Abstract
Background
The vertebrate nuclear receptor subfamily 2, group f (nr2f) genes encode orphan receptors that have the capacity to act as negative regulators of retinoic acid (RA) signaling.
Results
We describe embryonic and larval expression of four of the six zebrafish nr2f genes, nr2f1a, nr2f1b, nr2f2 and nr2f5. These genes show highly regulated patterns of expression within the CNS, including in the developing hindbrain, as well as in the mesoderm and endoderm. We also investigated the role of RA and Fgf signaling in regulating early nr2f gene expression. RA is not required for nr2f expression in the hindbrain; however, exogenous RA can repress this expression. Conversely, we find that RA positively regulates nr2f1a expression in trunk endoderm and mesoderm. Fgf signaling is not required for nr2f expression onset in the hindbrain; however, it may play a role in maintaining rhombomere-specific expression.
Conclusions
We report detailed expression analysis of four nr2f genes in all three germ layers. The onset of nr2f expression in the hindbrain does not require RA or Fgf signals. Our finding that RA positively regulates nr2f1a expression in the trunk supports the possibility that Nr2fs function in a negative feedback loop to modulate RA signaling in this region.
Keywords: zebrafish, nr2f, retinoic acid, Fgf, hindbrain, neural patterning, transcription factors, orphan nuclear receptors
INTRODUCTION
The nr2fs (nuclear receptor subfamily 2, group f genes), also known as COUP-TF (chicken ovalbumin upstream promoter-transcription-factor) in chick, mouse, and human, and svp (seven-up) in Drosophila, encode orphan receptors of the large steroid/thyroid nuclear receptor superfamily. The steroid/thyroid nuclear receptors are activated by their ligands (i.e. steroids, thyroid hormones and retinoids); however, many receptors in this superfamily are considered orphan receptors as no ligands have been identified (Pereira et al., 2000). Indeed, the ligand binding domain of the orphan receptor Nurr1 folds into the canonical ligand-bound structure and its stability affects transcriptional activity similar to ligand-bound nuclear receptors. However, the ligand binding site instead is blocked with bulky hydrophobic residues and it lacks the ‘classical’ cofactor interaction surface (Wang et al., 2003). Nr2fs are orphan receptors, and structure analysis of the ligand binding domain of human Nr2f2 also revealed occupation of the binding pocket by bulky hydrophobic and aromatic residues (Kruse et al., 2008). Nr2fs have been hypothesized to interfere with nuclear hormone receptor-mediated regulation of gene expression; specifically they have been suggested to function as negative regulators of Retinoic Acid (RA) signaling (for reviews, see Qiu et al., 1996; Tsai and Tsai, 1997; Pereira et al., 2000; Park et al., 2003).
RA, like other ligands of steroid/thyroid receptors, regulates gene expression by binding directly to its cognate receptor within the nucleus. The RA receptor (RAR) heterodimerizes with retinoid receptor (RXR), and this complex binds RA response element (RARE) consensus sequences within target genes. In the absence of RA ligand the RAR/RXR complex actively represses RA transcriptional targets, but in the presence of ligand, corepressors are displaced and replaced by coactivators, leading to activation of transcription of the RA target genes (Xu et al., 2009; reviewed by Linney et al., 2011). Nr2f receptors also bind to RAREs, and experiments in vitro have shown that they can negatively regulate RA response genes in at least three possible ways: (1) by competing for RARE binding with ligand-bound RAR/RXR heterodimers to inhibit transcription activation, (2) by binding RAREs and interacting with cellular corepressors such as silencing mediator for retinoid and thyroid hormone receptors (SMRT) or nuclear receptor corepressor (N-CoR) to actively silence transcription of RA target genes, and (3) by binding to and sequestering RXR molecules to leave a pool of inactive RARs (Tsai & Tsai, 1997). As RXRs are the universal heterodimerization partners for nuclear receptors, this mechanism can likely influence multiple signaling pathways. In addition to these repressive roles there are also multiple reported cases of Nr2fs acting as activators of transcription (Tsai & Tsai, 1997), suggesting complex roles for this gene family.
The Nr2f genes were first identified and purified from HeLa cell nuclear extracts, as factors that bind the COUP box consensus sequence, and were shown to be required for in vitro transcription of the ovalbumin gene (Sagami et al., 1986). Nr2f homologs have since been described in a wide array of vertebrates and invertebrates, and they show robust expression in multiple tissues, particularly in the developing central nervous system (CNS) and eye (Tsai and Tsai, 1997; Pereira et al., 2000; Gauchat et al., 2004). Functional analyses have been reported in C. elegans (Zhou et al., 1998), Drosophila (Mlodzik et al., 1990; Hoshizaki et al., 1994; Kerber et al., 1998), mouse (Qiu et al., 1997; Pereira et al., 1999; Lin et al., 2011), and Xenopus (van der Wees et al., 1996). Interestingly, gain-of-function experiments in Xenopus have suggested that ectopic Nr2f1 can inhibit RA-induced expression of the hindbrain genes krox20 and lim1 (Schuh and Kimelman, 1995), supporting the hypothesis that Nr2fs may function as negative regulators of RA signaling.
In zebrafish there are a total of 6 nr2f genes: nr2f1a, nr2f1b, nr2f2, nr2f5, nr2f6a and nr2f6b. The presence of two nr2f1 genes and two nr2f6 genes is the consequence of a teleost-specific whole genome duplication event (Bertrand et al., 2007). An initial study from Fjose et al. (1993) described expression of zebrafish nr2f1a and nr2f5 in the brain and mesoderm. Subsequently, Fjose and colleagues (Fjose et al., 1995) reported an intriguing graded expression pattern of nr2f2 in the hindbrain, and further showed that hindbrain and retinal expression of nr2f2 is modulated by exogenous RA treatment. They did not, however, establish whether expression is dependent on endogenous retinoid signals. More recently, a broad expression analysis of all six nr2fs was provided in a comparative study of the expression of all zebrafish nuclear receptor genes (Bertrand et al., 2007), indicating complex expression of nr2f genes in the CNS, eye and mesoderm.
RA signaling plays important patterning roles in multiple embryonic structures including hindbrain and foregut endoderm. During early development RA is synthesized by activity of Aldh1a2 in the anterior mesoderm (for a review on RA synthesis during development, see Duester 2008). These RA signals are critical for normal development of the adjacent posterior hindbrain (Dupe et al., 1999; Grandel et al., 2002; Begemann et al., 2004), with full patterning of the hindbrain rhombomeres (a series of seven segments along the anteroposterior (AP) extent of the hindbrain; r1–r7 from A to P) dependent on interplay between posterior RA signals and anterior fibroblast growth factor (Fgf) signals (Reifers et al., 1998; Maves et al., 2002; Wiellette and Sive, 2004; Maves and Kimmel, 2005; Nakamura et al., 2005; Schilling 2008). Regionalization of the foregut endoderm, in particular specification of the pancreas, is also reliant on mesoderm-derived RA signals (Stafford and Prince, 2002; Stafford et al., 2006). In both hindbrain and foregut endoderm, RA signaling is modulated by the Cyp26 RA-degrading enzymes. The Cyp26 enzymes ensure appropriate levels of RA signaling in individual organisms despite varying levels of the RA precursor Vitamin A (White et al., 2007a). As cyp26 genes are themselves positive targets of RA, they generate a negative feedback loop that is largely responsible for responding to and degrading RA signals to shape the RA gradient in both the hindbrain and foregut endoderm (Begemann and Meyer, 2001; White et al., 2007a; Hernandez et al., 2007; Kinkel et al., 2009). However, mathematical modeling has suggested that cyp26 modulation alone is insufficient to explain the full range of RA signals across the hindbrain (White et al., 2007b). It is therefore an appealing hypothesis that nr2fs, which like cyp26 genes can function to negatively regulate RA signaling albeit via an entirely different mechanism, may similarly play an important role in regulation of RA activity within the hindbrain region and elsewhere.
In this study, we report detailed expression analysis of zebrafish nr2f1a, nr2f1b, nr2f2 and nr2f5. We find that expression commences shortly after gastrulation is completed, and is dynamic through segmentation stages within anterior neural tissues, including the hindbrain, as well as in the trunk. nr2f expression at later developmental stages is restricted to the nervous system, as previously described (Bertrand et al., 2007; Thisse and Thisse, 2008). In a microarray experiment to identify targets of RA signaling in the developing zebrafish endoderm (Kinkel et al., 2009), we obtained evidence that nr2f1a, nr2f2, and nr2f5 are positively regulated by RA. As cyp26 genes are similarly RA-regulated in endoderm, we hypothesized that nr2f genes might also function to provide an additional layer of RA regulation via a negative feedback loop. Here we verify that RA signaling positively regulates nr2f1a in trunk endoderm and mesoderm. However, while exogenous RA signaling can negatively regulate nr2f expression in the hindbrain, normal expression in this region is not dependent on RA signals. We additionally asked whether Fgfs are upstream regulators of nr2f gene expression in the midbrain and hindbrain, as nr2f expression is adjacent to two major Fgf signaling centers in this region. Our results indicate that Fgf signaling is not required for early onset of nr2f gene expression, but may play a role in later rhombomere-specific expression. Together, our findings suggest that individual nr2f genes respond differently to RA signaling, and that expression is regulated differentially at particular AP levels. Overall, our results do not support the hypothesis that nr2f genes function in a negative feedback loop to modulate RA signals in the hindbrain, although our data do allow the possibility that nr2f1a may play such a role in the mesoderm and endoderm.
RESULTS AND DISCUSSION
Expression analysis of zebrafish nr2f genes
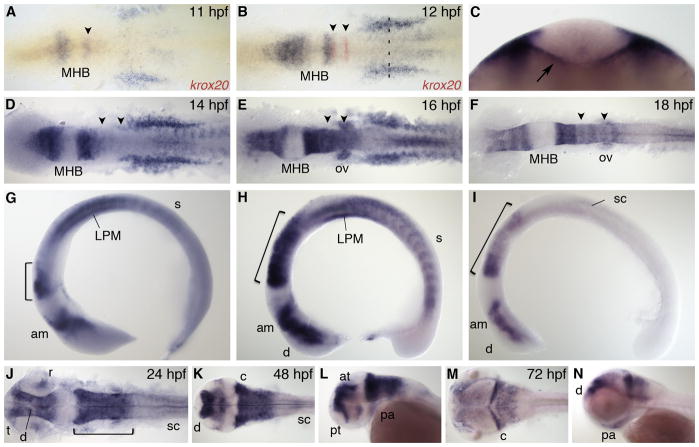
Zebrafish nr2f1a, also known as svp[44] or COUP-TFαA, is an ortholog of human and mouse COUP-TFI (also designated as EAR3) (Achatz et al., 1997). nr2f1a expression begins at 11 hours post fertilization (hpf) in structures within all three germ layers (Fig. 1A). In the anterior neural plate, expression originates in two domains separated by the midbrain-hindbrain boundary (MHB). The more posterior domain lies immediately anterior of r3 as visualized by krox20 expression, which is a marker of r3 and r5. In the trunk, nr2f1a is expressed in the lateral plate mesoderm (LPM), newly forming somites, and the underlying endoderm. By 12 hpf, neural expression of nr2f1a has expanded both anteriorly and posteriorly to the MHB; expression in the trunk increases and is clearly localized to the endoderm (Fig. 1B, C). The expansion of neural expression away from the MHB continues through 14 hpf (Fig. 1D, G) and by 16 hpf nr2f1a is expressed throughout the anterior midbrain and presumptive diencephalon and in r1–6 in the hindbrain (Fig. 1E, H). Within the hindbrain, transcript levels are graded with highest levels in r1 and r2 and lowest levels in r6. nr2f1a levels remain high in the LPM and somites, and starts to be expressed in the otic vesicle and spinal cord. By 18 hpf, nr2f1a is expressed within the presumptive diencephalon and anterior midbrain, in the hindbrain maintaining a high to low expression gradient from anterior to posterior, and in the spinal cord (Fig. 1F, I). nr2f1a is no longer expressed in the LPM or somites from this stage onwards.
Fig. 1.
nr2f1a expression. A,B: Dorsal view during early segmentation stages co-stained with r3 and r5 marker krox20 (red). C: Optical section of 12 hpf embryo at anteroposterior level indicated by dashed line in B. Arrow indicates endoderm expression. D–F: Dorsal view and G–I: lateral view during segmentation stages. J,K,M: Dorsal view. L,N: Lateral view. am, anterior midbrain; at, anterior tegmentum; c, cerebellum; d, diencephalon; LPM, lateral plate mesoderm; MHB, midbrain-hindbrain boundary; ov, otic vesicle; pa, pharyngeal arch; pt, prethalamus; r, retina; s, somite; sc, spinal cord; t, telencephalon. Arrowheads indicate r3 and r5. Bracket indicates hindbrain.
At 24 hpf, towards the end of segmentation, nr2f1a is expressed in the telencephalon, diencephalon, anterior midbrain and the hindbrain, with a complete absence of transcript in the posterior midbrain (Fig. 1J). Within the hindbrain, uniform high expression has replaced the graded, rhombomere-specific expression, and section analysis shows that this expression is mainly dorsal (data not shown). nr2f1a is also now expressed in the dorsal region of the retina, the pharyngeal arches, and in the spinal cord. By 48 hpf, the most anterior nr2f1a expression domain is now specific to the preventricular zone, prethalamus, dorsal diencephalon, and anterior tegmentum. Expression within the entire hindbrain persists, with highest transcript levels in the cerebellum. More ventral expression domains include the lens and pharyngeal arches. The most posterior expression is a small region within the anterior spinal cord (Fig. 1K, L). At 72 hpf, high levels of nr2f1a transcripts are found in the dorsal diencephalon, anterior tegmentum, and the cerebellum. There is also more diffuse expression within the tectum, hindbrain, and pharyngeal arches (Fig. 1M, N).
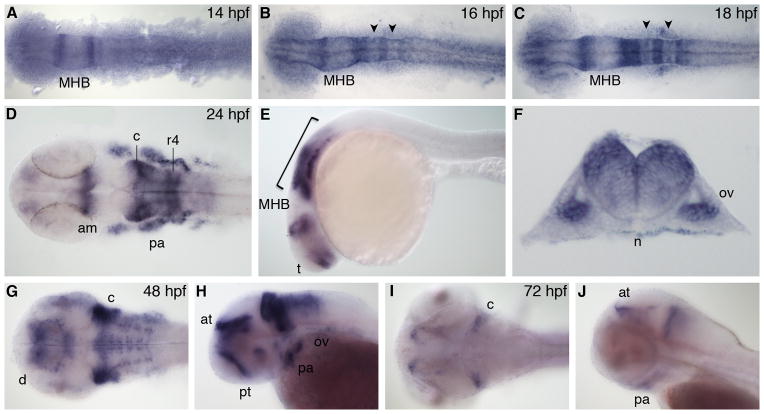
Zebrafish nr2f1b, also known as COUP-TFαB, is the duplicate of nr2f1a and therefore also an ortholog of human and mouse COUP-TFI. The nr2f1b gene is first expressed at 14 hpf in two domains just anterior and posterior to the MHB (Fig. 2A). At 16 hpf, there is expression in the presumptive telencephalon and anterior midbrain. More posteriorly, nr2f1b is expressed in r1, r2, r4, and r6 in the hindbrain, as well as the otic vesicle (Fig. 2B). This expression pattern persists in the 18 hpf embryo, but with more refined expression in the dorsal eye. Within the hindbrain, transcript levels in r3 and r5 are significantly lower than in other rhombomeres (Fig. 2C), and expression in the otic vesicle has increased. At 24 hpf, there is expression in the telencephalon, anterior midbrain, and predominantly r1–2 and r4 the hindbrain (Fig. 2D, E). Expression within the hindbrain is localized dorsally, as seen in a transverse section through r5 (Fig. 2F). nr2f1b is also now expressed robustly in the migratory neural crest moving out to the pharyngeal arches, as well as in the otic vesicle. At 48 hpf, we see expression in the prethalamus, dorsal diencephalon, and anterior tegmentum in the most anterior cranial regions. In the 48 hpf hindbrain nr2f1b is highly expressed in the lateral regions of the cerebellum, and in neurogenic regions (Fig. 2G). Other regions of expression include the retina, the trigeminal ganglia, pharyngeal arches and otic vesicle (Fig. 2G, H). By 72 hpf, expression of nr2f1b is reduced, with transcripts seen only in the anterior tegmentum, pharyngeal arches and lateral regions of the cerebellum (Fig. 2I, J).
Fig. 2.
nr2f1b expression. A–C: Dorsal view during segmentation stages, anterior to left. D,G,I: Dorsal view. E,H,J: Lateral view, anterior to left. F: Transverse section through r5. am, anterior midbrain; at, anterior tegmentum; c, cerebellum; d, diencephalon; MHB, midbrain-hindbrain boundary; n, notochord; ov, otic vesicle; pa, pharyngeal arch; pt, prethalamus; r4, rhombomere 4. Arrowheads indicate r3 and r5. Bracket indicates hindbrain.
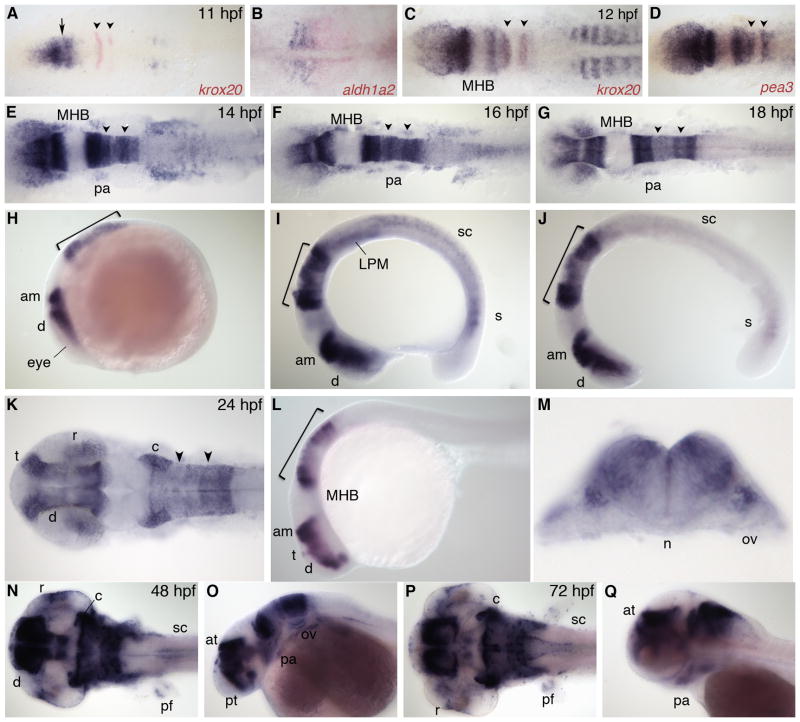
Zebrafish nr2f2, also known as svp[40] or COUP-TFβ, is the ortholog of mammalian COUP-TFII (also referred to as ARP-1) (Achatz et al., 1997). This gene is first expressed at 10 hpf throughout the anterior neural plate and the tailbud (data not shown). At 11 hpf, expression in the anterior neural plate is at high levels (Fig. 3A). Interestingly, there is a narrow gap separating this anterior domain into two (arrow); this gap will later separate an expression domain in the diencephalon from one in the anterior midbrain. The nr2f2 gene is also expressed in newly forming somites at this stage, overlapping with and anterior to the RA source, as visualized by aldh1a2 expression (Fig. 3B). By 12 hpf, there is high expression within r1–3 and r5 in the hindbrain (Fig. 3C, note overlap with krox20 expression in r3). The expression in r5 at this stage contrasts with the pattern of nr2f1a, and it is striking that expression is absent in both the MHB and r4, two areas of high Fgf signaling, as visualized by co-expression with the Fgf target gene pea3 (Fig. 3D). There is also low expression in the anterior LPM as well as in somites. By 14 hpf, anterior expression extends laterally to the future eye region, and there is now expression in the neural crest migrating into the first pharyngeal arch. In the hindbrain, levels of expression are rhombomere-specific and appear to be graded from high to low r1–4, then high in r5 and low in r6. Expression in the LPM is greatly up-regulated, and expression begins in the spinal cord (Fig. 3E, H). By 16 hpf, nr2f2 expression in the eye is located posteriorly, and there is expression in all cranial neural crest streams migrating out to the pharyngeal arches. Expression in the lateral plate mesoderm is reduced while transcript levels have increased in the anterior spinal cord (Fig. 3F, I). Similar to nr2f1a, expression in the trunk at 18 hpf is greatly reduced and there is still expression in the spinal cord and recently forming somites. Anteriorly, nr2f2 is expressed in the eye, presumptive diencephalon and anterior midbrain, as well as at lower levels in pharyngeal arches. Within the hindbrain, transcript levels are highest in r1–2, lowest in r3–4, and intermediate in r5–6 at this stage (Fig. 3G, J).
Fig. 3.
nr2f2 expression. A,C: Dorsal view, co-stained with krox20 (red). Arrow indicates gap between presumptive diencephalon and anterior midbrain. B: Dorsal view of trunk, anterior to left, co-stained with aldh1a2 (red). D: Dorsal view, co-stained with pea3 (red). E–G,K,N,P: Dorsal view. H–J,L,O,Q: Lateral view. M: Transverse section through r5. am, anterior midbrain; at, anterior tegmentum; c, cerebellum; d, diencephalon; LPM, lateral plate mesoderm; MHB, midbrain-hindbrain boundary; n, notochord; ov, otic vesicle; pa, pharyngeal arch; pf, pectoral fin; pt, prethalamus; r, retina; s, somite; sc, spinal cord; t, telencephalon. Arrowheads indicate r3 and r5. Bracket indicates hindbrain.
At 24 hpf, nr2f2 expression is very high in the posterior telencephalon, diencephalon and anterior midbrain (Fig. 3K, L). Within the hindbrain, expression is dorsal and high in r1, r2, r5 and r6, with lower expression in the two middle rhombomeres (Fig. 3M). Transcripts are also found in the pharyngeal arches and in the spinal cord. By 48 hpf, nr2f2 is expressed in the posterior telencephalon, dorsal diencephalon, midbrain tegmentum and anterior tectum. In the hindbrain, expression remains high in the cerebellum and posterior rhombomeres, with lower expression in r3 and r4. nr2f2 is also expressed in the retina, pharyngeal arches, otic vesicle, pectoral fin, and anterior spinal cord (Fig. 3N, O). Finally, by 72 hpf, nr2f2 transcripts are localized to the telencephalon, midbrain tegmentum, anterior tegmentum, and cerebellum. Within the remainder of the hindbrain expression is diffuse except within dorsal neurons in the posterior part of the hindbrain that also extend into the spinal cord. Transcripts are also seen in the retina, pharyngeal arches, and the pectoral fin (Fig. 3P, Q).
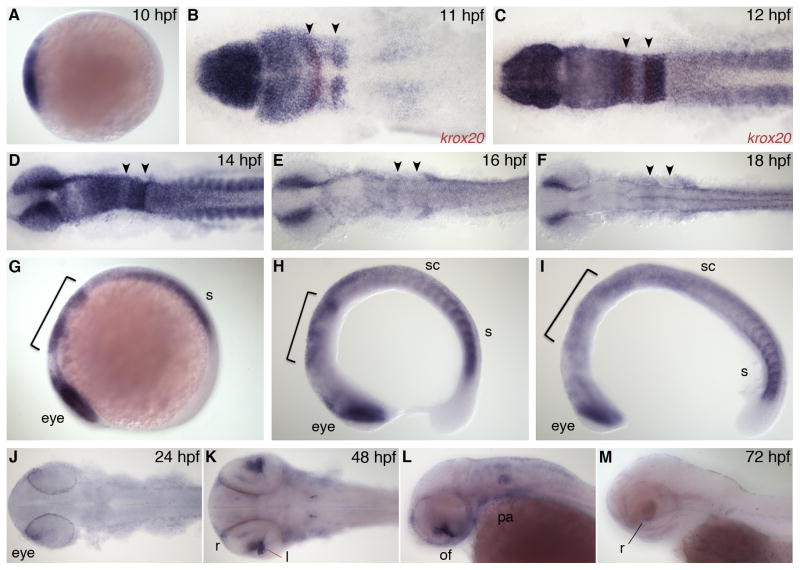
Zebrafish nr2f5, also known as svp[46] or COUP-TFγ, has a very dynamic expression pattern. It is first expressed at the bud stage (10 hpf), at very high levels throughout anterior neural plate (Fig. 4A). At 11 hpf, nr2f5 is highly expressed in the forebrain, midbrain, and r1–5 (Fig. 4B). There is no gap in expression at the MHB as seen with the previous nr2f genes at this stage; however, there is a small gap and differences in expression levels between the midbrain and forebrain regions, with the forebrain having higher expression levels. The midbrain expression domain extends posteriorly to r3 in the hindbrain. In r4 and r5, expression is restricted laterally. In the trunk there is expression in the paraxial and lateral plate mesoderm. In the 12 hpf embryo, the hindbrain expression extends posteriorly to r6, with very high expression in r5 and r6, lowest expression in r4, and diffuse expression in r7–r8. There is also strong expression within the somites (Fig. 4C). At 14 hpf, the most anterior expression in the forebrain as well as in the anterior midbrain start to diminish and we see strong expression in the dorsal eye regions. Within the hindbrain, there is high expression in r1–3 and r5–6, with lowest expression in r4. There is also broad expression in the anterior spinal cord and high levels in the somites (Fig. 4D, G). By 16 hpf, nr2f5 expression is much reduced in the forebrain and midbrain, and there is only diffuse expression throughout the hindbrain and anterior spinal cord (Fig. 4E, H). nr2f5 is expressed in cranial neural crest cells and there is high expression in the dorsal eye and newly forming somites (Fig. 4H); a similar expression pattern is retained at 18 hpf (Fig. 4F, I).
Fig. 4.
nr2f5 expression. A: Lateral view, anterior to left. B,C: Dorsal view, co-stained with krox20 (red). D–F: Dorsal view, and G–I: lateral view during segmentation stages. J,K: Dorsal view. L,M: Lateral view. l, lens; of, optic fissure; pa, pharyngeal arch; r, retina; s, somite; sc, spinal cord. Arrowheads indicate r3 and r5. Bracket indicates hindbrain.
At 24 hpf, nr2f5 is expressed in the optic fissure and there is a low level of expression in the anterior trunk (Fig. 4J). At 48 hpf, there is diffuse expression in the dorsal telencephalon and a small dorsal region of retina, and strong expression in the choroid fissure (Fig. 4K, L). There is also diffuse expression in the lateral regions of the hindbrain and pharyngeal arches, as well as strong expression in a small bilateral cluster of neurons in the hindbrain. By 72 hpf, nr2f5 is expressed only in a few retinal cells just under the lens (Fig. 4M).
RA regulation of nr2f gene expression
Previously, our lab performed a microarray experiment to identify genes that are targets of RA signaling in the 10 hpf zebrafish endoderm (Kinkel et al., 2009; Dalgin et al., 2011). In this experiment, three groups of embryos were compared: RA-treated, RA-deficient (aldh1a2 morpholino), and wild type, and GFP positive endoderm cells were isolated from Tg(sox17:GFP) embryos by fluorescence-activated cell sorting (FACS). This approach identified nr2f genes as positive targets of RA signaling in the endoderm. Our microarray data (in arbitrary units; Table 1) show that nr2f1a, nr2f6a, and nr2f6b are highly expressed in the endoderm at 10 hpf. In response to RA treatment from 8–9 hpf, nr2f1a was most significantly affected with transcript levels increased 10-fold. Further, nr2f1a expression was also down-regulated 11-fold in the absence of RA signals, suggesting that this endoderm expression is downstream of RA signaling. Although expression levels of nr2f5 are relatively unaffected in the absence of RA signaling, we find that endoderm expression is up-regulated nearly 4-fold in the presence of exogenous RA signals. Similarly, although nr2f2 is expressed at levels close to or below the limit of detection in the endoderm at this stage, it is up-regulated 2.4-fold in the presence of exogenous RA signals. Together, these data suggest that nr2f transcription in the endoderm is up-regulated when RA levels are high, consistent with a negative feedback model in this region. Interestingly, these findings contrast with a previous study of nr2f2 expression in the hindbrain. Fjose et al. (1995) showed that nr2f2 expression is reduced in response to RA treatment, implying that in this context nr2f2 is a negative target of RA signaling; the consequences of RA deficiency were not examined in this study. Together, these findings motivated us to further investigate the details of RA-regulation of nr2f1a, nr2f2 and nr2f5 expression in both the endoderm and hindbrain.
Table 1.
Summarized microarray data showing RA regulation of all six zebrafish nr2f genes. The microarray experiment was performed in triplicate; methodology is described in detail by Kinkel et al. (2009). Values indicate relative expression of nr2f genes in FACS-sorted endoderm cells from Tg(sox17:GFP) embryos. In cases where a gene was represented more than once on the array values are averaged. All numbers are in arbitrary units (a.u.), where values less than 60 indicate baseline (essentially undetectable) expression levels.
| Gene | Wild type | Aldh1a2 MO | RA-treated | Fold decrease Wt/MO | Fold increase RA/Wt |
|---|---|---|---|---|---|
| nr2f1a | 2,400 | 220 | 24,000 | 11 | 10 |
| nr2f1b | 45 | 46 | 46 | 1.0 | 1.0 |
| nr2f2 | 45 | 43 | 110 | 1.0 | 2.4 |
| nr2f5 | 850 | 600 | 3,300 | 1.4 | 3.9 |
| nr2f6a | 7,000 | 8,000 | 8,400 | 0.9 | 1.2 |
| nr2f6b | 3,500 | 2,600 | 7,000 | 1.3 | 2.0 |
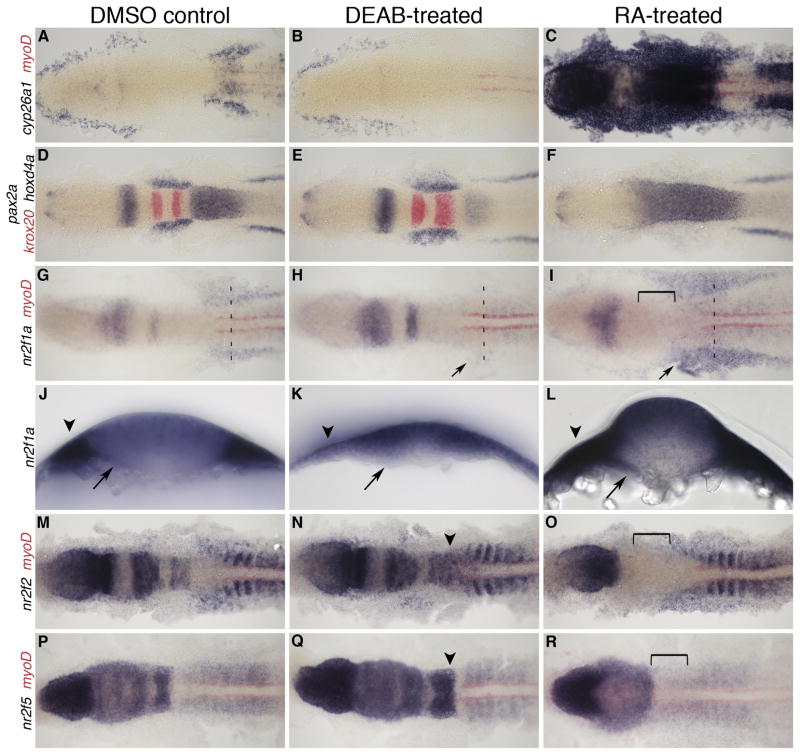
To examine whether RA signaling during gastrulation was upstream of nr2f expression, we either increased or abolished retinoid signaling prior to the onset of nr2f expression. Embryos were treated with exogenous RA to enhance RA signaling or with DEAB to block Aldh1a2 activity (Russo et al., 1988) and thus prevent RA synthesis starting at 8 hpf. Control embryos treated with dimethyl sulfoxide (DMSO) carrier alone were indistinguishable from untreated embryos. To verify our microarray data and focus our analysis on late gastrulation, the chemicals were washed out at 10 hpf, and the embryos fixed at 12 hpf for in situ hybridization analysis of nr2f expression. As a positive control for the treatment, we also followed the expression of the known RA target gene cyp26a1 (Kinkel et al., 2009) (Fig. 5A–C). We monitored changes in tissue specification with markers pax2a, which labels the eye field, MHB, otic placode and pronephric mesoderm; krox20 to label r3 and r5; and hoxd4a in the posterior hindbrain, which normally has an anterior limit at the r6/r7 boundary. We used the somite marker myoD to monitor changes in expression along the AP axis. As expected, we see an anterior expansion of hoxd4a up to the MHB in RA-treated embryos, and a truncation of hoxd4a in the posterior hindbrain with a slight expansion of krox20 in DEAB-treated embryos (Fig. 5D–F). In the absence of RA signaling, we find nr2f1a, nr2f2, and nr2f5 hindbrain expression is remarkably similar to controls, with only a very subtle posterior extension of nr2f2 and nr2f5 expression (Fig. 5G, H, M, N, P, Q). We see hindbrain expression even in embryos treated with DEAB from 50% epiboly to 12 hpf (data not shown), which suggests that the onset of nr2f expression in the hindbrain is not dependent on RA signaling. However, we do see an obvious reduction of nr2f1a in the trunk, including the LPM and endoderm (Fig. 5J, K), but no change in trunk expression of nr2f2 and nr2f5 in DEAB-treated embryos (Fig. 5H, N, Q). Exogenous RA treatment abolishes expression of all three genes in the hindbrain, supporting the previous observation that RA represses expression in this region (Fig. 5I, O, R; Fjose et al., 1995). While nr2f2 and nr2f5 expression in the trunk appear relatively unchanged in response to RA treatment (Fig. 5O, R), we see a substantial increase in nr2f1a trunk expression in the lateral plate and paraxial mesoderm, as well as the endoderm (Fig. 5G, I, J, L). This response of trunk nr2f1a expression to increased retinoid signaling is again consistent with its positive regulation by RA and our microarray results.
Fig. 5.
nr2f expression at 12 hpf with RA and DEAB treatment 8–10 hpf. A–C: Expression of RA target gene cyp26a1 to confirm treatment efficiency, co-stained with myoD (red) to mark the myotome. D–F: MHB and hindbrain markers pax2a, krox20 (red), and hoxd4a show changes in tissue specification. G–I: nr2f1a expression. Arrow indicates changes in LPM expression. J–L: transverse sections showing changes in nr2f1a transcript levels in the endoderm (arrows) and LPM (arrowheads) at anteroposterior levels designated by dashed lines in G–I. M–O: nr2f2 expression, and P–R: nr2f5 expression. Arrowheads (N,Q) indicate a posterior extension of hindbrain expression. Brackets indicate loss of hindbrain expression.
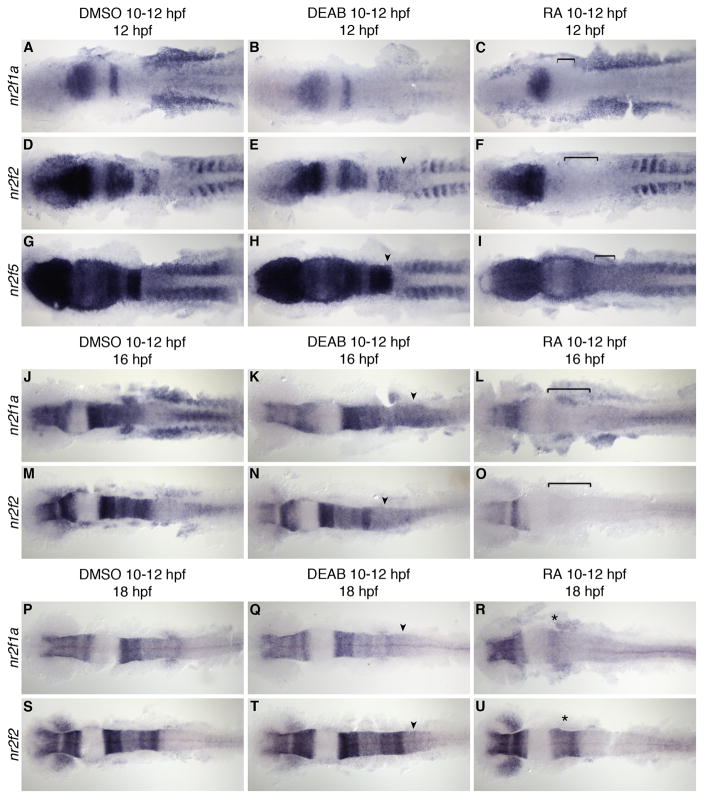
To further examine the effects of RA signaling on nr2f expression, we either increased or abolished retinoid signaling at the onset of nr2f expression between 10 and 12 hpf. Similar to previous experiments by Fjose et al. (1995), we treated embryos with exogenous RA to increase RA signaling during this time; however, to extend the previous analysis we again included treatment with DEAB to test if RA signaling is required for nr2f expression. The chemicals were washed out and embryos fixed at 12, 16, or 18 hpf for analysis of nr2f expression by in situ hybridization (Fig. 6). We again used the same panel of markers to monitor treatment efficacy and changes in CNS regionalization (data not shown). We find that at this stage, exogenous RA treatment induces a complete posteriorization of the hindbrain, with hoxd4a expression extending as far anterior as the MHB; whereas blocking RA signaling at this stage has no effect on this already specified tissue (as described in Maves and Kimmel, 2005).
Fig. 6.
nr2f expression at 12, 16, and 18 hpf with RA and DEAB treatment 10–12 hpf. A–I: nr2f1a, nr2f2, and nr2f5 expression at 12 hpf. J–O: nr2f1a and nr2f2 expression at 16 hpf after RA or DEAB treatment from 10–12 hpf. P–U: nr2f1a and nr2f2 expression at 18 hpf after RA or DEAB treatment from 10–12 hpf showing a return of anterior hindbrain expression (asterisk). Arrowheads indicate a posterior extension of hindbrain expression. Brackets indicate loss of hindbrain expression.
Treatment with DEAB from 10–12 hpf did not significantly disrupt the normal hindbrain expression pattern of nr2f1a, nr2f2 or nr2f5, but did cause a modest expansion of nr2f2 and nr2f5 expression into the posterior hindbrain (Fig. 6D, E, G, H). This suggests either that RA is necessary to limit the posterior expression of these genes, or that loss of RA signaling at this stage resulted in an expansion of the anterior hindbrain towards the posterior, consistent with described loss of posterior hindbrain tissues in RA-deficient embryos (Begemann et al., 2004). There is also a marked reduction of nr2f1a expression in the paraxial and lateral plate mesoderm of the trunk, and in the underlying endoderm (Fig. 6A, B). This suggests that RA signaling is not necessary for nr2f expression in the hindbrain, but is important for normal nr2f1a expression in the trunk at this stage. In RA-treated embryos, however, hindbrain expression of nr2f1a and nr2f2 was abolished, and nr2f5 expression was greatly reduced, again suggesting that exogenous RA signaling suppresses expression in the hindbrain (Fig. 6C, F, I). In these embryos forebrain expression was unchanged; however, exogenous RA increased nr2f1a expression in the LPM, while having little effect on nr2f2 or nr2f5. The loss of trunk nr2f1a expression in DEAB-treated embryos and its expansion in RA-treated embryos is consistent with the results of our microarray, confirming that in this context nr2f1a is a positive target of RA signaling. By contrast, nr2f2 and nr2f5 did not show obvious changes in trunk expression in response to RA. Further analysis of our microarray data indicates relatively low absolute expression levels of these genes in the endoderm, which are likely below the detection level of whole-mount in situ hybridization.
Since our results suggest RA negatively regulates nr2f expression in the hindbrain, and RA positively regulates nr2f1a in the trunk, we next tested if normal expression returns in embryos that continue to develop after exogenous RA signals or DEAB are washed out. We therefore let development of embryos treated with RA or DEAB from 10–12 hpf proceed to 16 or 18 hpf, and then analyzed nr2f expression. In DEAB-treated embryos, expansion of nr2f1a and nr2f2 expression in the posterior hindbrain was still found at 16 and 18 hpf (Fig. 6K, N, Q, T), similar to our findings at 12 hpf. Likewise, nr2f1a trunk expression was abolished (Fig. 6J, K, P, Q), whereas trunk expression of nr2f2 was unchanged (Fig. 6M, N, S, T). nr2f5 expression is unchanged in DEAB-treated embryos, and the low-level expression in r3 and r5 in controls is lost in RA-treated embryos (data not shown). Interestingly, we did find a temporal change in nr2f1a and nr2f2 hindbrain expression in embryos treated with exogenous RA. At 16 hpf, hindbrain expression of both genes was still abolished (Fig. 6L, O); however, at 18 hpf we found that both nr2f1a and nr2f2 expression return in the anterior hindbrain (Fig. 6R, U). This return of nr2f2 expression is not consistent with the results of Fjose et al. (1995), who reported that nr2f2 hindbrain expression continues to be absent in embryos at 24 hpf, 12 hours after RA was washed out. This discrepancy may be the result of differences in effectively washing all chemicals out before allowing development to continue to later stages. The reemergence of nr2f1a and nr2f2 expression in the anterior hindbrain suggests either that RA signaling can repress expression directly or, perhaps more likely given the usual role of RA as an activator, that a factor responsible for activating nr2f expression is inhibited in the presence of exogenous RA signals.
Since these chemical treatments expose the entire embryo to RA, our results suggest that nr2f expression can be divided into three separate response ‘territories’: forebrain, which is unaffected by RA signaling; hindbrain, which is negatively regulated by RA signaling; and trunk, which is positively regulated by RA signaling in the case of nr2f1a only. The patterns of nr2f expression in the hindbrain are not consistent with a negative feedback model, in which RA treatment should elevate nr2f expression, as we see for cyp26a1. In contrast, we find that RA treatment reduces nr2f expression in the posterior hindbrain, and blocking RA synthesis enhances this expression. The high-level expression domain of nr2f2 in r5 and r6 is very close to endogenous RA signals; nevertheless, this expression is also lost in response to sufficiently high RA treatments. Our results also confirm that nr2f expression in the forebrain, far from the RA source, is not a target of retinoid signaling. However, our analysis of nr2f1a expression in the trunk does reveal the up-regulation in response to RA treatment that is consistent with a negative feedback model. We conclude that nr2f1a may indeed function in trunk endoderm and mesoderm to limit the AP extent of RA signals by competing for RA signaling target genes.
Fgf regulation of nr2f gene expression
Since strong nr2f1a and nr2f2 expression commences on either side of the MHB, and then spreads both anteriorly and posteriorly in a graded fashion, it is possible that a diffusible signal emanating from the MHB may induce this expression. Fgf8 is a good candidate, as it is secreted from the MHB signaling center and required for proper hindbrain patterning (Reifers et al., 1998; Nakamura et al., 2005). Fgf8 is also secreted from the r4 signaling center to initiate r5 and r6 development (Wiellette & Sive, 2004). Thus the r4 source of Fgf8 could potentially also explain the elevated expression levels of nr2f2 in these rhombomeres.
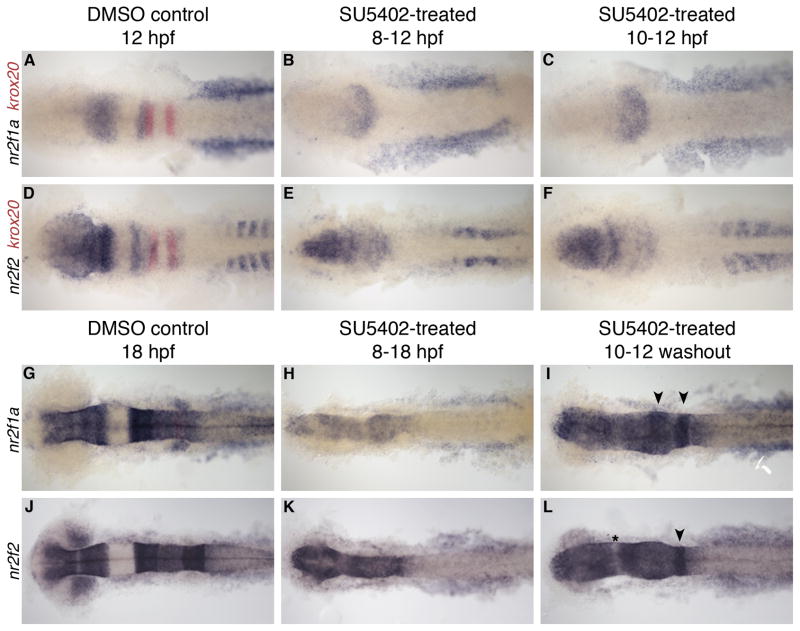
To examine whether Fgf signaling is required for expression of nr2f1a and nr2f2 genes we used the pharmacological inhibitor SU5402 starting at two different stages: prior to onset of nr2f gene expression (8 hpf) or at the onset of expression (10 hpf). We continued treatment until fixation at 12 hpf and observed nr2f expression by in situ hybridization (Fig. 7A–F). To monitor changes in tissue identity and the effectiveness of our treatments, we again analyzed the markers pax2a, krox20, and hoxd4a (data not shown). Fgf signaling is required for both pax2a and krox20 expression (Walshe et al., 2002); when SU5402 was applied from 8 hpf we observed the expected loss of these two markers in anterior regions, as well as a lack of tail elongation. When we applied the inhibitor at 10 hpf, expression of pax2a in the eye and tail extension was restored, but there remained a lack of these markers in the MHB, otic vesicle or rhombomeres. Evaluating nr2f1a expression in these treated embryos, we still observed expression in the neural tissue and normal levels of expression in the LPM (Fig. 7A–C). However, the expression levels were slightly lower and the characteristic gap between hindbrain and midbrain expression domains was not present, most likely due to the expected loss of MHB tissue. Expression of nr2f2 was also largely normal in SU5402 treated embryos (Fig. 7D–F). Although blocking Fgf signaling during these stages greatly hinders embryonic development, we can conclude that the establishment of nr2f gene expression does not require Fgf signals.
Fig. 7.
nr2f expression at 12 and 18 hpf after SU5402 treatment. A–F: nr2f1a and nr2f2 expression at 12 hpf. SU5402 treatment from 8–12 hpf (B, E) or 10–12 hpf (C,F). G–L: nr2f1a and nr2f2 expression at 18 hpf. SU5402 treatment from 8–18 hpf (H, K) or 10–12 hpf and washed out (I, L). Arrowheads indicate rhombomere-specific expression in the hindbrain. Asterisk indicates gap in expression, possibly between midbrain and hindbrain.
To further investigate the effect of Fgf signaling on nr2f1a and nr2f2 expression, we next observed the effects on expression at 18 hpf. Analysis of pax2a and krox20 expression showed that neither the MHB nor hindbrain expression of krox20 were present following this treatment (data not shown). We performed two treatments compared to DMSO controls, again starting at either 8 or 10 hpf. First, we treated the embryos with SU5402 between 8–18 hpf, and we find that this treatment did not completely block nr2f1a or nr2f2 expression (Fig. 7G, H, J, K). nr2f1a transcript levels are decreased, but we can conclude that Fgf signals are not important for nr2f1a or nr2f2 induction during this time. In our second group, we started treatment at 10 hpf but washed the chemical out at 12 hpf and let the embryos develop until 18 hpf (Fig. 7I, L). In these embryos there was a substantial increase of overall nr2f1a expression in both neural and mesodermal tissues compared to when the chemical was not washed out. There was also a restoration of rhombomere-specific expression patterns within the hindbrain, indicating that Fgf signaling is either important for maintaining the identity of these rhombomeres, or that Fgf signaling is directly required for maintenance of nr2f1a. Changes in expression of nr2f2 were not as dramatic as those observed for nr2f1a. In embryos treated from 8–18 hpf, there was strong expression of nr2f2 in neural tissues. In washout embryos, expression levels were still high overall, but we noted some restoration in rhombomere patterning and rhombomere-specific expression. Interestingly, there seems to be a slight return of the gap between the midbrain and hindbrain within the neural tissue in nr2f2 washout embryos (Fig. 7J–L, asterisk); suggesting that Fgf signaling may be required to repress nr2f expression in this region. In summary, since nr2f1a and nr2f2 expression are still present in the absence of Fgf signaling, we conclude that these genes do not require Fgf signaling for their expression. However, our observation that nr2f1a expression is more affected than nr2f2 by long-term changes in Fgf signaling suggests that these two genes respond differently to the broader consequences of blocking Fgf signaling.
In summary, we have carefully examined the complex and variable expression patterns of the nr2f gene family in zebrafish, particularly during early segmentation stages when the anteroposterior axis is being patterned. We provide the first evidence for nr2f1a expression within the endoderm at these stages as well as its regulation by RA signaling in the trunk endoderm and mesoderm. RA signaling plays many critical roles in early development, including patterning the anteroposterior axis of the hindbrain and establishment of pancreatic lineages in the developing endoderm. As RA is derived from the dietary precursor Vitamin A, its levels must be carefully regulated within the organism. Results from previous in vitro studies suggest Nr2fs function within a negative feedback loop to modulate RA signals. While we find that nr2f1a expression is positively regulated by RA signaling in trunk endoderm and mesoderm, consistent with such a feedback loop, our results do not support existence of an Nr2f/RA negative feedback loop model in the hindbrain. Finally, hindbrain expression relies on a close interplay of RA and Fgf signals; our analysis suggests that Fgf signaling is not required for the onset of nr2f expression but may play a role in maintaining rhombomere-specific expression.
EXPERIMENTAL PROCEDURES
Fish Maintenance
Zebrafish were maintained following standard procedures. Embryos were raised at 28.5°C and staged as described (Kimmel et al., 1995) under IACUC approved protocols. AB wild type embryos were used for in situ hybridization. 48 and 72 hpf embryos were incubated in 0.2 mM 1-phenyl 2-thiourea (PTU; Sigma) starting at 24 hpf to inhibit melanin synthesis. Embryos were fixed in 4% paraformaldehyde (PFA; Sigma) overnight at 4°C and dehydrated into methanol prior to in situ hybridization.
Probe Synthesis and Whole Mount In Situ Hybridization
Full-length cDNA constructs for nr2f1a (accession: NM_131180.1), nr2f2 (accession: NM_131183.1), and nr2f5 (accession: NM_131186.1) were obtained via PCR from 18 or 30 hpf zebrafish cDNA library, cloned into a pGEM-T vector (Promega) and sequenced. Primers used (brackets indicate added restriction enzyme sites for cloning):
-
Nr2f1a: For 5′ [AAAAGGCCT]ATGGCAATGGTAGTTAGCGTC 3′
Rev 5′ [AAAAGGCCT]TCATTGAATGGACATGTAGGG 3′
-
Nr2f2: For 5′ [AAAAGGCCT]ATGGCAATGGTAGTGTGGAGA 3′
Rev 5′ [AAAAGGCCT]CTACTGAATCGACATATAAGG 3′
-
Nr2f5: For 5′ [AAAAGGCCT]ATGGCAATGGTAGTAAATCAG 3′
Rev 5′ [AAAAGGCCT]TCAGGGCCCGTTCTCATTGTA 3′
The nr2f1b construct in pBluescript was kindly provided by B. and C. Thisse (MGC: 65854; Thisse and Thisse, 2008). Digoxigenin-labeled RNA probes were synthesized using the following templates: the nr2f1a plasmid was linearized with NcoI and transcribed using SP6 polymerase (Promega); the nr2f1b plasmid was linearized with SacI and transcribed using T7 polymerase (Promega); the nr2f2 plasmid was linearized with SpeI and transcribed using T7 polymerase; and the nr2f5 plasmid was linearized with SacII and transcribed using SP6 polymerase. The fluorescein-labeled krox20 riboprobe was used as described (Oxtoby and Jowett, 1993). Embryos were rehydrated and whole mount in situ hybridization was performed as described (Prince et al., 1998).
Pharmacological Treatments
RA and DEAB
Embryos were left in their chorions and treated with 1 μM RA in DMSO (Sigma) (as described by Fjose et al., 1995) or 10 μM DEAB (Aldrich) in DMSO diluted in embryo media in the dark for times indicated at 28.5°C. The embryos were rinsed 3x with embryo media and either fixed or left to develop to later stages.
SU5402
Embryos were left in their chorions and treated with 60 μM SU5402 (Tocris) in DMSO (Walshe et al., 2002), in the dark starting at 8 hpf or 10 hpf until fixation at 12 or 18 hpf or treated 10–12 hpf, washed out 3x in embryo medium and fixed at 18 hpf.
Image Analysis
Embryos were deyolked and flat-mounted in glycerol for dorsal imaging. Transverse sections were obtained by embedding in 3% agarose in PBS and sectioning at 150 μM intervals for 12 hpf embryos, and 100 μM intervals for 24 hpf embryos using a Vibratome Series 1000 Plus Tissue Sectioning System. All images were acquired using a Zeiss Axioskope microscope and Leica DFC490 camera.
Key findings.
Zebrafish nr2f1a, nr2f1b, nr2f2, and nr2f5 show dynamic expression during development
RA and Fgf signals are not required for the onset of nr2f hindbrain expression
RA positively regulates nr2f1a in the trunk endoderm and mesoderm
Acknowledgments
We thank B. Thisse for providing the nr2f1b construct and the C. Lowe lab for providing SU5402. We are very grateful to J. Clarke for discussions on zebrafish neural anatomy, S. Burns and A. Ng for help with fish care, and comments on the manuscript by G. Dalgin, S. Horne-Badovinac, and S. Wanner. This work was supported by the Developmental Biology Training Grant T32HD55164 and NIH grant RO1DK064973 to VEP.
References
- Achatz G, Hölzl B, Speckmayer R, Hauser C, Sandhofer F, Paulweber B. Functional domains of the human orphan receptor ARP-1/COUP-TFII involved in active repression and transrepression. Mol Cell Biol. 1997;17:4914–4932. doi: 10.1128/mcb.17.9.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Meyer A. Hindbrain patterning revisited: timing and effects of retinoic acid signaling. Bioessays. 2001;23:981–986. doi: 10.1002/bies.1142. [DOI] [PubMed] [Google Scholar]
- Begemann G, Marx M, Mebus K, Mayer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol. 2004;271:119–129. doi: 10.1016/j.ydbio.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escrivà H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3:e188. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgin G, Ward AB, Hao le T, Beattie CE, Nechiporuk A, Prince VE. Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development. 2011;138:4597–4608. doi: 10.1242/dev.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupe V, Ghyselinck NB, Wendling O, Chambon P, Mark M. Key roles of retinoic acid receptors alpha and beta in the patterning of the caudal hindbrain, pharyngeal arches and otocyst in the mouse. Development. 1999;126:5051–5059. doi: 10.1242/dev.126.22.5051. [DOI] [PubMed] [Google Scholar]
- Fjose A, Nornes S, Weber U, Mlodzik M. Functional conservation of vertebrate seven-up related genes in neurogenesis and eye development. EMBO J. 1993;12:1403–1414. doi: 10.1002/j.1460-2075.1993.tb05784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A, Weber U, Mlodzik M. A novel vertebrate svp-related nuclear receptor is expressed as a step gradient in developing rhombomeres and is affected by retinoic acid. Mech Dev. 1995;52:233–246. doi: 10.1016/0925-4773(95)00404-o. [DOI] [PubMed] [Google Scholar]
- Gauchat D, Escriva H, Miljkovic-Licina M, Chera S, Langlois MC, Begue A, Laudet V, Galliot B. The orphan COUP-TF nuclear receptors are markers for neurogenesis from cnidarians to vertebrates. Dev Biol. 2004;275:104–123. doi: 10.1016/j.ydbio.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Küchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signaling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshizaki DK, Blackburn T, Price C, Ghosh M, Miles K, Ragucci M, Sweis R. Embryonic fat-cell lineage in Drosophila melanogaster. Development. 1994;120:2489–2499. doi: 10.1242/dev.120.9.2489. [DOI] [PubMed] [Google Scholar]
- Kerber B, Fellert S, Hoch M. Seven-up, the Drosophila homolog of the COUP-TF orphan receptors, controls cell proliferation in the insect kidney. Genes Dev. 1998;12:1781–1786. doi: 10.1101/gad.12.12.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kinkel MD, Sefton EM, Kikuchi Y, Mizoguchi T, Ward AB, Prince VE. Cyp26 enzymes function in endoderm to regulate pancreatic field size. Proc Natl Acad Sci USA. 2009;106:7864–7869. doi: 10.1073/pnas.0813108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, Xu HE. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Qin J, Tang K, Tsai SY, Tsai MJ. Coup d’etat: an orphan takes control. Endocr Rev. 2011;32:404–421. doi: 10.1210/er.2010-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E, Donerly S, Mackey L, Dobbs-McAuliffe B. The negative side of retinoic acid receptors. Neurotoxicol Teratol. 2011;33:631–640. doi: 10.1016/j.ntt.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Katahira T, Matsunaga E, Sato T. Isthmus organizer for midbrain and hindbrain development. Brain Res Brain Res Rev. 2005;49:120–126. doi: 10.1016/j.brainresrev.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1109. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI, Tsai SY, Tsai MJ. Molecular mechanism of chicken ovalbumin upstream promoter-transcription factor (COUP-TF) actions. Keio J Med. 2003;52:174–181. doi: 10.2302/kjm.52.174. [DOI] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FA, Tsai MJ, Tsai SY. COUP-TF orphan nuclear receptors in development and differentiation. Cell Mol Life Sci. 2000;57:1388–1398. doi: 10.1007/PL00000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Krishnan V, Pereira FA, Tsai SY, Tsai MJ. Chicken ovalbumin upstream promoter-transcription factors and their regulation. J Steroid Biochem Mol Biol. 1996;56:81–85. doi: 10.1016/0960-0760(95)00225-1. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Pereira FA, DeMayo FL, Lydon JP, Tsai SY, Tsai MJ. Null mutation of mCOUP-TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 1997;11:1925–1937. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Böhli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Russo JE, Hauguitz D, Hilton J. Inhibition of mouse cytosolic aldehyde dehydrogenase by 4-(diethylamino)benzaldehyde. Biochem Pharmacol. 1988;37:1639–1642. doi: 10.1016/0006-2952(88)90030-5. [DOI] [PubMed] [Google Scholar]
- Sagami I, Tsai SY, Wang H, Tsai MJ, O’Malley BW. Identification of two factors required for transcription of the ovalbumin gene. Mol Cell Biol. 1986;6:4259–4267. doi: 10.1128/mcb.6.12.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF. Anterior-posterior patterning and segmentation of the vertebrate head. Integr Comp Biol. 2008;48:658–667. doi: 10.1093/icb/icn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh TJ, Kimelman D. COUP-TFI is a potential regulator of retinoic acid-modulated development in Xenopus embryos. Mech Dev. 1995;51:39–49. doi: 10.1016/0925-4773(94)00346-o. [DOI] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Stafford D, White RJ, Kinkel MD, Linville A, Schilling TF, Prince VE. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 2006;133:949–956. doi: 10.1242/dev.02263. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Expression from: unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. ZFIN Direct Data Submission. 2008 doi: 10.1371/journal.pgen.0030188. http://zfin.org. [DOI] [PMC free article] [PubMed]
- Tsai SY, Tsai MJ. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- van der Wees J, Matharu PJ, de Roos K, Destrée OH, Godsave SF, Durston AJ, Sweeney GE. Developmental expression and differential regulation by retinoic acid of Xenopus COUP-TF-A and COUP-TF-B. Mech Dev. 1996;54:173–184. doi: 10.1016/0925-4773(95)00471-8. [DOI] [PubMed] [Google Scholar]
- Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by fgf3 and fgf8. Curr Biol. 2002;12:1117–1123. doi: 10.1016/s0960-9822(02)00899-0. [DOI] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NPC, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007a;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007b;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiellette EL, Sive H. Early requirement for fgf8 function during hindbrain pattern formation in zebrafish. Dev Dyn. 2004;229:393–399. doi: 10.1002/dvdy.10464. [DOI] [PubMed] [Google Scholar]
- Xu F, Li K, Tian M, Hu P, Song W, Chen J, Gao X, Zhao Q. N-CoR is required for patterning the anterior-posterior axis of zebrafish hindbrain by actively repressing retinoid signaling. Mech Dev. 2009;126:771–780. doi: 10.1016/j.mod.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Zhou HM, Walthall WW. UNC-55, an orphan nuclear hormone receptor, orchestrates synaptic specificity among two classes of motor neurons in Caenorhabditis elegans. J Neurosci. 1998;18:10438–10444. doi: 10.1523/JNEUROSCI.18-24-10438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]