, Kosaka Y, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 5.Hand NJ, Master ZR, Eauclaire SF, et al. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hand NJ, Horner AM, Master ZR, et al. MicroRNA profiling identifies miR-29 as a regulator of disease-associated pathways in experimental biliary atresia. J Pediatr Gastroenterol Nutr. 2012;54:186–192. doi: 10.1097/MPG.0b013e318244148b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 8.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 11.Morimura R, Komatsu S, Ichikawa D, et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laterza OF, Lim L, Garrett-Engele PW, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 13.Zahm AM, Thayu M, Hand NJ, et al. Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2011;53:26–33. doi: 10.1097/MPG.0b013e31822200cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright K, Christie DL. Use of gamma-glutamyl transpeptidase in the diagnosis of biliary atresia. Am J Dis Child. 1981;135:134–136. doi: 10.1001/archpedi.1981.02130260026008. [DOI] [PubMed] [Google Scholar]
- 15.Platt MS, Potter JL, Boeckman CR, et al. Elevated GGTP/SGOT ratio. An early indicator of infantile obstructive cholangiopathy. Am J Dis Child. 1981;135:834–836. [PubMed] [Google Scholar]
- 16.Manolaki AG, Larcher VF, Mowat AP, et al. The prelaparotomy diagnosis of extrahepatic biliary atresia. Arch Dis Child. 1983;58:591–594. doi: 10.1136/adc.58.8.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggiore G, Bernard O, Hadchouel M, et al. Diagnostic value of serum gamma-glutamyl transpeptidase activity in liver diseases in children. J Pediatr Gastroenterol Nutr. 1991;12:21–26. doi: 10.1097/00005176-199101000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Malone JP, Gilmore PE, et al. Serum markers may distinguish biliary atresia from other forms of neonatal cholestasis. J Pediatr Gastroenterol Nutr. 2010;50:411–416. doi: 10.1097/MPG.0b013e3181cb42ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Faramawy AA, El-Shazly LB, Abbass AA, et al. Serum IL-6 and IL-8 in infants with biliary atresia in comparison to intrahepatic cholestasis. Trop Gastroenterol. 2011;32:50–55. [PubMed] [Google Scholar]
- 20.Ukarapol N, Wongsawasdi L, Ong-Chai S, et al. Hyaluronic acid: additional biochemical marker in the diagnosis of biliary atresia. Pediatr Int. 2007;49:608–611. doi: 10.1111/j.1442-200X.2007.02423.x. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida S, Nio M, Hayashi Y, et al. Serum insulinlike growth factor-I in biliary atresia. J Pediatr Surg. 2003;38:211–215. doi: 10.1053/jpsu.2003.50045. [DOI] [PubMed] [Google Scholar]
- 22.Lu BR, Brindley SM, Tucker RM, et al. Alpha-enolase autoantibodies cross-reactive to viral proteins in a mouse model of biliary atresia. Gastroenterology. 2010;139:1753–1761. doi: 10.1053/j.gastro.2010.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva J, Garcia V, Zaballos A, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 24.Zuo Z, Calin GA, de Paula HM, et al. Circulating microRNAs let-7a and miR-16 predict progression-free survival and overall survival in patients with myelodysplastic syndrome. Blood. 2011;118:413–415. doi: 10.1182/blood-2011-01-330704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong X, Du Y, Wang G, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.