Abstract
Recent interest in the process of vascularization within the biomedical community has motivated numerous new research efforts focusing on the process of angiogenesis. Although the role of chemical factors during angiogenesis has been well documented, the role of mechanical factors, such as the interaction between angiogenic vessels and the extracellular matrix, remain poorly understood. In vitro methods for studying angiogenesis exist, however measurements available using such techniques often suffer from limited spatial and temporal resolution. For this reason, computational models have been extensively employed to investigate various aspects of angiogenesis. This manuscript outlines the formulation and validation of a simple and robust computational model developed to accurately simulate angiogenesis based on length, branching, and orientation morphometrics collected from vascularized tissue constructs. Excellent agreement was observed between computational and experimental morphometric data over time. Computational predictions of microvessel orientation within an anisotropic matrix correlated well with experimental data. The accuracy of this modeling approach makes it a valuable platform for investigating the role of mechanical interactions during angiogenesis.
Keywords: Angiogenesis, computational model, tissue engineering, extracellular matrix, fiber orientation, matrix anisotropy
1. Introduction
Angiogenesis, the process by which new blood vessels sprout off from existing vasculature, is highly sensitive to both the chemical and the mechanical microenvironment [1 –4]. During angiogenesis, endothelial cells within existing blood vessels detach from the basement membrane, migrate into the ECM, and form sprouts that elongate and eventually mature into new vasculature. Externally applied and cell-generated traction forces affect motility, metabolism, proliferation and differentiation of all anchorage-dependent cells, including endothelial cells and pericytes that participate in angiogenesis [5 – 11]. Mechanical stimuli received by cells via mechanotransduction depends on the structure and composition of the extracellular matrix (ECM) [12, 13] and on cell receptor structures bound to ECM components [14].
The mechanism as to how mechanical forces, 3D boundary conditions and ECM structure/composition influence neovessel growth during angiogenesis is poorly understood. Investigating this mechanism is difficult as the ECM is constantly remodeled and reorganized during angiogenesis through protease activity, formation of new cell-matrix adhesions, and cellular force generation [15, 16]. Overcoming these challenges and characterizing the mechanical interactions between angiogenic microvessels and the ECM would not only provide new insight into the driving forces behind morphogenic processes, but would also lead to new design considerations for engineering patterned microvasculature.
The role of mechanical factors during angiogenesis has been previously investigated in vitro using a novel three-dimensional culture method [3, 16 – 19]. Using this method, sprouting occurs within microvessel fragments in a spontaneous and consistent manner. Sprouts elongate as patent tubes, branching and forming anastomoses with other vessels [20, 21]. In free floating constructs, microvascular networks were found to have no preferred orientation. When the vascularized constructs were subject to an applied strain or a boundary constraint, microvessels and collagen fibers were found aligned along the constrained axis [3]. It is unclear if this alignment arises from microvessel growth being directed along aligned collagen fibers, from microvessels being reoriented due to contraction of the matrix and internal remodeling, or from some combination of both mechanisms.
Computational models can be utilized to supplement experimental efforts, often providing investigators with the ability to view systems at time points not sampled during the experiment or test hypotheses in ways not possible in the lab. Growth models have proven useful to investigators studying angiogenesis [22 – 27]. Computational frameworks can be categorized into different classes depending on how the system of interest is represented. Continuous models are typically governed by differential equations based on physical laws, while discrete models assemble a collection of discrete geometric units that behave according to a particular set of rules. Continuous-discrete models, or hybrid models, combine both approaches, often through determining the behavior of discrete units by solving a problem governed by physical laws
A computational model capable of simulating the interaction between microvessels and the ECM during angiogenesis would be a valuable platform for studying the role of mechanical forces, matrix boundary conditions, and ECM composition. However, the intricate nature of neovessel morphology prevents use of simple meshing schemes found in traditional methods in computational solid mechanics such as the Finite Element (FE) method. A recent study used a particle-based method known as the Material Point Method (MPM) [28, 29] to describe the state of stress in three-dimensional vascularized constructs using distributions of particles generated using confocal microscopy image datasets on a specimen-specific basis [30, 31].
Vascular geometry can be captured in two ways: Imaging data from vascularized constructs (specimen-specific data) or from computational predictions of microvessel growth. Since geometry obtained using the first method is constrained to the time point at which the culture was imaged, the second method was chosen for the MPM algorithm as it provides a generalized description of microvascular geometry at any desired point in time. This manuscript proposes such a computational model of vessel growth, designed to provide accurate, up-to-date microvascular geometry during in vitro angiogenesis. To facilitate coupling the simulations of microvessel growth with the MPM, the model was designed in a similar continuous-discrete framework. This involves explicit representation of the microvessels as discrete structures with the ability to simulate elongation, branching, and anastomosis over a regular computational grid. The continuous component of the model is in the differential equations used to govern field variables that influence neovessel growth. The growth model will also need to accurately predict the changes in microvessel growth resulting from ECM anisotropy and imposed boundary conditions as seen in the laboratory. The objectives of this study were to develop such a growth model and to demonstrate its ability to describe experimental data on microvessel length, branching, and orientation obtained from vascularized constructs.
2. Methods
2.1 Cell culture experiments - In vitro model of angiogenesis
Morphometric data was collected from in vitro cultures of angiogenic microvessels in order to calibrate the computational growth model and to determine if predictions from the simulation framework were valid. Based on previously described methods [20], 38 microvessel cultures were prepared from 5 separate dissections. Microvessel fragments were isolated from rat epydidimal fat pads. Sterile rat tail type I collagen (BD Biosciences, Franklin Lakes, NJ) was mixed with concentrated Dulbecco’s modified Eagle medium (DMEM, GIBCO-Invitrogen, Carlsbad, CA) to a density of 3.0 mg/ml collagen in 1X DMEM. Microvessel fragments were suspended in collagen at a density of 15,000 fragments/ml, and the solution was transferred to circular culture wells (diam. ~ 15mm). The collagen was allowed to polymerize at 37°C, 95% humidity for 30 minutes. Constructs were incubated in serum-free media [32] supplemented with rhVEGF (10 ng/ml, VEGF 165, PeproTech, Rocky Hill, NJ). The constructs cast within the circular culture wells were free-floating with no applied stretch or imposed boundary conditions, which created collagen fibers with no preferred orientation (random) (Figure 1 A, B). Constructs were incubated at 37°C and 95% humidity for 7 days, after which the microvascular network within each construct was imaged using confocal microscopy.
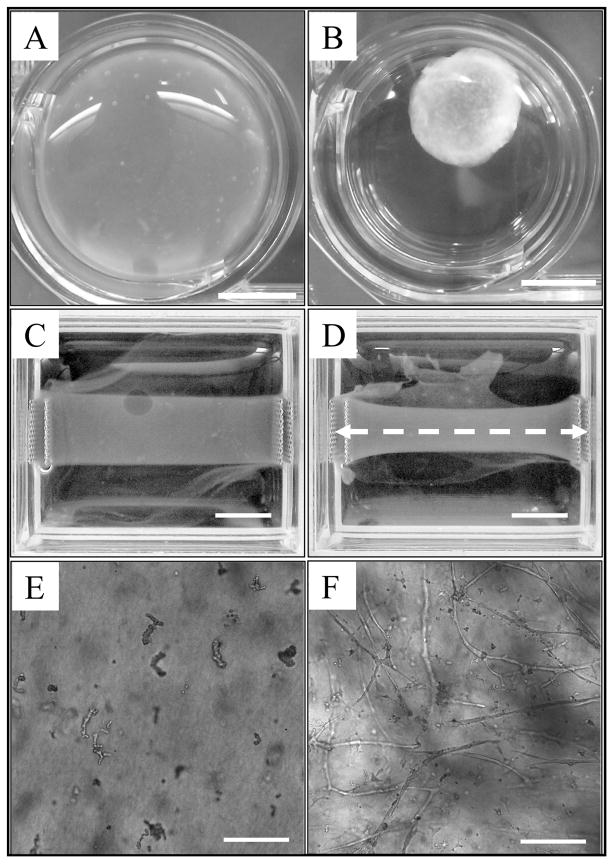
Figure 1.
Microvessel cultures provided an in vitro model of angiogenesis used for validating the computational model. Collagen gels serving as the ECM were subjected to two different boundary conditions. (A) Round, free-floating culture at day 0. (B) After 6 days, free-floating gels were uniformly contracted by cellular traction forces. Scale bar = 5 mm. (C) Rectangular collagen gel with the long axis constrained at day 0. (D) After 6 days, the fixed-edge constructs contracted along the short axis, with the most contraction found at the center of the gel (furthest point away from the boundary conditions). Microvessels and collagen fibers within these constructs were found predominately orientated along the constrained axis, shown as a white dashed line. Scale bar = 5 mm. (E) A 10X light micrograph displaying initial microvessel fragments within a free-floating collagen gel at day 0. (F) Representative growth profile seen within a free-floating vascularized construct after 6 days of culture. Scale bar = 200 μm.
To create the anisotropic ECM condition, 5 rectangular vascularized constructs were also prepared. These constructs were subjected to fixed-edge boundary conditions that prevented contraction of the collagen gel along the long axis but left the short axis free to contract (Figure 1 C, D). This boundary condition produces microvessel and collagen fibril alignment along the long axis of the constructs [3]. Microvessel orientation data was collected after 6 days of growth to compare with the simulation results. Collagen fiber orientation data was also collected to provide the fiber orientation information for the model when simulating the anisotropic matrix condition.
2.2 Image acquisition and processing
At the end of each culture period, the vascularized constructs were fixed with 4% formaldehyde overnight and then washed with phosphate-buffered saline (PBS) containing 0.1% Triton-X-100. Endothelial cells were stained with a 2 μg/ml solution of Isolectin IB4-Alexa 488 conjugate (Invitrogen, Carlsbad, CA). 3D image data sets of the stained microvessels were obtained using laser scanning confocal microscopy (Olympus FV1000 CLSM) utilizing a 488 nm excitation laser and a 10X objective. Six adjacent fields (1.27×1.27 mm) were acquired about the construct center through a depth of 300 μm from the bottom surface of the gel with a 2.5 μm z-step interval. Individual image stacks were acquired at a resolution of 512 × 512 × 300 voxels.
The image stacks for the 6 adjacent fields were stitched together using custom software. Unless specified, all other image processing was performed through Amira™ (Mercury Computer Systems, Carlsbad, CA). A blind deconvolution (10 iterations) was performed on image stacks using a point spread function based on the numerical aperture (NA = 0.4, 10X – air), wavelength of light (λ = 520 nm), and an estimate of the refractive index of the collagen gel (n = 1.35, [33]) to eliminate out-of-plane blur. Volume data was filtered to remove fragments smaller than 600 μm3. Microvessel volumes were then reduced to line segments by a skeletonization algorithm as previously described [3, 16]. A custom application was used to analyze the skeletonized data and collect the total vessel length and number of branches in each culture [34].
2.3 Morphometric Data - Vascular Length
A function describing microvessel growth over time was created from experimental data to define growth within the computational model. For each time point sampled, the length of all microvessels within the field and normalized by the initial number of microvessel fragments. The number of initial fragments was calculated from the seeding density used when preparing the cultures (15,000 fragments/ml). This normalization ensures that the growth mechanisms within the model are independent from the initial microvessel seeding density. Length metric data from each experiment was averaged across each of the free-floating cultures, and this data was fit with a 4-parameter sigmoid curve:
| (1) |
This function describes the total vascular length within the domain at any point in time. A sigmoid curve was chosen to describe microvessel growth as such a curve is often used to describe population growth restricted by limited resources (carrying capacity). In Eq. 1, g0 is the initial microvessel length (bottom of the sigmoid curve), a1 is the range of the function (top minus bottom), t1/2 is the time at which g(t) is halfway in between the top and bottom of the sigmoid curve, and b1 is the slope of the curve. The carrying capacity of the system can be described as g0 + a1.
2.4 Morphometric Data - Vascular Branching
The computational model also required a function describing microvessel branching over time. For each time point, the number of branch points within the field were averaged together and normalized. The branching b(t) was calculated by fitting the average branching data with a 3-parameter exponential curve:
| (2) |
In this equation, y0 describes the initial number of branches, a2 scales the exponential term, and b2 describes the rate of branch formation. An exponential function was used to describe branch formation as branching metric data taken from experimental cultures does not appear to approach any limit during the 7 day culture period.
2.5 Collagen Fiber Orientation
The culture and imaging procedure described above was repeated for the 5 fixed-edge constructs at day 6 of culture. Measurements from these constructs included the angles for all microvessel segments relative to the constrained axis (long axis). The structural organization of the underlying collagen matrix within the fixed-edge constructs was quantified by imaging collagen fibrils using confocal reflectance microscopy with a 60X water objective and a 633 nm laser. For each construct, two stacks were acquired through a depth of 120 μm with a 1 μm z-step interval. An orientation of collagen fibers was determined from the images by employing a 2D Fourier Transform technique on each image [35, 36]. The angle distributions from the 5 cultures were averaged together and fit with a Gaussian distribution. This distribution was used to seed a vector field that represents an anisotropic matrix for the computational growth model.
2.6 Computational model - Overview
The computational model of vessel growth during in vitro angiogenesis was calibrated and validated with data obtained using the methods described above. The formulation and implementation was three-dimensional, although for simplicity the simulations presented in this manuscript were only performed in two dimensions. The simulation domain, or virtual ECM, was discretized with a regular quadrilateral mesh. Local field information such as ECM collagen fibril orientation and microvessel density was specified at each of the grid nodes. At any position (x,y), the field described at the nodes could be mapped to that point via bilinear shape functions:
| (3) |
where ϕ was a field variable and Si(x,y) were the values of the shape functions for each of the four nodes of the cell, evaluated at the point (x,y) (Figure 2). This approach can be applied to include any field information deemed pertinent by the user. The dimensions of the domain were 1.27×1.27 mm for all simulations, corresponding to the 10X field of view on the confocal microscope. Periodic boundary conditions were imposed on all edges of the simulation domain.
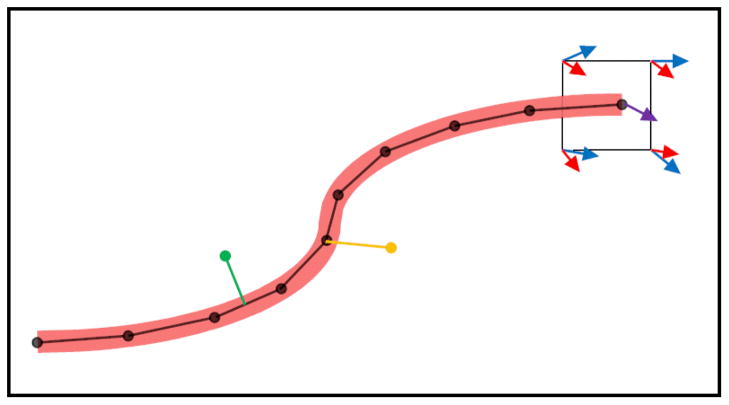
Figure 2.
Microvessels were represented as a series of line segments. At each time step, neovessel growth occurred through the addition of a new line segment at the active tips of existing segments. The orientation of new segments was determined from information stored at the nodes of the grid cell. In this figure, the red and blue arrows represent directional cues determined by field information stored at the nodes. The red arrows are longer than the blue to represent the uneven weighting of various factors, i.e. collagen fibril direction was more influential than vessel density gradient. The direction of the new segment (purple arrow) was determined by interpolation of red and blue field variables via bilinear shape functions (Eq. 3). At each time step, a segment formed a branch if a random number was below a branching probability parameter. The yellow segment demonstrates a branch formation. Anastomoses can form anywhere along a microvessel where an intersection occurs, as shown by the green line segment.
2.7 Computational model - Vessel Elongation
Initial microvessel fragments were represented as discrete independent line segments with lengths corresponding to the value of the sigmoid growth curve g(t) at time t = 0 (Eq. 1). Twenty-five fragments (average number in the field of view of a confocal image) were seeded throughout the domain, each at a random position and with a random orientation.
At each time step, microvessel growth was represented by the addition of new segments to the ends of existing segments. A variable time step was implemented to limit the growth of any vessel segment to half a grid cell. The length of each new segment calculated by referencing the growth curve g(t) and determining the change in length over the time step size (Eq. 1). The direction of new segments was determined by a linear combination of several directional cues. The orientation vector v for a newly created segment was found using the following expression:
| (4) |
In Eq. 4, θcoll was the directional component pointing in the direction of local collagen fibers, θvdens was the directional component that caused microvessels to grow away from regions of high microvessel density, and θrand was a random-walk component. w1, w2, and w3 were the respective weights for each of these components. Each of the directional components was determined by mapping field information stored at the nodes of the grid to the position of the new segment (Eq. 3).
2.8 Computational model - Branching
Microvessel branching was modeled as a stochastic process. During each time step, each segment was assigned a random number between 0 and 1. Branching occurred when a segment’s random number was less than a branching probability value:
| (5) |
The initial branching probability, B0, value was determined by using a golden section, single variable, bounded optimization algorithm to minimize the RMS error between simulated and experimental length and branch metrics. Multiplying by the time step Δt eliminated dependence of branching behavior on time step size.
2.9 Computational model - Anastomosis
Growth tips within close proximity to other microvessels were capable of forming anastomoses. At the end of each time step, a search for intersections between all active vessel tips and existing segments was conducted. If the intersection found was in between two segments within the same vessel, the intersection was ignored to avoid the formation of a terminal loop. If the intersecting segments were not from the same microvessel, a procedure was used to determine if they would intersect in three dimensions. This process is necessary because the model represented a 2D projection of vessels in 3D. In 3D, a microvessel could be positioned at any point throughout the culture thickness of 300 μm. To account for the three dimensional geometry, a test was performed based on the probability of the two microvessels being offset by less than 1 microvessel diameter in the third spatial dimension. This involved taking the ratio between the microvessel diameter and the culture thickness. If the random number was less than this ratio, the microvessels formed an anastomosis; otherwise, the intersection was ignored.
2.10 Computational Model - Optimization of Branching
A golden-section line search was used to determine the optimal value of B0 within the branching mechanism (Eq. 5) by minimizing the RMS error between simulated and experimental branching data:
| (6) |
Here, ψe(ti) was branching metric value from experimental data at a given time point ti, ψm(ti) was the branching metric from the computational simulations, and n was the number of time steps taken.
2.11 Computational Model - ECM Anisotropy
The computational model was designed with the ability to simulate microvessel growth under various matrical boundary conditions. Different boundary conditions were modeled by changing the orientation of collagen fibrils within the matrix. For simulations involving a free-floating collagen gel, a random collagen fiber orientation angle between −90° and 90° was generated at each node of the grid. When simulating angiogenesis within the anisotropic matrix condition, nodal collagen fiber orientation values were seeded using angle distributions collected from the fixed-edge constructs. The orientation of microvessels from these simulations was compared to orientation data from the fixed-edge constructs to verify the model’s ability to predict angiogenesis within an anisotropic matrix.
2.12 Computational Model - Optimization of Vessel Orientation
Microvessel orientation within the model was optimized to match experimental data. An routine similar to Eq. 6 was used to determine the optimal values of the weights describing the strength of directional cues when determining the orientation of new microvessel segments (w1, w2, w3) (Eq. 4). These weights were adjusted to minimize the RMS error between microvessel orientation in the fixed-edge constructs on day 6 and microvessel orientation from the anisotropic ECM simulations:
| (7) |
where φe(i) was the percentage of microvessels whose orientation angle fell into discrete angle bin i (0°–10°, 10°–20°, …, 80°–90°) measured off the constrained axis. φm (i) was the percentage of microvessels found in bin i from the simulations, and n was the number of discrete angle bins.
2.13 Computational Model - Variation Due to Stochastic Processes
While microvessel elongation within the model was a deterministically-governed process, the mechanisms branching and anastomosis were stochastically-based. Since branch formation and anastomosis directly affect the number of active growth tips, stochastic variations between different simulations can be passed from these random mechanisms to length metric data. To eliminate stochastic variations, growth rate was normalized by the number of active growth tips currently in the model. This adjustment ensures that the total length in each simulation will not vary too far away from the value prescribed by g(t), reducing significant variation in length metric data over multiple simulations.
The seed used to initialize the random number generator algorithm prior to each simulation was varied based on the system clock. In order to assess the degree of variance across simulations caused by the stochastic processes, fifty simulations of microvessels growing within a randomly-orientated ECM were conducted, each using a unique seed. The variance in the length and branch metrics between these trials was then evaluated using a 1-way ANOVA.
Variations in the simulation results due to the stochastic components of the model prevented convergence to strict tolerances (ε = 1e-7) during the optimization routines. Therefore, a constant seed was used to initialize the C++ library function, rand(), during the optimization routines to facilitate convergence.
3. Results
Microvessel growth profiles were consistent across all in vitro experiments. Very little growth occurred before the 3–4 day mark, at which point sprouting was observed. Microvessels grew with at a high rate until day 6–7, after which growth rate began to slow as the system neared its carrying capacity (Figure 3 – top). The sigmoid curve fit to the averaged length metric data (g(t), Eq. 1) resulted in an R2 value of 0.998.
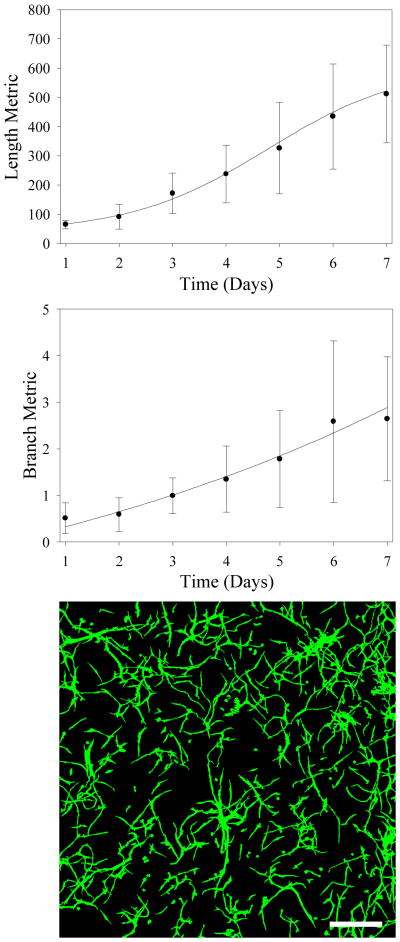
Figure 3.
Morphometric data describe microvessel growth and branching during angiogenesis. (Top) Length metric data (current vessel length/number of vessels at day 1) versus time. The function g(t) was fit to this data (Eq. 1). (Middle) Microvessel branch metric data (current number of branches/number of vessels at day 1) versus time. The function b(t) was fit to this data (Eq. 2). In both the top and middle figure, points represent experimental data (mean ± standard deviation) while solid curves represent regression fits. (Bottom) Z-projection of a representative microvessel culture. Image data was collected by imaging vascularized constructs at day 6 using a confocal microscope. Scale bar = 400 μm.
The number of branch points measured in the vascularized constructs increased exponentially throughout time with no observable limit during the 7 day culture period (Figure 3 – bottom). The exponential curve fit to the averaged branch metric data (b(t), Eq. 2) achieved an R2 value of 0.968. The parameters determined from the curve fits can be found compiled in Table 1. Variance in length and branch metric data across in vitro cultures was high, with maximum standard deviations of 179.7 and 1.74 respectively.
Table 1.
Parameters for the growth model were determined using an optimization routine comparing morphometric data from experimental cultures against computational results. Tolerance for the optimization routine was ε = 1e-7.
| Growth function g(t) | |
| a1 | 568.6 |
| g0 | 38.8 |
| b1 | 1.3 |
| t1/2 | 4.8 |
| Branching function b(t) | |
| y0 | −2.50 |
| a2 | 2.62 |
| b2 | 0.105 |
| Branching Probability | |
| B0 | 0.038 |
| Strength of Directional Cues | |
|
w1 Fiber Orientation |
0.508 |
|
w2 Vessel Density |
0.238 |
|
w3 Random Walk |
0.254 |
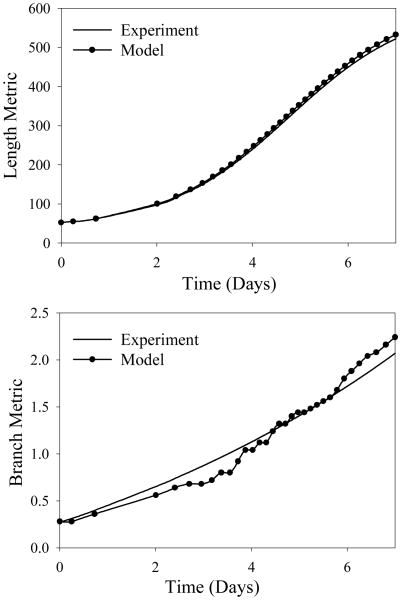
Microvessel growth resulting from computational simulations resembled growth profiles seen in vitro (Figure 4). The length of microvascular networks from the computational simulations correlated well with experimental data, with approximately 1% normalized RMS error between computational and experimental length metric data (Figure 5 – top). The optimization routine used to determine the branching chance parameter returned a value of B0 = 0.038 (Eq. 5). This branching parameter minimized the normalized RMS error between computational and experimental branching metric data to roughly 6% (Figure 5 – bottom).
Figure 4.
Images from computational simulations presented for comparison with experimental data. Confocal images from vascularized constructs are on the left, while data from simulations of microvessel growth can be found on the right. The time points at which these images were obtained are as follows: Day 1 (A, B), Day 4 (C, D), and Day 7 (E, F). Scale bar = 300 μm. Predictions of microvasculature from the computational model were similar to images taken from vascularized constructs. Notice that growth predicted in the simulations followed a more tortuous path compared to microvessels in vitro.
Figure 5.
Computational simulations accurately predict experimental length and branching metric data taken from experimental cultures. (Top) Comparison of computational and experimental length metric data versus time. There was a normalized RMS error of approximately 1% between the two data sets. (Bottom) Comparison of simulated and experimental branch metric data (number of current branches divided by initial number of microvessels in culture) versus time. There was a normalized RMS error of approximately 6% between the two data sets.
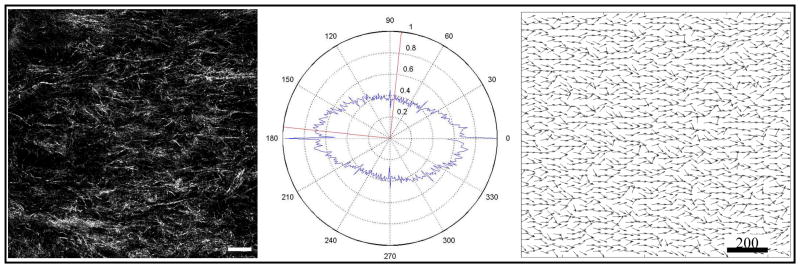
Microvessels within the fixed-edge constructs were found preferentially aligned along the constrained axis, consistent with what has been seen in past experiments [3]. The majority of fibers in the fixed-edge constructs were found aligned along the constrained axis. The distributions of fiber angles for all 5 of the fixed-edge constructs were averaged together and fit to a Gaussian distribution which was used to generate a collagen fiber orientation at each node of the grid node (Figure 6).
Figure 6.
Anisotropic collagen fibril orientation was observed within the fixed-edge constructs. (Left) Confocal reflection microscopy image of collagen fibrils within a representative fixed-edge construct at day 6 in vitro. (Middle) FFT analysis of collagen fibril data from constrained gels. Collagen fibrils were found to be predominately aligned along the constrained axis (0°–180°). (Right) A field of collagen fibril angles based on angle distribution extracted from confocal reflectance microscopy images. This vector field was stored at the nodes of the grid in simulations of angiogenesis subject to the anisotropic ECM condition. Scale bar = 200 μm.
Simulations of angiogenesis within a randomly-oriented ECM produced microvessels with no preferential alignment, similar to what has been seen in free-floating vascularized constructs [3] (data not shown). The computational model was also successful at predicting angiogenesis within an anisotropic ECM. The angle between each microvessel and the long axis of the vascularized construct was measured from the simulations and constrained gel experiments. In both the simulations and experiments, the majority of microvessels were found aligned within 20° of the constraint axis (Figure 7). Although significant differences exist between computational and experimental data for some of the angle bins, the correlation coefficient between the two data sets was calculated to be R2 = 0.98. The weights of the three directional components described in Eq. 4 were determined by optimizing the orientation of microvessel segments within the computational model. The optimized values of w1, w2, and w3 can be found in Table 1.
Figure 7.
Computational simulations successfully predicted microvessel orientation within an anisotropic ECM. (Top) Microvessels cultured within the fixed-edge constructs were found predominately aligned along the constrained axis. (Middle) Computational simulations of angiogenesis occurring with an anisotropic ECM accurately predicted experimental findings. (Bottom) The angle between each microvessel segment and the horizontal axis was collected and sorted into discrete bins. Orientation data from the computational model correlated well with data from the fixed-edge vascularized constructs. Scale bar = 400 μm.
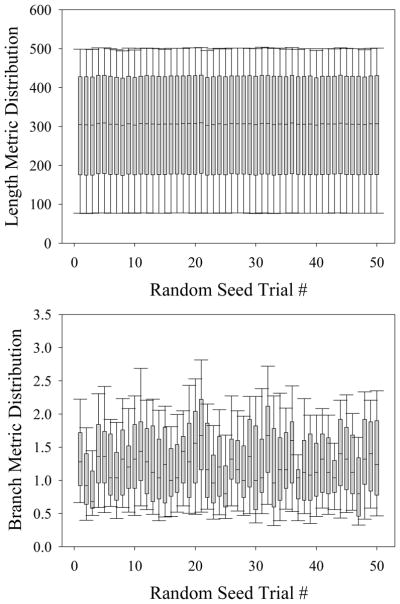
To render length data from the simulations immune to these stochastic variations, the growth function g(t) was normalized by the number of active growth tips within the simulation. This method was tested by observing the final length metric value over 50 simulations. For each of the 50 simulations, the final length metric value did not significantly vary from the value prescribed by g(t) (Figure 8 – top). In contrast, when the same test is performed for branch metric data, the number of branching points within the simulation varied significantly across 50 simulations (Figure 8 – bottom).
Figure 8.
Stochastic variation within the computational simulations. Box-whisker plots of length (top) and branch (bottom) metric collected over 50 simulations, each with a unique P-RNG seeds. A 1-way ANOVA was performed to check for significant variance between trials. No significant variation in the length metric was seen while varying the P-RNG seed (p = 1.0). Branching was modeled as a stochastic process, and therefore the branch metric significantly varied between trials (p < 0.001).
4. Discussion
The continuous-discrete framework outlined in this manuscript was successful at predicting length and branching behavior exhibited by angiogenic microvessels in vitro. Measurements of growth, branching, and alignment metrics provided by simulations had excellent statistical agreement with experimental data. This accuracy was obtained through optimization of only four parameters: the branching probability constant and the three weights influencing growth direction. Introducing additional parameters into the optimization routine would lead to even greater predictive performance from the model, but the simple approach employed in this work satisfied the objectives of this study.
Orientation data from simulations of the anisotropic ECM condition correlated well with microvessel orientation observed in the fixed-edge vascularized constructs. However, angle data from the simulations tended to favor the acute angle bins more than the corresponding experimental data. Approximately 90% of microvessels from the simulations were found orientated between 0° and 45° off of the constrained axis. Only 80% of microvessels within the fixed-edge constructs fell within this same range. Although this difference is seemingly trivial, it is important to account for all inconsistencies between the model and in vitro findings. This particular inconsistency is important since the cornerstone assumption for this simulation framework is that microvessel growth follows collagen fiber orientation.
The inconsistency in microvessel orientation during simulations of the anisotropic ECM condition may be due to the assignment of an aligned ECM on day 0 of each simulation. Within the fixed-edge constructs, angiogenesis begins in a random ECM which is remodeled during vessel growth, resulting in an aligned matrix. In future simulations supplying a fibril orientation field that can vary over time will allow the model to capture the transition to anisotropy that results from matrix remodeling. The eventual coupling of the growth model with the MPM framework will allow mechanical deformations of the ECM to be modeled explicitly, eliminating the need to specify fibril orientation past initialization. It should be noted that the growth model does not consider any deformation of the matrix when determining microvessel orientation. Realistically, the microvessel orientation seen in vascularized constructs is most likely due to both directional cues from aligned ECM fibers and displacement of vessels during remodeling and contraction of the matrix. The inclusion of the MPM into the simulation framework will allow both alignment mechanisms to be considered, creating a more realistic simulation of in vitro angiogenesis.
Although the simulations and experiments agreed statistically, there were qualitative discrepancies in the computational predictions of microvessel morphology. Simulated microvessel growth followed a more tortuous path compared to microvessels cultured in the lab. This discrepancy resulted from the mechanism used to determine the orientation of new segments within the computational model. New segments are allowed to form any angle with the parent vessel segment, leading to kinked microvessel morphology during elongation. Implementation of a persistence component into the vessel growth mechanism will make the microvessels resistant to changes in direction, ideally eliminating this morphological discrepancy.
The stochastic processes within the model led to minimal variation between simulations. Microvessel length was unaffected by the random numbers generated during the simulations. This independence from the random processes was expected as microvessel growth rate was normalized to account for the random formation of branches and anastomoses. However, some aspects of microvascular topology were sensitive to variations in the P-RNG seed. Branching was governed by a stochastic process, and branches would form at different times and in different locations after re-seeding the random number generator. Likewise, the position and orientation of initial microvessel fragments would vary between random number generations as well. The conclusion is that these stochastic variations are small enough to not cause instabilities, yet pronounced enough to ensure that each simulation returns a unique microvascular network.
Future development of the computational framework will involve the replacement of stochastic components with deterministic mechanisms. For example, past mathematical models have suggested that steep gradients in collagen density local to a tip cell can induce the formation of a branch [24]. Likewise, with further analysis of confocal image data it may be possible to determine the degree of persistence involved during microvessel elongation. During the simulation, ECM collagen fibril orientation was supplied as an input parameter. Therefore, future simulations could easily predict in vitro angiogenesis for a given matrix orientation by simply mapping the desired fibril orientation field to the grid.
Chemical factors such as VEGF, TGF-β, and PDGF play a vital role during angiogenesis [1]. In order to isolate and study the impact of mechanical/structural stimuli during angiogenesis, the current model implementation does not include any chemotactic guidance during neovessel outgrowth. The in vitro system being modeled was uniformly immersed in VEGF, leaving only local synthesis of chemical factors as a possible source of chemical inhomogeneity. Rat tail type I collagen used for the experimental model does not contain trapped cytokines which can be released upon digestion [37, 38]. Therefore, it was possible to simulate in vitro angiogenesis accurately without including chemotaxis since significant chemotactic gradients do not develop within the vascularized constructs.
The algorithms described in this study were highly robust and efficient, with simulations completing within seconds. The model demonstrates forward compatibility through the ability to simulate different matrical boundary conditions and simple coupling to optimization algorithms. The growth model will provide the MPM algorithm with an accurate and current microvessel geometry that can be used to determine the state of stress/strain within microvessels and the surrounding matrix for any given point in time [30]. Information from the MPM simulations can then be used to direct vessel growth in the next time step, and this process will repeat as both models step throughout time.
In summary, the simulation framework outlined in this work was capable of producing an accurate description of microvascular length, branching, and orientation metrics over time for both an isotropic and anisotropic ECM fibril orientation field. The extent of angiogenesis can be tightly controlled by adjusting the input parameters, giving the model the ability to simulate a wide range of problems. Finally, the shape functions within the grid provide a basis for expanding the model to include additional field variables that influence neovessel growth and orientation. For example, stress/strain fields could be mapped to the nodes of the grid within the vessel growth model, effectively coupling stress and strain fields within the matrix to angiogenesis. These features allow this framework to serve as an effective platform for exploring how mechanical interactions between neovessels and the ECM regulate angiogenesis.
Acknowledgments
Financial support from NIH grants R01HL077683 and R01EB007556 is gratefully acknowledged.
References
- 1.Shiu YT, Weiss JA, Hoying JB, Iwamoto MN, Joung IS, Quam CT. The role of mechanical stresses in angiogenesis. Crit Rev Biomed Eng. 2005;33:431–510. doi: 10.1615/critrevbiomedeng.v33.i5.10. [DOI] [PubMed] [Google Scholar]
- 2.Ingber DE. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ Res. 2002;91:877–87. doi: 10.1161/01.res.0000039537.73816.e5. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan L, Underwood CJ, Maas S, Ellis BJ, Kode TC, Hoying JB, Weiss JA. Effect of mechanical boundary conditions on orientation of angiogenic microvessels. Cardiovasc Res. 2008;78:324–32. doi: 10.1093/cvr/cvn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S, Huang NF, Hsu S. Mechanotransduction in endothelial cell migration. J Cell Biochem. 2005;96:1110–26. doi: 10.1002/jcb.20614. [DOI] [PubMed] [Google Scholar]
- 5.Carosi JA, Eskin SG, McIntire LV. Cyclical strain effects on production of vasoactive materials in cultured endothelial cells. J Cell Physiol. 1992;151:29–36. doi: 10.1002/jcp.1041510106. [DOI] [PubMed] [Google Scholar]
- 6.Stamatas GN, McIntire LV. Rapid flow-induced responses in endothelial cells. Biotechnol Prog. 2001;17:383–402. doi: 10.1021/bp0100272. [DOI] [PubMed] [Google Scholar]
- 7.McCormick SM, Frye SR, Eskin SG, Teng CL, Lu CM, Russell CG, Chittur KK, McIntire LV. Microarray analysis of shear stressed endothelial cells. Biorheology. 2003;40:5–11. [PubMed] [Google Scholar]
- 8.Patrick CW, Jr, McIntire LV. Shear stress and cyclic strain modulation of gene expression in vascular endothelial cells. Blood Purif. 1995;13:112–24. doi: 10.1159/000170194. [DOI] [PubMed] [Google Scholar]
- 9.Chien S, Li S, Shyy YJ. Effects of mechanical forces on signal transduction and gene expression in endothelial cells. Hypertension. 1998;31:162–9. doi: 10.1161/01.hyp.31.1.162. [DOI] [PubMed] [Google Scholar]
- 10.Joung IS, Iwamoto MN, Shiu YT, Quam CT. Cyclic strain modulates tubulogenesis of endothelial cells in a 3D tissue culture model. Microvasc Res. 2006;71:1–11. doi: 10.1016/j.mvr.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Ives CL, Eskin SG, McIntire LV. Mechanical effects on endothelial cell morphology: in vitro assessment. In Vitro Cell Dev Biol. 1986;22:500–7. doi: 10.1007/BF02621134. [DOI] [PubMed] [Google Scholar]
- 12.Deroanne CF, Lapiere CM, Nusgens BV. In vitro tubulogenesis of endothelial cells by relaxation of the coupling extracellular matrix-cytoskeleton. Cardiovasc Res. 2001;49:647–58. doi: 10.1016/s0008-6363(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 13.Vernon RB, Sage EH. A novel, quantitative model for study of endothelial cell migration and sprout formation within three-dimensional collagen matrices. Microvasc Res. 1999;57:118–33. doi: 10.1006/mvre.1998.2122. [DOI] [PubMed] [Google Scholar]
- 14.Jalali S, del Pozo MA, Chen K, Miao H, Li Y, Schwartz MA, Shyy JY, Chien S. Integrin-mediated mechanotransduction requires its dynamic interaction with specific extracellular matrix (ECM) ligands. Proc Natl Acad Sci U S A. 2001;98:1042–6. doi: 10.1073/pnas.031562998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manoussaki D, Lubkin SR, Vernon RB, Murray JD. A mechanical model for the formation of vascular networks in vitro. Acta biotheoretica. 1996;44:271–282. doi: 10.1007/BF00046533. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan L, Hoying JB, Nguyen H, Song H, Weiss JA. Interaction of angiogenic microvessels with the extracellular matrix. Am J Physiol Heart Circ Physiol. 2007;293:H3650–8. doi: 10.1152/ajpheart.00772.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan L, Hoying JB, Das R, Weiss JA. Alterations in the material properties of collagen by angiogenesis. Proc 49th Annual Orthopaedic Research Society Meeting; 2003. p. 278. [Google Scholar]
- 18.Krishnan L, Ngyuen H, Song H, Hoying JB, Weiss JA. Gene expression in a three-dimensional model of angiogenesis: Relation to matrix mechanical properties. Proc ASME Summer Bioengineering Conference abstract #b0290109; Vail, CO. June 22–26.2005. [Google Scholar]
- 19.Krishnan L, Utzinger U, Maas SA, Reese SP, Weiss JA, Williams SK, Hoying JB. Extacellular matrix stiffness modulates microvascular morphology during early sprouting angiogenesis in vitro. Proceedings of the ASME Summer Bioengineering Conference; 2009. p. 206782. [Google Scholar]
- 20.Hoying J, Boswell C, Williams S. Angiogenic potential of microvessel fragments established in three-dimensional collagen gels. In Vitro Cell Dev Biol Anim. 1996;32:409–19. doi: 10.1007/BF02723003. [DOI] [PubMed] [Google Scholar]
- 21.Shepherd B, Chen H, Smith C, Gruionu G, Williams S, Hoying J. Rapid Perfusion and Network Remodeling in Microvascular Construct After Implantation. Arterioscler Throm Vasc Biol. 2004;24:898–904. doi: 10.1161/01.ATV.0000124103.86943.1e. [DOI] [PubMed] [Google Scholar]
- 22.Chaplain MA. Mathematical modelling of angiogenesis. J Neurooncol. 2000;50:37–51. doi: 10.1023/a:1006446020377. [DOI] [PubMed] [Google Scholar]
- 23.Namy P, Ohayon J, Tracqui P. Critical conditions for pattern formation and in vitro tubulogenesis driven by cellular traction fields. J Theor Biol. 2004;227:103–20. doi: 10.1016/j.jtbi.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Bauer AL. A Cell-Based Model Exhibiting Branching and Anastomosis During Tumor-Induced Angiogenesis. Biophys J BioFAST. 2007;92:3105–3121. doi: 10.1529/biophysj.106.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capasso V, Micheletti A, Morale D. Stochastic geometric models, and related statistical issues in tumour-induced angiogenesis. Math Biosci. 2008;214:20–31. doi: 10.1016/j.mbs.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Mantzaris NV, Webb S, Othmer HG. Mathematical modeling of tumor-induced angiogenesis. J Math Biol. 2004;49:111–87. doi: 10.1007/s00285-003-0262-2. [DOI] [PubMed] [Google Scholar]
- 27.Qutub AA, Popel AS. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst Biol. 2009;3:13. doi: 10.1186/1752-0509-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulsky D, Zhou S, Schreyer HL. Application of a particle-in-cell method to solid mechanics. Computer Physics Communications. 1995;87:236–252. [Google Scholar]
- 29.Sulsky D, Chen Z, Schreyer HL. A particle method for history dependent materials. Computer Methods in Applied Mechanics and Engineering. 1994;118:179–196. [Google Scholar]
- 30.Guilkey JE, Hoying JB, Weiss JA. Computational modeling of multicellular constructs with the material point method. J Biomech. 2006;39:2074–86. doi: 10.1016/j.jbiomech.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 31.Guilkey JE, Hoying JB, Weiss JA. Large-scale modeling of the mechanical behavior of multicellular constructs. Proc ASME Summer Bioengineering Conference; Vail, CO. June 22–26.2005. [Google Scholar]
- 32.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc Natl Acad Sci U S A. 1979;76:514–7. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Griffith M, Watsky MA, Forrester JV, Kuffova L, Grant D, Merrett K, Carlsson DJ. Properties of porcine and recombinant human collagen matrices for optically clear tissue engineering applications. Biomacromolecules. 2006;7:1819–28. doi: 10.1021/bm060160o. [DOI] [PubMed] [Google Scholar]
- 34.WinFiber3D. available at http://mrl.sci.utah.edu/
- 35.Pourdeyhimi B, Dent R, Davis H. Measuring Fiber Orientation in Nonwovens Part III: Fourier Transform. Textile Res J. 1997;67:143–151. [Google Scholar]
- 36.Sander EA, Barocas VH. Comparison of 2D fiber network orientation measurement methods. J Biomed Mater Res A. 2009;88:322–31. doi: 10.1002/jbm.a.31847. [DOI] [PubMed] [Google Scholar]
- 37.Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB. Transforming growth factor-beta in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem. 1983;258:7155–60. [PubMed] [Google Scholar]
- 38.Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, et al. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci U S A. 1986;83:4167–71. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]