Abstract
We aimed to identify novel molecular prognostic markers to better predict relapse risk estimate for children with high-risk (HR) metastatic neuroblastoma (NB). We performed genome- and/or transcriptome-wide analyses of 129 stage 4 HR NBs. Children older than 1 year of age were categorized as “short survivors” (dead of disease within 5 years from diagnosis) and “long survivors” (alive with an overall survival time ≥ 5 years). We reported that patients with less than three segmental copy number aberrations in their tumor represent a molecularly defined subgroup with a high survival probability within the current HR group of patients. The complex genomic pattern is a prognostic marker independent of NB-associated chromosomal aberrations, i.e., MYCN amplification, 1p and 11q losses, and 17q gain. Integrative analysis of genomic and expression signatures demonstrated that fatal outcome is mainly associated with loss of cell cycle control and deregulation of Rho guanosine triphosphates (GTPases) functioning in neuritogenesis. Tumors with MYCN amplification show a lower chromosome instability compared to MYCN single-copy NBs (P = .0008), dominated by 17q gain and 1p loss. Moreover, our results suggest that the MYCN amplification mainly drives disruption of neuronal differentiation and reduction of cell adhesion process involved in tumor invasion and metastasis. Further validation studies are warranted to establish this as a risk stratification for patients.
Introduction
Neuroblastoma (NB) is the second most common solid tumor in children with a markedly heterogeneous clinical behavior, varying from spontaneous regression to rapid progression [1]. About 40% of children onset with high-risk (HR) disseminated disease. The current therapies have led to a modest improvement in long-term survival rates of this group of patients [2–4], despite intensified therapeutic regimens [5]. A poor rate survival is frequently observed in patients more than 1 year of age, mainly because of poor responses or disease relapse in primary and metastatic sites, particularly bone marrow. Moreover, about 40% of survivors with advanced NB have sequelae of aggressive multimodal therapies, i.e., endocrine disturbances, cataracts, hypertension, bronchiolitis, blindness, peripheral neuropathy, nonfunctioning kidney, cholelithiasis, and thyroid nodules. Hence, a risk-adapted therapy according to disease status and molecular profile may decrease incidence of cytotoxic deaths because of high-dose therapy and morbidity among surviving patients.
Patient's age, stage, MYCN oncogene amplification, histology, and ploidy of tumor are associated with poor prognosis [1]. In the last decades, microarray-based high-throughput studies identified genome and transcriptome signatures that predict patient outcome [6–15]. Tumors with near-triploid karyotypes and numerical aberrations (i.e., whole-chromosome gains and losses) have a good prognosis, whereas near-diploid or near-tetraploid tumors with segmental rearrangements, including deletions of parts of chromosome arms 1p or 11q, gain of 17q, and MYCN amplification, have a poor prognosis [16,17]. Furthermore, a 59-gene expression (GE) signature proved to be helpful as an accurate outcome predictor in patients with NB [18,19].
Recently, the application of a pan-genomic approach has been validated in the ongoing multicenter Low- and Intermediate-Risk Neuroblastoma Study [20]. The treatment stratification in the Low- and Intermediate-Risk Neuroblastoma Study is based on recent trials, suggesting that the risk of relapse in patients with MYCN not amplified low-risk tumors may be defined by the presence of any structural genetic abnormalities [21]. By contrast, for patients with stage 4 NB more than 1 year of age, the MYCN amplification and other biomarkers fail in predicting risk of progression/recurrence.
Previously, Bilke et al. [14] reported that whole chromosome alterations predict survival in stage 4 NBs without MYCN amplification. However, this was a pilot study and no further validations established this risk stratification for these patients.
The present study is the largest one performed on children older than 1 year of age at diagnosis with stage 4 NB and with the longest follow-up time as yet. We describe the results of an integrated genomic and RNA analysis using both array-based comparative genomic hybridization (aCGH) and GE profiling in stage 4 NB tumors of children older than 1 year of age at diagnosis. We also investigated the role of MYCN oncogene amplification in disease progression of these patients.
Materials and Methods
Tumor Samples
Tumor specimens were collected at the onset of disease from 129 patients who were diagnosed with a primary NB between 1988 and 2006. The study was approved by the Institutions' Ethical Committees. All patients were classified as stage 4 according to the International Neuroblastoma Staging System [22], and they were older than 1 year of age at time of diagnosis (average age, 47 months). Regarding the clinical course, patients were categorized into two subgroups, namely, “short survivors” (dead of disease within 60 months from diagnosis; deaths because of toxicity were censored) and “long survivors” [alive with an overall survival (OS) time ≥ 60 months]. No significant differences over time were recorded in OS of patients in relation to enrollment year (data not shown). Patients' and tumor characteristics are summarized in Table W1.
Analyses were performed in cohorts of patients defined as follows: 1) For aCGH profiling, all patients with available genomic tumor DNA were selected (total 91: 55 Italian and 36 German patients; short survivors, 46; long survivors, 45). Median age of patients at diagnosis was 43 months. The median follow-up of alive patients was 100 months (range, 61–164 months) and median event-free survival (EFS) time was 96 months (range, 22–164 months). None but three patients of the long-survivor group had relapse or progression of disease. The short survivors showed a median follow-up of 22 months (range, 1–57 months) and a median EFS of 15 months (range, 1–40 months). Amplification of MYCN oncogene was present in tumors of 22 short- and 4 long-surviving patients. 2) For GE profiling, we selected all available tumors from the previous study by Oberthuer et al. [23], which fitted to the clinical criteria described above (total 73: short survivors, 50; long survivors, 23). Median age at diagnosis was 54 months. The median follow-up of alive patients was 93 months (range, 60–156 months) and median EFS time was 90 months (range, 7–156 months). The short-survivor group showed a median follow-up of 28 months and a median EFS time of 16 months. Amplification of MYCN oncogene was present in tumors of 20 short and 2 long-surviving patients.
All tumor samples were classified as Schwannian stroma-poor NB according to the International Neuroblastoma Pathology Committee [24], and they have at least 60% of neuroblasts.
Array-based Comparative Genomic Hybridization
High-resolution aCGH was performed using either 44K or 105K oligonucleotide microarrays (Agilent Technologies, Santa Clara, CA) as described elsewhere [17,25]. Images were extracted by Feature Extraction 9.5 and analyzed by CGH Analytics 3.5.14 software (Agilent Technologies), as specified previously [26]. Fisher exact test [27] was used to select individual chromosomes showing nonrandom associations between groups and aberrations and to select individual chromosomes showing nonrandom associations among groups and aberrations. Chromosomes with P values smaller than .05 after Benjamini-Hockberg multiple comparison correction were selected. Minimal common regions (MCRs) were defined as loci with significant copy number aberrations (CNAs) in all samples. The aCGH data have been deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and they are available by Gene Expression Omnibus Series Accession Nos. GSE14109, GSE25771, and GSE35953 (for details, see Table W1).
GE Profiling
GE profiling experiments were carried out using a customized NB-related oligonucleotide microarray (Agilent Technologies) that comprised 10,163 probes covering 8155 Unigene clusters [23,28]. Expression profiles were analyzed as described elsewhere [26]. Microarray data are available on Array Express database (http://www.ebi.ac.uk/arrayexpress; Accession Nos. E-TABM-38 and E-MTAB-161).
Integrated Copy Number and Expression Analyses
We used two approaches to identify genes whose transcription levels were potentially affected by DNA alterations. In a “targeted study,” we identified the significant targeted genes (STGs) differentially expressed among NB groups [false discovery rate (FDR) < 0.05], located in the most frequent aberrant chromosome region, and concordant to the corresponding CNAs, i.e., we selected overexpressed and underexpressed genes located in a chromosome region of gain and loss, respectively. In a “correlated study,” we identified the genes with expression levels correlated with MCRs but not necessarily with a significant differential expression in all NBs. For this purpose, chromosome coordinates of all probe sets were extracted from Feature Extraction export files by R scripts and we selected those probe sets located in MCR and concordant to the corresponding CNAs, i.e., we selected those probe sets located in a region of gain or loss and overexpressed or underexpressed, respectively.
Statistical and Bioinformatics Analyses
MedCalc (Mariakerke, Belgium) software was used for statistical analyses. Mann-Whitney and Fisher exact tests compared the frequency of CNA between long- and short-surviving individuals. All significance tests were two tailed. To analyze the effect of changing the chromosome aberration number threshold providing the best separation between long and short survivors, we dichotomized the number of CNAs according to receiver operating characteristic (ROC) curves [29]. These threshold values were employed in subsequent survival analyses.
The OS was defined as the time elapsed from diagnosis to cancer-related death or to the date of the last follow-up if the patient survived. Patients who survived were censored at the last date they were reported to be alive. The EFS was calculated from diagnosis to the date of tumor progression or relapse or to the date of the last followup if no event occurred. Survival curves were computed according to the Kaplan-Meier method. The Cox proportional hazards regression model was used to study in multivariate settings the effect of the known prognostic markers in NB (MYCN amplification, 1p and 11q losses, and 17q gain) as potential confounders. An enter selection approach was chosen and only variables with P < .05 were retained in the model.
Gene Ontology and Canonical Pathway Analyses
Functional annotation analyses were performed by Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) [30]. A Gene Ontology (GO) category was considered statistically significant at significance threshold set on P < .05. Functional annotation analyses were performed by taking into account underexpressed and overexpressed groups separately.
Results
Specific Genomic Aberrations Characterize Clinical HR NB Subgroups
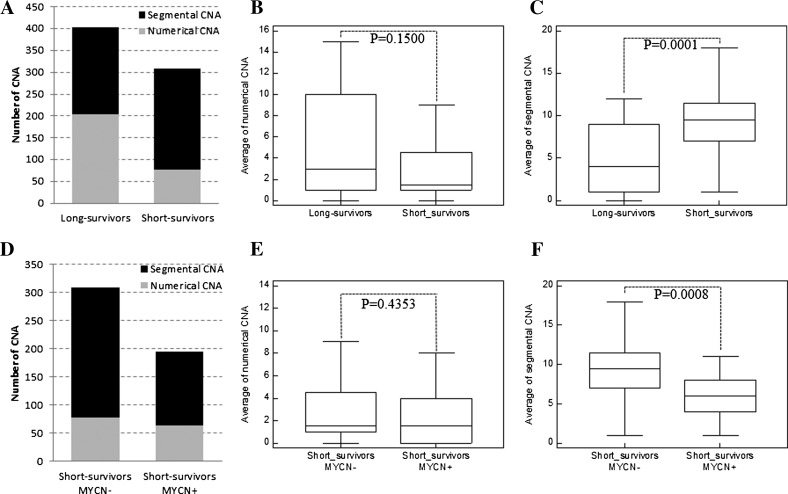
First, we investigated CNAs in tumors of long- and short-surviving patients. For this purpose, tumors with MYCN amplification were intentionally excluded from the analysis because of their potential bias. According to this criterion, 65 samples (24 short and 41 long survivors) were selected. All tumors showed CNAs (Table W1). Tumors of long survivors showed 204 numerical aberrations (average 5.0/case; range, 0–15) and 200 segmental aberrations (average 4.9/case; range, 0–12). The most frequent abnormalities were 7 gain (24 of 41, 58.5%), 17q gain (23 of 41, 56%), 11q loss (19 of 41, 46%), 17 gain (17 of 41, 41.5%), 18 gain (17 of 41, 41.5%), and 2p gain (16 of 41, 39%). All tumors of short-surviving group had at least one segmental aberration (average 9.7/case; range, 1–20). Recurrent changes were 17q gain (22 of 24, 92%), 11q loss (15 of 24, 62.5%), 3p loss (11 of 24, 46%), 11q gain (11 of 24, 46%), 1p loss (11 of 24, 46%), 2p gain (9 of 24, 37.5%), and 11p gain (9 of 24, 37.5%). Numerical aberrations were present with a lower frequency in tumors of patients with an adverse clinical outcome (average 3.2/case; range, 0–14; Figure 1, A and B), mainly represented by gain of whole chromosome 7 (11 of 24, 45.8%). Although all tumors were characterized by complex aberration patterns focused on chromosomes 1, 2, 3, 11, and 17, the number of segmental alterations was significantly higher (P = .0001, Mann-Whitney test) in the short than in long survivors (Figure 1, A–C).
Figure 1.
Summary of aCGH results. (A) Bar chart and box-and-whisker graphs of the number of (B) numerical and (C) segmental CNAs in the two groups of long and short survivors without MYCN amplification. (D) Bar chart and box-and-whisker graphs of the number of (E) numerical and (F) segmental CNAs in the two groups of short survivors with and without MYCN amplification.
Genomic instability of each tumor was also evaluated by taking into account the number of intrachromosome segmental aberrations (ICSAs). Our results showed that the number of ICSA was significantly higher (P = .0005) in the short than in the long survivors (Figure W1A).
Chromosome Aberrations Associated with MYCN Amplification in HR NB Tumors
To investigate the association between MYCN amplification and chromosome instability of HR NB, we compared genome profiles of 22 MYCN-amplified [MYCN+; median follow-up of patients: 16 months (range, 1–57 months); median EFS time: 13 months (range, 1–40 months)] and 24 MYCN single-copy [MYCN-; median follow-up of patients: 28 months (range, 6–48 months); median EFS time: 18 months (range, 5–29 months)] tumors from short-surviving patients. The MYCN- tumors showed a wide variety of segmental CNAs dominated by 17q gain (22 of 24, 92%), 11q loss (15 of 24, 62.5%), 1p loss (11 of 24, 46%), 3p loss (11 of 24, 46%), and 11q gain (11 of 24, 46%). In contrast, the MYCN+ tumors showed a less complex segmental CNA pattern compared to the MYCN- group (P = .0008, Mann-Whitney test; Figure 1, D–F). The lower degree of genomic instability of MYCN+ tumors was defined also by the smaller number of ICSA than the MYCN- group (P = .0231; Figure W1B). The most recurrent changes in MYCN+ cases were 17q gain (18 of 22, 82%) and 1p loss (17 of 22, 77%).
Correlation of Genomic Instability with Clinical Outcome
Five segmental genomic aberrations of MYCN- tumors revealed a close association with the course of the disease: adverse clinical outcome was associated with 1p loss (P = .009, Fisher exact test), 11p gain (P = .010), 11q gain (P = .021), 17q gain (P = .002), and 19q loss (P = .0005). As shown in Figure W2, these CNAs were negative predictors of OS (P = .0008, P = .0110, P = .0185, P = .0055, and P = .0001, respectively) and EFS (P = .0010, P = .0097, P = .0067, P = .0067, and P = .0001, respectively).
Therefore, although we confirmed the inverse correlation between MYCN amplification and 11q loss, in contrast to previous reports [31,32], we found 11q loss (MRC: 11q23.1–q24.1) as one of the most frequent abnormalities in MYCN- tumors of both long and short survivors without any significant relationship with clinical outcome (P = .3477 for OS and P = .2395 for EFS). Intriguingly, in our cohort of MYCN- tumors, 11q loss was significantly frequently associated with 11q gain (MCR: 11q13.1–q13.3) and/or 11p gain (MCR: 11p11.12–p12) in short survivors with respect to long survivors (10 of 24, 42% vs 6 of 41, 15%, P = .019 for 11q gain; 8 of 24, 42% vs 4 of 41, 15%, P = .024 for 11p gain). The 11q gain resulted a negative predictor of poor survival. We argue that it may be because of the high chromosome instability found in short survivors that leads, in case of chromosome 11, to several chromosome breakpoints causing losses/gains of different portions of short and long arms. We cannot exclude the presence of isochromosome 11 [i.e., idic(11)(pter→q13.4:: q13.4→pter), idic(11)(pter→q14.1::q14.1→pter)] as result of 11p and 11q arm rearrangements.
The predictive value of CNA on patients' outcome was then evaluated by univariate and multivariate models within the cohort of all 91 patients analyzed by aCGH. The Kaplan-Meier curves confirmed that 1p loss, 17q gain, and 19q loss were associated with a poorer outcome (P values for the log-rank tests for OS and EFS were <.04 for the three CNAs; Figure W3). The Cox regression analysis (Table 1), after the adjustment for the effect of MYCN amplification, showed the predictive value on OS and EFS of 19q loss (P < .03) but not of 1p loss and 17q gain likely because of the association between these two CNAs and MYCN amplification. Because the role of 17q is still somewhat controversial [33,34], it is noteworthy that the stratified analysis using MYCN- tumors confirmed that 17q gain is independently predictive of poor outcome (Figure W2).
Table 1.
Cox Regression Analysis in 91 Stage 4 HR NB Patients.
| Segmental CNA | Covariate | EFS | OS | ||||||||
| b | SE* | P | Exp(b) | Hazard Ratio (95%) CI† | b | SE* | P | Exp(b) | Hazard Ratio (95%) CI† | ||
| 1p loss | MYCN ampl | 0.638 | 0.369 | .084 | 1.893 | 0.921 to 3.892 | 0.587 | 0.390 | .133 | 1.798 | 0.839 to 3.849 |
| 11q loss | MYCN ampl | 0.515 | 0.349 | .141 | 1.674 | 0.846 to 3.310 | 0.413 | 0.323 | .201 | 1.512 | 0.805 to 2.840 |
| 17q gain | MYCN ampl | 0.529 | 0.434 | .223 | 1.698 | 0.728 to 3.956 | 0.543 | 0.464 | .242 | 1.722 | 0.696 to 4.257 |
| 19q loss | MYCN ampl | 1.49 | 0.449 | .001 | 4.427 | 1.846 to 10.668 | 1.513 | 0.461 | .001 | 4.542 | 1.849 to 11.153 |
| ≥3 CNA | MYCN ampl | 0.952 | 0.390 | .015 | 2.592 | 1.211 to 5.547 | 1.015 | 0.414 | .014 | 2.758 | 1.231 to 6.181 |
| ≥3 CNA | 1p loss | 0.701 | 0.416 | .033 | 2.016 | 0.896 to 4.538 | 0.782 | 0.435 | .038 | 2.185 | 0.934 to 5.109 |
| ≥3 CNA | 11q loss | 1.304 | 0.400 | .001 | 3.685 | 1.688 to 8.044 | 1.383 | 0.425 | .001 | 3.987 | 1.740 to 9.137 |
| ≥3 CNA | 17q gain | 0.891 | 0.420 | .034 | 2.437 | 1.073 to 5.531 | 0.936 | 0.448 | .037 | 2.550 | 1.064 to 6.110 |
| ICSA | MYCN ampl | 0.910 | 0.249 | .0003 | 2.485 | 1.526 to 4.045 | 0.995 | 0.262 | .0002 | 2.705 | 1.621 to 4.513 |
| ICSA | 1p loss | 0.730 | 0.261 | .005 | 2.075 | 1.246 to 3.455 | 0.818 | 0.272 | .003 | 2.266 | 1.332 to 3.855 |
| ICSA | 11q loss | 1.268 | 0.275 | <.0001 | 3.553 | 2.078 to 6.073 | 1.359 | 0.287 | <.0001 | 3.892 | 2.225 to 6.809 |
| ICSA | 17q gain | 0.803 | 0.257 | .002 | 2.233 | 1.353 to 3.686 | 0.859 | 0.273 | .002 | 2.363 | 1.388 to 4.024 |
Hazard ratios were adjusted for MYCN amplification, 1p loss, 11q loss, and 17q gain.
SE, standard error.
CI, confidence interval.
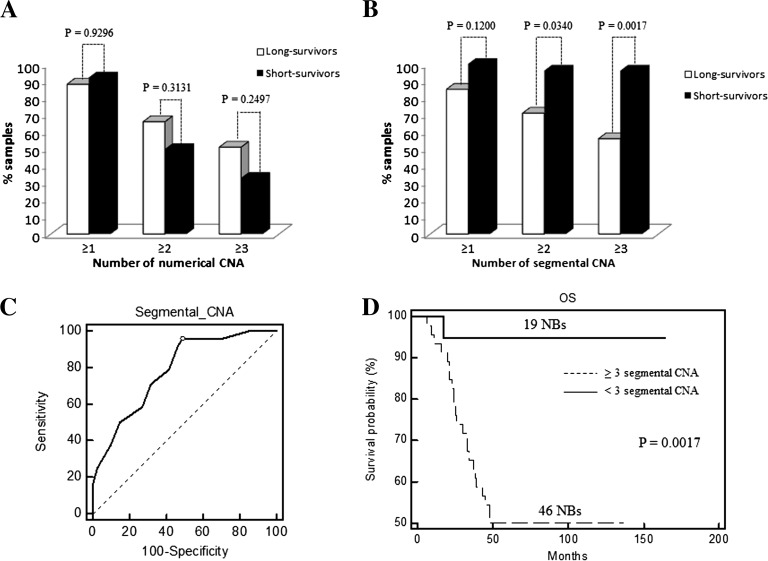
Figure 2A plots the frequency of MYCN- samples with at least one, two, or three whole chromosome changes, regardless of which specific chromosome are affected. The distinction of the positive and negative outcome groups was poor (Chi-square test, P > .2). Conversely, a significant distinction was obtained by counting samples with at least two or three segmental chromosome changes (P = .0340 and P = .0017, respectively; Figure 2B). The effect of further changing the chromosome number threshold is demonstrated by the ROC analysis (Figure 2C). ROC and Kaplan-Meier curves (Figure 2D) showed that at least three segmental CNAs are needed to discriminate (P = .0017) long from short survivors at the highest sensitivity (95.8%) and specificity (51.2%).
Figure 2.
Histogram charts showing frequency of long and short survivors without MYCN amplification showing ≥1, ≥2, and ≥3 (A) numerical and (B) segmental CNAs. (C) Sensitivity and specificity in predicting survival based on different numbers of segmental CNAs are above the thresholds defined by ROC curve. The point marked with dot represents the highest average of sensitivity and specificity at the cutoff value of 3 (AUC = 0.785, Z statistic = 5.024, P < .0001). (D) Kaplan-Meier analysis for OS of 65 MYCN- NB patients with ≥3 or <3 segmental CNAs.
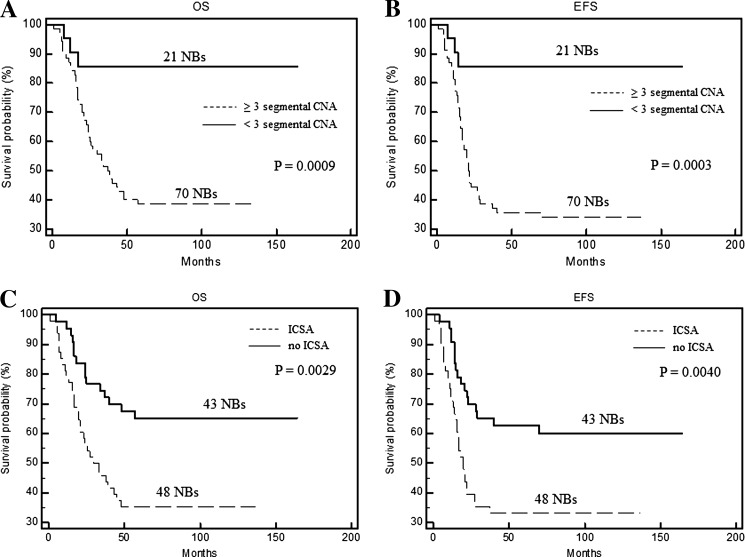
In the cohort of all 91 patients analyzed by aCGH, both univariate (Figure 3, A and B) and multivariate (Table 1) models confirmed the predictive value of segmental CNA above the threshold on OS and EFS (P = .0009 and P = .0003, respectively), regardless of other NB-associated chromosomal aberrations, i.e., MYCN amplification, 1p and 11q losses, and 17q gain. Accordingly, NBs with high number of ICSA were associated with a poor prognosis (P < .005; Figure 3, C and D). The prognostic impact of structural variations per chromosome was independent of MYCN amplification, 1p loss, 11q loss, and 17q gain in multivariate analyses (Table 1).
Figure 3.
Kaplan-Meier OS and EFS estimates in 91 HR NB patients according to the segmental CNA threshold (A, B) and the presence of ICSA (C, D).
Identification of Region-Specific Candidate Genes by Integration of aCGH and GE Data Sets
To determine the influence of chromosomal alterations on locus-specific GE, we integrated the aCGH and transcriptome data sets. We first focused on a gene-by-gene basis analyses of copy number and expression data throughout the genome of 51 MYCN- NBs (21 long and 30 short survivors). Tumors from the two clinical subgroups differed in 169 genes (Table W2). Transcription levels were correlated to genomic patterns for 36 STGs, representing 22% of the genes significantly associated with adverse outcome. The STGs were located on chromosomes 1p, 3p, 11q, and 17q (Table W3). Functional annotation highlighted GO terms mostly associated with cell junction, neuritogenesis, and Rho GTPase-mediated signal transduction (Table W4).
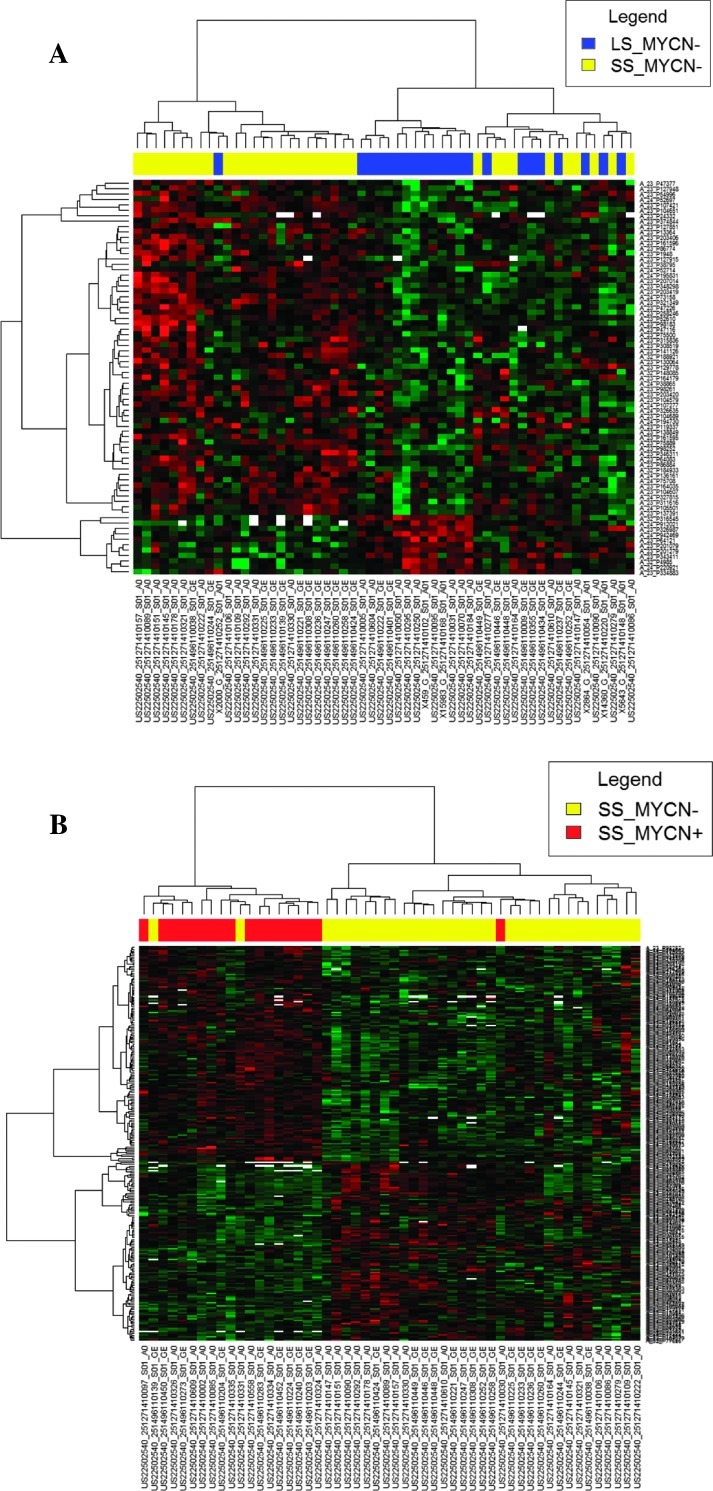
Then, we combined the aCGH and GE data sets to focus only on the genes restricted to MCRs for which the expression was affected by aberrant copy number variations. We selected the 646 probe sets mapping at MCRs of the CNA significantly associated with adverse outcome in MYCN- tumors. Most patterns of gene amplification and increased GE were concordant, that is, a significant fraction of the amplified genes were correspondingly highly expressed. Such concordance was also found for part of the genes located in losses. The “correlated study” showed that DNA copy number influenced GE across a range of MCRs corresponding to 57 correlated genes that were differentially expressed between tumors from the two clinical subgroups and concordant with MCR patterns (Figure 4A and Table W5). Fifty-two genes (91%) were overexpressed in NBs of short survivors and 77% of them mapped at 11q gain (MCR: 11q12–q13.5). The most enriched GO terms were mRNA processing (AIP, DRAP1, SART1, and SF3B2) and cell cycle process (ARL2, SAC3D1, SSSCA1, CDCA5, PSMC3, PSMD13, and SART1). Intriguingly, 11p15.5 gain was found associated with overexpression of HRAS gene, known to be involved in activation of cell proliferation. Moreover, the deletions of 1p36 and 19q13.43 affected expression of three genes (CAMTA1, PRDM2, and ZSCAN22) involved in regulation of transcription (Table W5).
Figure 4.
Correlated studies. Hierarchically clustered heat maps of probe sets mapped on MCR with significant differential expression in (A) MYCN- tumors from long survivors versus MYCN- tumors from short survivors and (B) MYCN- tumors from short survivors versus MYCN+ tumors from short survivors. Each color patch represents the expression level of genes (row) in that sample (column), with a continuum of expression levels from bright green (lowest) to bright red (highest).
Transcription profile differences associated with MYCN overexpression were highlighted by comparing 20 MYCN+ and 30 MYCN- tumors of short survivors (Table W2). The “targeted study” showed that DNA copy number directly influenced expression of 39 STGs representing 27% of the genes significantly associated with MYCN amplification and located on chromosomes 1p, 2p, 3p, 11q, and 17q (Table W3). The 31 STGs (56%) down-expressed in regions frequently lost in MYCN+ tumors were related to angiogenesis (VEGFB), cell adhesion (CDC42 and PIK3CD), neuritogenesis of NB cells (CDC42) [35], and regulation of small GTPase-mediated signal transduction (CDC42, ASAP3, ARHGEF10L, and MFN2; Tables W3 and W4).
Finally, we selected 591 probe sets mapping at gained or lost MCRs significantly associated with MYCN amplification (Figure 4B). GE analysis identified 169 genes differentially expressed between the two groups of NBs (Table W5). A significant number of amplified and correspondingly highly expressed genes in MYCN+ tumors were active in proliferative (i.e., CUL5, H2AFX, MRE11A, RBM7, and ZW10 mapping at 11q21–q23.3) and transcriptional processes (ZNF350, ZNF135, TRIM28, ZNF473, RUVBL2, ZNF587, ZNF525, and CRX at 19q13.3–q13.4). Conversely, down-expressed correlated genes in MYCN+ NBs are mostly associated with apoptosis (HRAS, CD44, MADD, ILK, SMPD1, MAPK8IP1, and APBB1), cell-matrix and cell-cell adhesion (CD44, FXC1, ILK, and PARVA), and neuronal differentiation (CD44, TH, ILK, and APBB1) pathways at 11p11.2-pter. Interestingly, MYCN oncogene overexpression was also associated with upregulation of its neighbor genes (DDX1, FAM49A, and NAG) and of MRE11A target gene. Moreover, the “correlated study” showed the decrease of DDB2 expression, one of the known genes directly downregulated by MYCN, and RHOG, a member of the Rac subfamily of the Rho family of small G proteins. Finally, MYCN+ NBs showed a lower expression of two functionally interesting genes at 11q13.1–q23, namely, RARRES3, which encodes a retinoic acid receptor responder, and BRMS1, which is a cancer metastasis suppressor gene involved in breast cancer progression.
Discussion
In the present paper, we report integrative DNA copy number and GE analyses in HR metastatic NB. During the last decade, several articles reported cytogenetic aberrations able to affect abnormal GE profiles in NB [6–19]. Moreover, large-scale genomic imbalances play a role in the dysregulation of miRNA expression [36]. Correlations of specific patterns of copy number alterations with patient survival have been identified in a broad set of NB samples covering all risk categories. Differently, here we focused on stage 4 HR NB with follow-up for at least 5 years, and this is the largest study for this HR subtype.
It is noteworthy that although tumor specimens were collected from patients who were diagnosed with a primary NB between 1988 and 2006, no significant differences over time were recorded in OS of patients in relation to enrollment year. This is in agreement with the recent report by Haupt et al. [4] showing that no OS improvement was seen for stage 4 patients in 1985 to 2005 period of diagnosis.
First, we excluded tumors with MYCN oncogene amplification. The resulting association between average of segmental aberrations and fatal outcome was therefore not affected by MYCN bias. Our analysis indicates that patients with CNA < 3 in their tumors have a significantly better prognosis with respect to those with a higher number of chromosomal alterations, including ICSA, both in terms of EFS and OS. This supports speculations of multiple molecular subtypes of NB [13,37]. The significant higher chromosome instability in tumors of short survivors may be because of a loss of function of mitotic checkpoint control mechanisms [38].
Then, we highlighted CNA associated with MYCN amplification in patients with fatal outcome. Our data show two subtypes of NBs with different genomic profiles: whereas MYCN+ cases involve 2p gain and often 11q loss and 17q gain but very few other segmental aberrations, the MYCN- tumors generally have a complex pattern of CNA, mainly focused on chromosomes 1, 2, 3, 11, 17, and 19. This higher DNA instability observed in MYCN- NBs may explain why children older than 12 months of age with a stage 4 tumor have a poor outcome independently by MYCN status.
As regarding transcript analysis, we used a dual strategy that delineated genes with either significantly concordant or correlated changes in copy number and in expression. The major advantage of choosing such approach was that it bypasses limitations because of the small number of cases and intertumor heterogeneity. Moreover, such strategy allowed us to delineate two sets of genes with distinct biologic relevance. The STGs show expression alterations in almost all samples, and therefore, they correspond to changes essential for aggressiveness and tumor progression. In contrast, the genes identified by the “correlated study” have an expression directly linked to the DNA copy number status, representing functionally significant events associated with chromosome gains and losses and related to different clinical subclasses of stage 4 HR NBs.
The widespread homogeneous STG down-expression in tumors from short survivors involves cell adhesion processes, angiogenesis, and neuron differentiation defect. In particular, the disruption of neuronal differentiation mirrors a more primitive phenotype in aggressive NBs, regardless MYCN amplification. Functional annotation analyses show that the two clusters of long and short survivors are mainly dependent on deregulation of STGs involved in Rho/Ras-mediated signal transduction. The Rho family of small GTPases, in particular Rho, Rac (i.e., RhoG), and CDC42, are important regulators of signaling pathways controlling cell morphology, migration, and cell cycle progression [39–41]. Recent studies using knockout mouse models provided evidence of the primary role played by Rho signaling during the development of the nervous system [42]. As with all small G proteins, Rho family is able to signal to downstream effectors when bound to guanosine triphosphate (GTP) and inactive when bound to guanosine diphosphate (GDP). Among the proteins interacting with Rho to regulate GTP/GDP loading, guanine nucleotide exchange factors (GEFs) facilitate the exchange of GDP for GTP so as to induce subsequent Rho-mediated signaling, and GTPase-activating proteins promote GTP hydrolysis, thus terminating the Rho cascade. There are a number of GEFs reported to interact with RhoG. Well-characterized GEFs include TRIO that is able to promote nucleotide exchange on RhoG, Rac, and RhoA [43,44]. Activation of RhoG by TRIO has been shown to promote nerve growth factor (NGF)-induced neurite outgrowth in PC12 cells [45] and to coordinate cell-matrix and cytoskeletal rearrangements necessary for cell migration and cell growth. Our “targeted study” shows the involvement of deregulation of Rho GTPase pathway and genes related to cell motility in loss of cell cycle control and progression of tumor, regardless MYCN status. On the one hand, this explains the increase of tumor chromosome instability in patients with fatal outcome and why MYCN gene amplification is not a prognostic marker in stage 4 patients. However, we argue that defects in regulation of neuritogenesis represent important tumor-driving events in HRNBs, as recently reported by Molenaar et al. [46]. Actually, our findings indicate that tumors without MYCN amplification but with deregulation of Rho GTPase signaling genes functioning in neuritogenesis mostly are aggressive HR NBs. Intriguingly, in MYCN+ tumors, the inhibition of neuritogenesis may be because of the MYCN-driven downregulation of CDC42 and RHOG expression.
Moreover, high-level expression of angiogenic factors in tumors of short survivors confirms that angiogenic factors are associated with advanced tumor stage in human NB [47]. Notably, our data show that expression level of VEGFB was not associated with MYCN amplification. Therefore, one may speculate that up-regulation of VEGFB is a mechanism used by MYCN-aggressive NB tumors to attract vascular endothelial cells.
The “correlated study” highlights other signatures related to regulatory mechanisms for NB progression, such as proliferative and transcriptional processes, and specific MYCN-driven pathways. For example, in MYCN+ tumors, the expression of cell-matrix and cell-cell adhesion genes is significantly suppressed. Recent experimental results indicate that changes in the expression or function of cell adhesion molecules can contribute to tumor progression and promote tumor invasion and metastasis [48]. Finally, the top-ranked enriched sets in MYCN+ tumors contain MYCN-responsive genes and genes involved in ribosome biogenesis and assembly, as recently reported [49].
In conclusion, children with less than three segmental CNA in their tumor represent a molecularly defined subgroup with a high survival probability within the current HR group of patients, regardless MYCN status. The complex genomic pattern is a prognostic marker independent of NB-associated chromosomal aberrations, i.e., MYCN oncogene amplification, 1p and 11q losses, and 17q gain. Integrative analysis of genomic and expression signatures demonstrated that fatal outcome is mainly associated with loss of cell cycle control and deregulation of Rho GTPase pathway. Moreover, the MYCN amplification mainly drives the disruption of neuritogenesis and the reduction of cell adhesion process involved in tumor invasion and metastasis. Further validation studies are warranted to establish this as a risk stratification for patients.
Supplementary Material
Acknowledgments
We are grateful to surgeons, clinicians, pathologists of the Associazione Italiana Ematologia Oncologia Pediatrica, and the Tumor Bank of Cologne (University Children's Hospital of Cologne, Cologne, Germany) for providing tumor samples and to Dr Silvia De Luca (Fondazione Italiana per la Lotta al Neuroblastoma, Genoa, Italy) for language revision.
Abbreviations
- aCGH
array-based comparative genomic hybridization
- CNAs
copy number aberrations
- EFS
event-free survival
- FE
Feature Extraction
- GE
Gene Expression
- GO
Gene Ontology
- HR
high-risk
- ICSAs
intrachromosome segmental aberrations
- MCRs
minimal common regions
- MYCN+
MYCN-amplified
- MYCN-
MYCN single copy
- NB
neuroblastoma
- OS
overall survival
- ROC
receiver operating characteristic
- STGs
significant targeted genes
Footnotes
This study was funded by Fondazione Italiana per la Lotta al Neuroblastoma, UICC Yamagiwa-Yoshida Memorial International Cancer Study Grant (grant YY2/07/002), European Association for Cancer Research (Travel Fellowship No. 322), Associazione Italiana per la Ricerca sul Cancro, and Ministero dell'Istruzione, dell'Università e della Ricerca.
This article refers to supplementary materials, which are designated by Tables W1 to W5 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Pearson AD, Pinkerton CR, Lewis IJ, Imeson J, Ellershaw C, Machin D. European Neuroblastoma Study Group, and Children's Cancer and Leukaemia Group High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: a randomised trial. Lancet Oncol. 2008;9:247–256. doi: 10.1016/S1470-2045(08)70069-X. [DOI] [PubMed] [Google Scholar]
- 3.Matthay KK, Reynolds CP, Seeger RC, Shimada H, Adkins ES, Haas-Kogan D, Gerbing RB, London WB, Villablanca JG. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a Children's Oncology Group Study. J Clin Oncol. 2009;27:1007–1013. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haupt R, Garaventa A, Gambini C, Parodi S, Cangemi G, Casale F, Viscardi E, Bianchi M, Prete A, Jenkner A, et al. Improved survival of children with neuroblastoma between 1979 and 2005: a report of the Italian Neuroblastoma Registry. J Clin Oncol. 2010;28:2331–2338. doi: 10.1200/JCO.2009.24.8351. [DOI] [PubMed] [Google Scholar]
- 5.Ora I, Eggert A. Progress in treatment and risk stratification of neuroblastoma: impact on future clinical and basic research. Semin Cancer Biol. 2011;21:217–228. doi: 10.1016/j.semcancer.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Hiyama E, Hiyama K, Yamaoka H, Sueda T, Reynolds CP, Yokoyama T. Expression profiling of favorable and unfavorable neuroblastomas. Pediatr Surg Int. 2004;20:33–38. doi: 10.1007/s00383-003-1077-3. [DOI] [PubMed] [Google Scholar]
- 7.Takita J, Ishii M, Tsutsumi S, Tanaka Y, Kato K, Toyoda Y, Hanada R, Yamamoto K, Hayashi Y, Aburatani H. Gene expression profiling and identification of novel prognostic marker genes in neuroblastoma. Genes Chromosomes Cancer. 2004;40:120–132. doi: 10.1002/gcc.20021. [DOI] [PubMed] [Google Scholar]
- 8.Wei JS, Greer BT, Westermann F, Steinberg SM, Son CG, Chen QR, Whiteford CC, Bilke S, Krasnoselsky AL, Cenacchi N, et al. Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 2004;64:6883–6891. doi: 10.1158/0008-5472.CAN-04-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosse YP, Greshock J, Weber BL, Maris JM. Measurement and relevance of neuroblastoma DNA copy number changes in the post-genome era. Cancer Lett. 2005;228:83–90. doi: 10.1016/j.canlet.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 10.Ohira M, Oba S, Nakamura Y, Isogai E, Kaneko S, Nakagawa A, Hirata T, Kubo H, Goto T, Yamada S, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7:337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Scaruffi P, Valent A, Schramm A, Astrahantseff K, Eggert A, Tonini GP. Application of microarray-based technology to neuroblastoma. Cancer Lett. 2005;228:13–20. doi: 10.1016/j.canlet.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67:976–983. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 13.Warnat P, Oberthuer A, Fischer M, Westermann F, Eils R, Brors B. Cross-study analysis of gene expression data for intermediate neuroblastoma identifies two biological subtypes. BMC Cancer. 2007;7:89. doi: 10.1186/1471-2407-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilke S, Chen QR, Wei JS, Khan J. Whole chromosome alterations predict survival in high-risk neuroblastoma without MYCN amplification. Clin Cancer Res. 2008;14:5540–5547. doi: 10.1158/1078-0432.CCR-07-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, Garaventa A, Bonassi S, Tonini GP. Transcribed-ultra conserved region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer. 2009;9:441. doi: 10.1186/1471-2407-9-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spitz R, Betts DR, Simon T, Boensch M, Oestreich J, Niggli FK, Ernestus K, Berthold F, Hero B. Favorable outcome of triploid neuroblastomas: a contribution to the special oncogenesis of neuroblastoma. Cancer Genet Cytogenet. 2006;167:51–56. doi: 10.1016/j.cancergencyto.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Spitz R, Oberthuer A, Zapatka M, Brors B, Hero B, Ernestus K, Oestreich J, Fischer M, Simon T, Berthold F. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer. 2006;45:1130–1142. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- 18.Vermeulen J, De Preter K, Laureys G, Speleman F, Vandesompele J. 59-Gene prognostic signature sub-stratifies high-risk neuroblastoma patients. Lancet Oncol. 2009;10:1030. doi: 10.1016/S1470-2045(09)70325-0. [DOI] [PubMed] [Google Scholar]
- 19.Vermeulen J, De Preter K, Naranjo A, Vercruysse L, Van Roy N, Hellemans J, Swerts K, Bravo S, Scaruffi P, Tonini GP, et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009;10:663–671. doi: 10.1016/S1470-2045(09)70154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambros IM, Brunner B, Aigner G, Bedwell C, Beiske K, Benard J, Bown N, Combaret V, Couturier J, Defferrari R, et al. A multilocus technique for risk evaluation of patients with neuroblastoma. Clin Cancer Res. 2011;17:792–804. doi: 10.1158/1078-0432.CCR-10-0830. [DOI] [PubMed] [Google Scholar]
- 21.Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 22.Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP, De Bernardi B, Evans AE, Favrot M, Hedborg F. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 23.Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, Ernestus K, Konig R, Haas S, Eils R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 24.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B. Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer. 1999;86:349–363. [PubMed] [Google Scholar]
- 25.Scaruffi P, Coco S, Cifuentes F, Albino D, Nair M, Defferrari R, Mazzocco K, Tonini GP. Identification and characterization of DNA imbalances in neuroblastoma by high-resolution oligonucleotide array comparative genomic hybridization. Cancer Genet Cytogenet. 2007;177:20–29. doi: 10.1016/j.cancergencyto.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A, Valdora F, Fischer M, Gallo F, Hero B, et al. Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer. 2012;131:1591–1600. doi: 10.1002/ijc.27432. [DOI] [PubMed] [Google Scholar]
- 27.Fisher RA. The logic of inductive inference. J R Stat Soc. 1935;98:39–54. [Google Scholar]
- 28.Oberthuer A, Hero B, Berthold F, Juraeva D, Faldum A, Kahlert Y, Asgharzadeh S, Seeger R, Scaruffi P, Tonini GP, et al. Prognostic impact of gene expression-based classification for neuroblastoma. J Clin Oncol. 2010;28:3506–3515. doi: 10.1200/JCO.2009.27.3367. [DOI] [PubMed] [Google Scholar]
- 29.Griner PF, Mayewski RJ, Mushlin AI, Greenland P. Selection and interpretation of diagnostic tests and procedures. Principles and applications. Ann Intern Med. 1981;94:557–592. [PubMed] [Google Scholar]
- 30.Huang W, Brad T, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 32.Carén H, Kryh H, Nethander M, Sjöberg RM, Träger C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci USA. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spitz R, Hero B, Ernestus K, Berthold F. Gain of distal chromosome arm 17q is not associated with poor prognosis in neuroblastoma. Clin Cancer Res. 2003;9:4835–4840. [PubMed] [Google Scholar]
- 34.Buckley PG, Alcock L, Bryan K, Bray I, Schulte JH, Schramm A, Eggert A, Mestdagh P, De Preter K, Vandesompele J, et al. Chromosomal and microRNA expression patterns reveal biologically distinct subgroups of 11q-neuroblastoma. Clin Cancer Res. 2010;16:2971–2978. doi: 10.1158/1078-0432.CCR-09-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentijin LJ, Koppen A, van Asperen R, Root HA, Haneveld F, Versteeg R. Inhibition of a new differentiation pathway in neuroblastoma by copy number defects of N-myc, Cdc42, and nm23 genes. Cancer Res. 2005;65:3136–3145. doi: 10.1158/0008-5472.CAN-04-2469. [DOI] [PubMed] [Google Scholar]
- 36.Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, Alcock L, Mestdagh P, Vandesompele J, Speleman F, et al. Widespread dysregulation of miRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: association of miRNA expression with survival. PLoS One. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandesompele J, Speleman F, Van Roy N, Laureys G, Brinskchmidt C, Christiansen H, Lampert F, Lastowska M, Bown N, Pearson A, et al. Multicentre analysis of patterns of DNA gains and losses in 204 neuroblastoma tumors: how many genetic subgroups are there? Med Pediatr Oncol. 2001;36:5–10. doi: 10.1002/1096-911X(20010101)36:1<5::AID-MPO1003>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 38.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 39.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;250:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahai E, Olson MF, Marshall CJ. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001;20:755–766. doi: 10.1093/emboj/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark EA, King WG, Brugge JS, Symons M, Hynes RO. Integrin-mediated signals regulated by members of the rho family of GTPases. J Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antoine-Bertrand J, Villemure JF, Lamarche-Vane N. Implication of rho GTPases in neurodegenerative diseases. Curr Drug Targets. 2011;12:1202–1215. doi: 10.2174/138945011795906543. [DOI] [PubMed] [Google Scholar]
- 43.Bellanger J, Lazaro J, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- 44.Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouvière C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of RhoG. J Cell Sci. 2000;113:729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- 45.Estrach S, Schmidt S, Diriong S, Penna A, Blangy A, Fort P, Debant A. The human Rho-GEF trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr Biol. 2002;12:307–312. doi: 10.1016/s0960-9822(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 46.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 47.Eggert A, Ikegaki N, Kwiatkowski J, Zhao H, Brodeur GM, Himelstein BP. High-level expression of angiogenic factors is associated with advanced tumor stage in human neuroblastomas. Clin Cancer Res. 2000;6:1900–1908. [PubMed] [Google Scholar]
- 48.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 49.Chen QR, Song YK, Yu LR, Wei JS, Chung JY, Hewitt SM, Veenstra TD, Khan J. Global genomic and proteomic analysis identifies biological pathways related to high-risk neuroblastoma. J Proteome Res. 2010;9:373–382. doi: 10.1021/pr900701v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.