Abstract
Background
The early repolarization pattern (ERP) is common and associated with risk of sudden cardiac death. ERP is heritable and mutations have been described in syndromatic cases.
Objective
To conduct a meta-analysis of genome-wide association studies (GWAS) to identify common genetic variants influencing ERP.
Methods
We ascertained ERP based on electrocardiograms in three large community-based cohorts from Europe and the US: the Framingham Heart Study, the Health 2000 Study, and the KORA F4 Study. We analyzed GWAS in participants with and without ERP by logistic regression assuming an additive genetic model and meta-analyzed individual cohort results. We then sought to strengthen support for findings that reached p≤1×10−5 in independent individuals by direct genotyping or in-silico analysis of genome-wide data. We meta-analyzed the results from both stages.
Results
Of 7482 individuals in the discovery stage, 452 showed ERP (ERP positive: mean age 46.9±8.9 years, 30.3% women; ERP negative: 47.5±9.4 years, 54.2% women). After meta-analysis, eight single nucleotide polymorphisms reached p≤1×10−5: The most significant finding was intergenic rs11653989 (odds ratio 0.47; 95% confidence interval 0.36–0.61; p=6.9×10−9). The most biologically relevant finding was intronic to KCND3: rs17029069 (odds ratio 1.46; 95% confidence interval 1.25–1.69; p=8.5×10−7). In the replication step (7151 individuals), none of the eight variants replicated, and combined meta-analysis results failed to reach genome-wide significance.
Conclusions
In a GWAS, we were not able to reliably identify genetic variants predisposing to ERP, presumably due to insufficient statistical power and phenotype heterogeneity. The reported heritability of ERP warrants continued investigation in larger well-phenotyped populations.
Keywords: Early repolarization, Sudden cardiac death, Arrhythmia, GWAS, Meta-analysis, Electrocardiogram
Introduction
The electrocardiographic early repolarization pattern (ERP) has long been considered a benign normal ECG variant.1, 2 Recent case-control studies, however, have demonstrated an association of inferior and lateral ERP to idiopathic ventricular fibrillation,3, 4 and subsequent large-scale community-based studies have suggested an increased risk of all-cause and cardiac mortality, and sudden death.5, 6 Most recently, a subtype of ERP, defined by horizontal or descending ST-segment morphology, was shown to confer the highest risk for an arrhythmic death, whereas an ascending ST-segment morphology, common in young male athletes, was associated with benign prognosis.7 The population prevalence of ERP across recent studies ranged between 3 and 13%.5, 6, 8, 9
Mutations in KCNJ810, 11, CACNA1C, CACN2B, and CACNA2D112 have been reported in individuals with ERP and sudden cardiac death, but the genetic determinants of ERP at the population level remain undefined. ERP appears to be heritable. An investigation in families from the UK reported a heritability estimate of 0.498 and an odds ratio (OR) of 2.5 for ERP in the offspring of at least one affected parent.8 Findings from the Framingham Heart Study (FHS) reported a sibling recurrence risk of 1.89 for ERP.9
Here we sought to identify common genetic determinants of ERP by performing a meta-analysis of genome-wide association studies (GWAS) in individuals of European descent followed by independent replication, comprising a total of 940 subjects with ERP and 13,693 controls drawn from population- and community-based study samples.
Methods
Description of Study Cohorts
All study procedures were approved by the local institutional review boards of each center. Study participants provided written informed consent, including consent to use DNA for genetic analyses of cardiovascular disease. Individuals were considered eligible for the GWAS stage if they had both genome-wide genotyping data and electrocardiograms (ECGs) available. We excluded participants of non-European ancestry, with a QRS-duration >120ms, <18 years of age (and ≥80 years of age in the Health 2000 Study (H2K)), presenting with non-sinus rhythm or with a heart rate >100/min. A summary of excluded subjects is presented in Supplemental Table 1. Detailed descriptions of cohorts can be found in the Supplemental Methods.
The Framingham Heart Study (FHS) is a prospective community-based study started in 1948 to evaluate potential risk factors for coronary heart disease. In 2002, a third generation of participants was recruited (Generation 3, n=4,095), and here, 3,726 participants with both GWAS and ECG data were studied.
The Health 2000 Study (H2K) is an epidemiological survey conducted in Finland in 2000–2001 and represents the Finnish adult population aged ≥30 years. Of the total H2K population (n=8028), 2082 subjects with both GWAS data and ECGs were investigated here.
The Cooperative Health Research in the Augsburg Region (KORA) F4 Study (KORA F4) was conducted in 2006–2008 in men and women aged 32–81 years. After exclusions, of 3080 participants, we studied 1674 with available GWAS and ECG data.
Replication cohorts
Participants of H2K not contributing to the GWAS analysis were included in the replication analyses. After exclusions, the replication sample consisted of 2996 individuals.
Participants of KORA F4 not included in the GWAS analysis, but with DNA samples available (n=1226) were recruited for replication (n=1140).
Further individuals for replication were available from the MONItoring CArdiovascular Disease (MONICA) / KORA S1 and S2 Study Subcohort (MONICA/KORA S1/S2). Here, 1351 participants (644 from S1 and 707 from S2) were eligible for study.
Individuals were also included from the Young Finns Study (YFS), a population-based, ongoing prospective multicentre study between five Finnish university hospitals. After exclusions, the replication sample from YFS consisted of 466 subjects.
From the South Italian village of Carlantino, 1668 individuals had been recruited from 1998–2005, and 285 individuals were available for analysis. A second Italian cohort was derived from the project Genetic Park of Friuli Venezia Giulia (FVG), where 913 individuals remained in the dataset for analysis.
Ascertainment of ERP
ERP was determined on 12-lead ECGs by specifically trained physicians experienced in cardiac electrophysiology. The identical coding protocol was applied at each participating site. Measurements were performed for each lead separately. From among the leads, the one with the earliest offset of the QRS complex was defined as the reference point for J-point elevations. Criteria for the diagnosis of ERP required a J-point elevation of ≥0.1 mV compared to the baseline (defined as the T-P segment) in ≥2 leads within at least one regional territory. ERP was considered in inferior (leads II, III, aVF) or lateral (leads I, aVL, V4, V5, V6) regional territories, and was categorized as showing either slurring, notching, discrete, or mixed morphology. We assessed the morphology and amplitude of the ST segment at 100 ms after the J-point. ST-segments with an amplitude >0.1 mV were considered ascending; for amplitudes ≤0.1 mV, we distinguished between horizontal and descending morphologies.
Genotyping and replication SNP selection
GWAS Genotyping platforms included: Affymetrix 5.0 and a 50,000 SNP custom Illumina chip (FHS), Illumina Human610-Quad (H2K), and Affymetrix 6.0 (KORA F4). Cohort-based information on the genotyping process and quality control criteria are provided in Supplemental Table 2. Replication genotyping was performed using iPlex single base primer extension with matrix-assisted laser desorption / ionization time-of-flight (MALDI-TOF) mass spectrometry (Sequenom Inc, San Diego, California, USA) for H2K, KORA F4, and MONICA/KORA S1/S2. All sites used identical genotyping assays. All genotyping was performed according to the manufacturer’s instructions. For replication in Carlantino, FVG and YFS, we used in-silico results available from previous genome-wide genotyping.
Statistical Analysis
For GWAS, imputation of >2.5 million HapMap SNPs based on the CEU population was performed using Mach v1.0.1x13 in all three cohorts (Supplemental Table 2). In each study, we adjusted for population structure applying the principal components method,14 or stratified by identity-by-descent cluster groups as implemented in PLINK.15 The primary analysis was conducted separately at each site using logistic regression assuming an additive genetic model. Models were adjusted for age at ECG, sex, and RR interval. Prior to meta-analysis, we used genomic control methods to account for remaining population stratification: site-specific results with a genomic inflation factor γ >1.0 were corrected by multiplying the standard error of the SNP regression parameter with the square root of the study-specific γ (Supplemental Figure 1). We used the ratio of the observed to expected variance of the imputed SNP genotype counts as a quality control metric for imputed SNPs. For each SNP, we included all studies with variance ratios >0.3. Only SNPs with a minor allele frequency ≥0.01 (H2K, KORA F4) or ≥0.05 (FHS) were considered. After these quality control procedures, we analyzed 2,523,555 SNPs. The meta-analysis of site-specific association results was performed simultaneously in H2K and MONICA/KORA using the meta-analysis tool METAL.16 We performed fixed-effects meta-analysis using inverse variance weights. Results from both centers were compared and inconsistencies were eliminated. Finally, one single version was used for all subsequent analyses. To account for multiple comparisons, we applied a pre-specified Bonferroni adjustment for 1 million independent tests (p<5×10−8). We attempted to replicate SNPs that did not reach genome-wide significance: We included all SNPs with a p-value <1×10−5, if they had a minor allele frequency ≥0.05 and showed homogeneity across individual studies as assessed by an I2 <0.5 and a non-significant p-value for heterogeneity. All SNPs were clustered into loci according to their genomic position, and the top SNP of each locus was selected for replication. Replication results were calculated separately by replication site. Site-specific replication results were then meta-analyzed and this final result was used to assess positive replication. For an overall effect estimate, results of the GWAS and replication stage were finally combined. All reported p-values are 2-sided.
Results
The GWAS stage combined 7482 individuals. Their mean age across cohorts was 46.9±8.9 years in participants with ERP, and 47.5±9.4 years in participants without ERP. Women accounted for 30.3% of ERP positive subjects and 54.2% of ERP negative subjects. Further clinical characteristics by cohort are shown in Table 1. Details with respect to ERP and ST-segment morphology in ERP-positive individuals are presented in Table 2. Overall, 452 (6.0%) individuals demonstrated ERP, of whom 57 (12.6%) showed ERP in >1 regional territory. A horizontal or descending ST-segment morphology was noted in 70.9% of all ERP-positive regional territories.
Table 1.
Baseline characteristics of the GWAS study sample FHS H2K KORA F4
| Study | FHS | H2K | KORA F4 | |||
|---|---|---|---|---|---|---|
|
| ||||||
| ERP + | ERP − | ERP + | ERP − | ERP + | ERP − | |
| Number, total | 3726 | 2082 | 1674 | |||
| Females, n (%) | 52 (23%) | 1946 (56%) | 16 (26%) | 1054 (52%) | 69 (42%) | 809 (54%) |
| Age, years (mean ± SD) | 37 ± 9.0 | 40 ± 8.8 | 46 ± 10.2 | 50 ± 10.9 | 60 ± 8.4 | 61 ± 8.8 |
| Age, range | 19 to 63 | 19 to 72 | 31 to 72 | 30 to 75 | 44 to 78 | 31 to 81 |
| RR interval, ms (mean ± SD) | 1060 ± 184.5 | 985 ± 158.2 | 1045 ± 180.3 | 977 ± 157.1 | 964 ± 159.9 | 944 ± 143.3 |
GWAS – genome wide association study; ERP – early repolarization pattern; FHS – Framingham Heart Study; H2K – Health 2000 Study; n – number; SD – standard deviation.
Table 2.
Early repolarization pattern by GWAS cohorts
| FHS | H2K | KORA F4 | |
|---|---|---|---|
| ERP positive ECGs, n (%) | 227 (6.1%) | 61 (2.9%) | 164 (9.8%) |
| ERP positive regional territories, n | 243 | 66 | 200 |
| Regional territories | |||
| Inferior, n (%) | 81 (33.3%) | 19 (28.8%) | 109 (54.5%) |
| Lateral, n (%) | 162 (66.7%) | 47 (71.2%) | 91 (45.5%) |
| ERP morphology across regional territories | |||
| Notching, n (%) | 85 (35.0%) | 23 (34.8%) | 33 (16.5%) |
| Slurring, n (%) | 73 (30.0%) | 18 (27.3%) | 70 (35.0%) |
| Discrete, n (%) | 24 (9.9%) | 3 (4.5%) | 0 (0%) |
| Mixed, n (%) | 61 (25.1%) | 22 (33.3%) | 97 (48.5%) |
| ST-segment analysis across regional territories | |||
| Ascending, n (%) | 87 (35.8%) | 38 (57.6%) | 23 (11.5%) |
| Horizontal, n (%) | 156 (64.2%) | 28 (42.4%) | 146 (73.0%) |
| Descending, n (%) | 0 (0%) | 0 (0.0%) | 31 (15.5%) |
In an ERP-positive ECG, ERP can occur in more than one regional territory (inferior or lateral). A mixed ERP morphology indicates different morphologies across leads. ERP morphologies and ST-segment information is provided across regional territories. GWAS – genome wide association study; ERP – early repolarization pattern; FHS – Framingham Heart Study; H2K – Health 2000 Study; n – number.
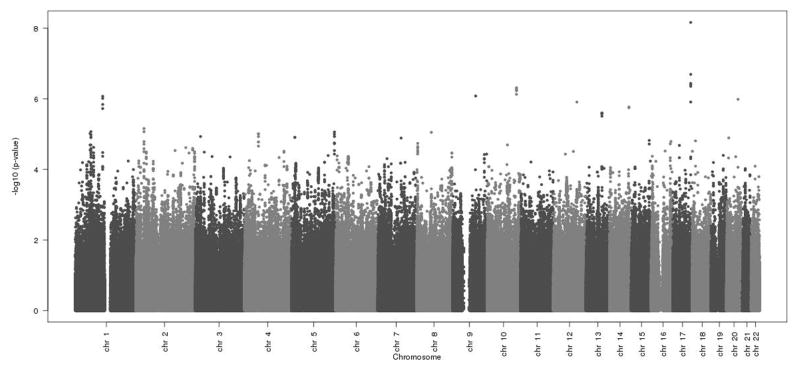
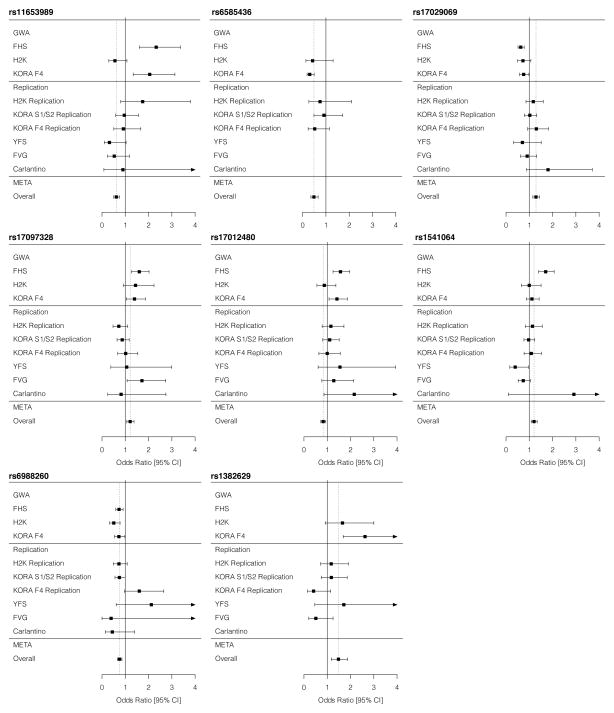
Following genotyping and quality control measures (Supplemental Table 1; Supplemental Figure 1), 2,523,555 SNPs were analyzed for association with ERP at the GWAS stage. The association results of the GWAS stage are presented in Figure 1, and the eight most significant SNPs, reaching p-values of <1×10−5 are shown in Table 3. We subsequently attempted replication of our results in 7151 independent samples, in which the prevalence of ERP was 6.8% (488 ERP-positive). None of the eight SNPs tested in the replication cohorts reached significance when all replication cohorts were meta-analyzed (Table 4). In the final meta-analysis, combining the results of the GWAS and the replication stages, none of the SNPs reached genome-wide significance (Table 4, Figure 2).
Figure 1. Genome-wide association results from GWAS meta-analysis for association with ERP.
The physical chromosomal position of each SNP is plotted on the x-axis; the −log10(p value) of the SNP-ERP association is plotted on the y-axis. chr – chromosome.
Table 3.
GWAS association results for the most significant SNPs
| SNP | Chr | Minor/major allele | MAF | Location | Closest gene(s) | Cohort specific | Meta-analysis | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| FHS OR (95% CI) | H2K OR (95% CI) | KORA F4 OR (95% CI) | OR | 95% CI | p-value | ||||||
| rs11653989 | 17 | A/T | 8.3% | Intergenic | MGAT5B, SEC14L | 0.43 (0.30–0.62) | 0.56 (0.29–1.07) | 0.49 (0.32–0.75) | 0.47 | 0.36–0.61 | 6.87×10−09 |
| rs6585436 | 10 | T/C | 1.7% | Intergenic | PDZD8, EMX2 | - | 0.43 (0.14–1.31) | 0.30 (0.19–0.49) | 0.32 | 0.21–0.50 | 4.88×10−07 |
| rs17029069 | 1 | T/C | 35.6% | Intronic | KCND3 | 1.58 (1.28–1.94) | 1.37 (0.94–2.02) | 1.32 (1.02–1.71) | 1.46 | 1.25–1.69 | 8.50×10−07 |
| rs17097328 | 14 | T/C | 19.0% | Intergenic | C14orf177 | 1.60 (1.27–2.02) | 1.44 (0.93–2.24) | 1.40 (1.04–1.86) | 1.51 | 1.27–1.78 | 1.71×10−06 |
| rs17012480 | 2 | A/G | 13.6% | Intronic | LTBP1 | 0.64 (0.51–0.80) | 0.88 (0.56–1.37) | 0.70 (0.54–0.93) | 0.69 | 0.58–0.81 | 6.97×10−06 |
| rs1541064 | 1 | A/G | 38.1% | Intronic | UBE2U | 1.70 (1.40–2.07) | 1.00 (0.66–1.51) | 1.11 (0.87–1.42) | 1.38 | 1.20–1.59 | 8.54×10−06 |
| rs6988260 | 8 | A/G | 21.0% | Intronic | FAM110B | 0.73 (0.59–0.91) | 0.51 (0.33–0.78) | 0.73 (0.54–0.98) | 0.69 | 0.59–0.81 | 8.92×10−06 |
| rs1382629 | 4 | A/G | 2.7% | Intronic | KIAA1211 | - | 1.66 (0.91–3.01) | 2.62 (1.69–4.09) | 2.23 | 1.56–3.18 | 9.82×10−06 |
Results sorted by ascending p-value. All results adjusted for age, sex, and RR-interval. Chr – chromosome; CI – confidence interval; GWAS – genome wide association study; ERP – early repolarization pattern; FHS – Framingham Heart Study; H2K – Health 2000 Study; MAF – minor allele frequency; OR – odds ratio per minor allele; SNP – single nucleotide polymorphism.
Table 4.
Replication and combined analysis
| SNP | Chr | Closest gene(s) | GWAS Meta-analysis | Replication Meta-analysis | Overall | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| OR (95% CI) | p-value | OR (95% CI) | p-value | OR (95% CI) | p-value | |||
| rs11653989 | 17 | MGAT5B, SEC14L | 0.47 (0.36–0.61) | 6.87×10−09 | 0.97 (0.70–1.34) | 0.84 | 0.62 (0.50–0.76) | 2.80×10−06 |
| rs6585436 | 10 | PDZD8, EMX2 | 0.32 (0.21–0.50) | 4.88×10−07 | 0.74 (0.47–1.15) | 0.18 | 0.48 (0.35–0.66) | 6.11×10−06 |
| rs17029069 | 1 | KCND3 | 1.46 (1.25–1.69) | 8.50×10−07 | 1.11 (0.94–1.31) | 0.23 | 1.29 (1.15–1.44) | 8.28×10−06 |
| rs17097328 | 14 | C14orf177 | 1.51 (1.27–1.78) | 1.71×10−06 | 0.87 (0.71–1.07) | 0.19 | 1.21 (1.06–1.38) | 4.03×10−03 |
| rs17012480 | 2 | LTBP1 | 0.69 (0.58–0.81) | 6.97×10−06 | 1.11 (0.90–1.37) | 0.31 | 0.83 (0.73–0.94) | 3.45×10−03 |
| rs1541064 | 1 | UBE2U | 1.38 (1.20–1.59) | 8.54×10−06 | 1.01 (0.86–1.18) | 0.93 | 1.20 (1.08–1.34) | 6.80×10−04 |
| rs6988260 | 8 | FAM110B | 0.69 (0.59–0.81) | 8.92×10−06 | 0.87 (0.71–1.08) | 0.20 | 0.75 (0.66–0.86) | 1.72×10−05 |
| rs1382629 | 4 | KIAA1211 | 2.23 (1.56–3.18) | 9.82×10−06 | 1.09 (0.80–1.49) | 0.58 | 1.48 (1.17–1.87) | 9.12×10−04 |
All results adjusted for age, sex, and RR-interval. CHR – chromosome; CI – confidence interval; GWAS – genome wide association study; OR – odds ratio per minor allele; SNP – single nucleotide polymorphism.
Figure 2. Association results by cohort for SNPs selected for replication.
For each SNP we attempted to replicate, ORs and 95% CIs are plotted by cohort and for the meta-analyzed results. For each SNP panel, the upper section represents cohorts included in the GWAS stage, the middle section those included in the replication stage, and the lower section depicts meta-analysis results. CI – confidence interval; META – meta-analysis; OR – odds ratio.
Of the eight SNPs we initially identified, five are located in introns of genes (Table 3). The initially most significant intronic SNP (rs17029069) is on chromosome 1p13.2, and maps to intron 2 of KCND3. KCND3 encodes the voltage-gated potassium channel Kv4.3. Other intronic SNPs are rs17012480 (intron 3 of LTBP1), rs1541064 (intron 6 of UBE2U), rs6988260 (intron 3 of FAM110B), and rs1382629 (intron 2 of KIAA1211).
Discussion
In our GWAS, we included almost 15,000 individuals from community-based cohorts of European ancestry. Close to 1000 participants were identified to be ERP-positive applying the identical, standardized diagnostic criteria by investigators at all contributing sites. While the initial results of our GWAS stage suggested promising association signals, none of the signals could be replicated.
Several independent investigations have suggested that ERP is a heritable ECG phenotype. In 1877 individuals from 505 UK families, the prevalence of ERP was 5.9%, and thus in keeping with the ERP prevalence in our current analysis.8 The authors reported a heritability estimate of h2=0.49 and an OR of 2.5 for offspring to demonstrate ERP, if ≥1 parent presented with ERP.8 Another investigation in FHS and H2K found an ERP prevalence of 6.1%.9 Heritability estimates for ERP fall within a similar range as other heritable ECG parameters such as the PR interval,17 the QT interval,17 or diseases identifiable by ECG, such as atrial fibrillation.18 GWAS have been successful in identifying chromosomal loci associated with each of these heritable phenotypes.19–22 Our inability to replicate even our pathophysiologically promising initial findings warrants some exploration.
A first explanation may be false-positive initial findings, which failed to replicate. In our study, <10% of the 15,000 participants were ERP-positive. A post-hoc power analysis revealed that we had 80% power to detect an OR of ≥1.68. We anticipate that with an increased sample size and thus increased statistical power, susceptibility loci for ERP will eventually be identified. While increased sample size can help to identify true loci associated with disease, it was still worth pursuing the current analysis as loci for other traits have been identified with similar sample sizes. Specifically, the first GWAS for the QT interval consisted of an initial stage of only 200 individuals, yet it successfully identified the locus most significantly associated with the QT interval.23 Similarly, for atrial fibrillation a cohort of only 636 cases and 804 controls identified the 4q25 locus for the arrhythmia.24
A second explanation for the non-replication of genetic loci might be that ERP is a more heterogeneous phenotype than initially anticipated. The prevalence of ERP appears variable. Despite a very homogeneous definition across participating cohorts, our prevalence ranged from 3%–13%. In a Japanese study, the prevalence reached almost 24%.25 ERP can be sub-classified based on the affected regional territory, ST segment morphology, amplitude, and J point morphology, a process that results in fragmentation of the phenotype into distinct and individually rare patterns. Also, there may be interactions with other clinical factors such as sex and race. Community-based investigations in Europe and Japan indicated that ERP in inferior or lateral territories is associated with the highest risk of mortality.5, 6, 25 A Finnish study found that only a horizontal or descending ST-segment was associated with adverse outcomes.26 Subsequently, in patients with idiopathic ventricular fibrillation a combination of ERP and a horizontal ST segment was associated with a markedly increased odds of arrhythmias compared to the presence of ERP alone.27 Higher ERP amplitudes also conferred an increased risk of death.5 The importance of sex has been controversial; one study suggested that the ERP-effect on mortality is more pronounced in males,6 whereas others have shown that women are particularly at risk.28 An increased SCD risk has been demonstrated in white, but not in black individuals.28 ERP might be subjected to temporal changes as well.29 In conclusion, the non-replication of our GWAS findings could be due to a too un-specific definition of ERP. Future investigations will need to focus on the subtypes with the strongest heritability and the strongest evidence for clinical implications. Presently, we lacked sufficient statistical power to adequately examine subtypes.
Although not genome-wide significant, one particularly interesting locus was on chromosome 1 intronic to KCND3. The gene encodes the voltage-gated potassium channel Kv4.3, a key channel underlying the cardiac transient outward current Ito.30 The current is of great importance for the early phase of repolarization,31 and reported to be the predominant contributor to phase I of the cardiac action potential.32 Its transmural dispersion leads to different shapes of the cardiac action potential in endo- vs. epicardial layers, and results in the ERP notch or slur on the surface ECG.32 Genetic variants in KCND3 possibly lead to altered channel properties that could underlie an increased arrhythmogenic potential. A mutation in KCND3 has been described to cause the Brugada syndrome, in which the early phase of cardiac repolarization is critically involved as well.33 In contrast, KCND3 mutations were described to be rare in patients with long QT syndrome, where late phases of cardiac repolarization are important.34 However, since the association of common variants at this locus to ERP was not significant considering the burden of testing, these potential functional links remain purely speculative.
Strengths and limitations
Strengths of our study are the highly standardized assessment of ERP status and a large collaborative effort to provide maximally achievable statistical power. However, sample size considerations limited our ability to explore the heterogeneity of ERP in subgroup analyses. Also, successful replication of variants predisposing to ERP might have been prevented by residual confounding. It will require further studies to refine ERP phenotype in more detail. Despite our intention to increase statistical power, our study still did not yield enough participants to reliably identify common genetic variants associated with ERP. All investigated ECGs stem from a single time point. As ERP showed temporal variability in prior studies, we cannot exclude that we have missed ERP positive individuals. All participants were of European descent. We therefore cannot comment on genetic predisposition to ERP in different ethnicities. Carlantino and FVG are isolated populations that could have influenced our genetic analyses if genetic substructure exists. However, the heterogeneity of the results of these two cohorts appeared to be similar to other cohorts in our study.
Conclusion
In conclusion, we have performed a multicenter meta-analysis of GWAS attempting to identify SNPs associated with ERP. Limited statistical power and potentially unrecognized phenotypic heterogeneity are potential explanations for our non-replication results. Future investigations will require many collaborating centers and high phenotype resolution based on unified diagnostic criteria to adequately identify common variant associations to ERP.
Supplementary Material
Acknowledgments
We thank all volunteers for their generous participation in the studies.
Expert assistance is gratefully acknowledged: Genotyping (H2K): Minna Suvela; development of ECG analysis software (H2K): Heikki Väänänen; statistical analyses (YFS): Ville Aalto, Irina Lisinen; technical assistance (Carlantino / FVG): Milla Davanzo, Laura Esposito, Angela D’Eustacchio; municipal administration / logistical support (Carlantino / FVG).
List of abbreviations
- CI
confidence interval
- ECG
electrocardiogram
- ERP
early repolarization pattern
- FHS
Framingham Heart Study
- FVG
Genetic Park of Friuli Venezia Giulia
- GWAS
genome-wide association study
- H2K
Health 2000 Study
- KORA F4
Cooperative Health Research in the Augsburg Region (KORA) F4 Study
- MONICA / KORA S1/S2
MONItoring Cardiovascular Disease (MONICA) / KORA S1 and S2 Study Subcohort
- MALDI-TOF
Matrix-assisted laser desorption / ionization time of flight
- OR
odds ratio
- SCD
sudden cardiac death
- SNP
single nucleotide polymorphism
- YFS
Young Finns Study
Footnotes
Disclosures
Dr. Newton-Cheh reports to be on the scientific advisory board of Merck, Inc. No other relationships to disclose.
Support through the German Heart Foundation (M.F.S.), the Finnish Foundation for Cardiovascular Research (K.P., L.O.), the Orion-Farmos Research Foundation (K.P.), the Max Schaldach Fellowship in Cardiac Pacing and Electrophysiology (P.A.N.), NIH grants 1RO1HL092577, 5R21DA027021, 1RO1HL104156, 1K24HL10578 (P.T.E.), the German National Genome Research Network NGFN-Plus (01GS0838) (S.K.), the German Federal Ministry for Education and Research (BMBF) / French Agence National de la Recherche (ANR) (SCD-Gene: 01KU0907) (S.K.), the BMBF cluster of excellence “personalized medicine M4” (S.K.), the NIH and the Burroughs Wellcome Fund (C.N.C.), and the Academy of Finland grants 129494 and 139635 (V.S.).
This research used data and resources from the NHLBI’s and Boston University School of Public Health’s Framingham Heart Study (N01-HC-25195), its contract with Affymetrix, Inc for genotyping services (Contract N02-HL-6-4278), and involved researchers participating in the SNP Health Association Resource (SHARe) project utilizing the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Funding for analyses was provided by the NHLBI Division of Intramural Research (C.O.D, G.P., S.J.H.). The Health 2000 Study was funded by the National Institute for Health and Welfare (THL), the Finnish Centre for Pensions (ETK), The Social Insurance Institution of Finland (KELA), The Local Government Pensions Institution (KEVA) and other organizations listed on the website of the survey (http://www.terveys2000.fi). GWAS genotyping was supported by the Wellcome Trust Sanger Institute. The MONICA/KORA Augsburg studies were financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany and supported by grants from the BMBF and the state of Bavaria. Additional support was provided from BMBF in the context of NGFN plus (01GS0838), the LMU Excellence Initiative, and the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. The Young Finns Study was financed by the Academy of Finland: grants 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi), the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9M048 and 9N035 for TeLeht), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation and Emil Aaltonen Foundation (TL). The project of “Genetic Park of FVG” was supported by Regione FVG (L.26.2008) and the Associazione Amici del Cuore.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liebman J. The early repolarization syndrome is a variation of normal. J Electrocardiol. 2007;40:391. doi: 10.1016/j.jelectrocard.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Shipley RA, Hallaran WR. The four lead electrocardiogram in 200 normal men and women. Am Heart J. 1936;11:325–345. [Google Scholar]
- 3.Haïssaguerre M, Derval N, Sacher F, et al. Sudden cardiac arrest associated with early repolarization. N Engl J Med. 2008;358:2016–2023. doi: 10.1056/NEJMoa071968. [DOI] [PubMed] [Google Scholar]
- 4.Rosso R, Kogan E, Belhassen B, et al. J-point elevation in survivors of primary ventricular fibrillation and matched control subjects: incidence and clinical significance. J Am Coll Cardiol. 2008;52:1231–1238. doi: 10.1016/j.jacc.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Tikkanen JT, Anttonen O, Junttila MJ, et al. Long-Term Outcome Associated with Early Repolarization on Electrocardiography. N Engl J Med. 2009;361:2529–2537. doi: 10.1056/NEJMoa0907589. [DOI] [PubMed] [Google Scholar]
- 6.Sinner MF, Reinhard W, Müller M, et al. Association of early repolarization pattern on ECG with risk of cardiac and all-cause mortality: a population-based prospective cohort study (MONICA/KORA) PLoS Med. 2010;7:e1000314. doi: 10.1371/journal.pmed.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tikkanen JT, Junttila MJ, Anttonen O, et al. Early Repolarization: Electrocardiographic Phenotypes Associated With Favorable Long-Term Outcome. Circulation. 2011;123:2666–2673. doi: 10.1161/CIRCULATIONAHA.110.014068. [DOI] [PubMed] [Google Scholar]
- 8.Reinhard W, Kaess BM, Debiec R, et al. Heritability of early repolarization: a population-based study. Circ Cardiovasc Genet. 2011;4:134–138. doi: 10.1161/CIRCGENETICS.110.958298. [DOI] [PubMed] [Google Scholar]
- 9.Noseworthy PA, Tikkanen JT, Porthan K, et al. The early repolarization pattern in the general population clinical correlates and heritability. J Am Coll Cardiol. 2011;57:2284–2289. doi: 10.1016/j.jacc.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haïssaguerre M, Chatel S, Sacher F, et al. Ventricular fibrillation with prominent early repolarization associated with a rare variant of KCNJ8/KATP channel. Journal of Cardiovascular Electrophysiology. 2009;20:93–98. doi: 10.1111/j.1540-8167.2008.01326.x. [DOI] [PubMed] [Google Scholar]
- 11.Medeiros-Domingo A, Tan B, Crotti L, et al. Gain-of-function mutation S422L in the KCNJ8-encoded cardiac K(ATP) channel Kir6. 1 as a pathogenic substrate for J-wave syndromes. Heart Rhythm. 2010;7:1466–1471. doi: 10.1016/j.hrthm.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burashnikov E, Pfeiffer R, Barajas-Martinez H, et al. Mutations in the cardiac L-type calcium channel associated with inherited J-wave syndromes and sudden cardiac death. Heart Rhythm. 2010;7:1872–1882. doi: 10.1016/j.hrthm.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Willer CJ, Ding J, Scheet P, Abecasis G. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.METAL - Meta Analysis Helper. [Accessed May 28, 2012.]; http://www.sph.umich.edu/csg/abecasis/metal/index.html.
- 17.Hanson B, Tuna N, Bouchard T, et al. Genetic factors in the electrocardiogram and heart rate of twins reared apart and together. Am J Cardiol. 1989;63:606–609. doi: 10.1016/0002-9149(89)90907-7. [DOI] [PubMed] [Google Scholar]
- 18.Fox CS, Parise H, D’Agostino RB, Sr, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 19.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newton-Cheh C, Eijgelsheim M, Rice K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeufer A, Sanna S, Arking DE, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arking DE, Pfeufer A, Post W, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 24.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 25.Haruta D, Matsuo K, Tsuneto A, et al. Incidence and prognostic value of early repolarization pattern in the 12-lead electrocardiogram. Circulation. 2011;123:2931–2937. doi: 10.1161/CIRCULATIONAHA.110.006460. [DOI] [PubMed] [Google Scholar]
- 26.Tikkanen JT, Junttila MJ, Anttonen O, et al. Early repolarization: electrocardiographic phenotypes associated with favorable long-term outcome. Circulation. 2011;123:2666–2673. doi: 10.1161/CIRCULATIONAHA.110.014068. [DOI] [PubMed] [Google Scholar]
- 27.Rosso R, Glikson E, Belhassen B, et al. Distinguishing “benign” from “malignant early repolarization”: The value of the ST-segment morphology. Heart Rhythm. 2012;9:225–229. doi: 10.1016/j.hrthm.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Olson KA, Viera AJ, Soliman EZ, Crow RS, Rosamond WD. Long-term prognosis associated with J-point elevation in a large middle-aged biracial cohort: the ARIC study. Eur Heart J. 2011;32:3098–3106. doi: 10.1093/eurheartj/ehr264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adhikarla C, Boga M, Wood AD, Froelicher VF. Natural History of the Electrocardiographic Pattern of Early Repolarization in Ambulatory Patients. Am J Cardiol. 2011;108:1831–1835. doi: 10.1016/j.amjcard.2011.07.055. [DOI] [PubMed] [Google Scholar]
- 30.Dixon JE, Shi W, Wang HS, et al. Role of the Kv4.3 K+ channel in ventricular muscle. A molecular correlate for the transient outward current. Circ Res. 1996;79:659–668. doi: 10.1161/01.res.79.4.659. [DOI] [PubMed] [Google Scholar]
- 31.Tseng GN. Molecular structure of cardiac Ito channels: Kv4.2, Kv4.3, and other possibilities? Cardiovasc Res. 1999;41:16–18. doi: 10.1016/s0008-6363(98)00282-x. [DOI] [PubMed] [Google Scholar]
- 32.Gussak I, Antzelevitch C. Early repolarization syndrome: clinical characteristics and possible cellular and ionic mechanisms. J Electrocardiol. 2000;33:299–309. doi: 10.1054/jelc.2000.18106. [DOI] [PubMed] [Google Scholar]
- 33.Delpón E, Cordeiro JM, Núñez L, et al. Functional effects of KCNE3 mutation and its role in the development of Brugada syndrome. Circ Arrhythm Electrophysiol. 2008;1:209–218. doi: 10.1161/CIRCEP.107.748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank-Hansen R, Larsen LA, Andersen P, Jespersgaard C, Christiansen M. Mutations in the genes KCND2 and KCND3 encoding the ion channels Kv4.2 and Kv4. 3, conducting the cardiac fast transient outward current (ITO, f), are not a frequent cause of long QT syndrome. Clin Chim Acta. 2005;351:95–100. doi: 10.1016/j.cccn.2004.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.