Abstract
New copolyesters derived from poly(β,L-malic acid) have been designed to serve as nanoconjugate platforms in drug delivery. 25% and 50% methylated derivatives (coPMLA-Me25H75 and coPMLA-Me50H50) with absolute molecular weights of 32 600 Da and 33 100 Da, hydrodynamic diameters of 3.0 nm and 5.2 nm and zeta potential of −15mV and −8.25mV, respectively, were found to destabilize membranes of liposomes at pH 5.0 and pH 7.5 at concentrations above 0.05mg/mL. The copolymers were soluble in PBS (half life of 40 hours) and in human plasma (half life of 15 hours) but they showed tendency to aggregate at high levels of methylation. Fluorescence-labeled copolymers were internalized into MDA-MB-231 breast cancer cells with increased efficiency for the higher methylated copolymer. Viability of cultured brain and breast cancer cell lines indicated moderate toxicity that increased with methylation. The conclusion of the present work is that partially methylated poly(β,L-malic acid) copolyesters are suitable as nanoconjugate platforms for drug delivery.
1. Introduction
Biodegradable polymers are suitable materials for the manufacture of various devices which, because of their biocompatibility, are widely applied in medicine and pharmacology. Examples are poly(lactic acid) and (PLA), poly(glycolic acid) and their copolymers, with a successful 30 years history in surgical settings [1].
The relatively new biopolymer, poly (β,L-malic acid) (PMLA), is a carboxylic-functionalized polyester that can be produced by either chemical synthesis or biological fermentation from the slime mold Physarum polycephalum. Both α- and β-structures, either racemic or optically pure, may be obtained by chemical methods whereas microorganisms exclusively generate PMLA of extremely high optical purity [2, and references therein]. The attractive properties in nanobiotechnology and biomedicine are the lack of in vitro and in vivo toxicity and non-immunogenicity. PMLA is a completely biodegradable polymer that is metabolized to water and carbon dioxide in the citric acid cycle. It is biocompatible with regard to its limited stability in the bloodstream thus prohibiting physiologically adverse host responses after injection. It is chemically convenient to handle due to its solubility in water and certain organic solvents and to the reactivity of its pendant carboxylic groups [2-9].
So far, the most advanced application reported for PMLA, is the development of “Polycefin”, a nanoconjugate prototype vehicle for the cellular delivery of antisense oligonucleotides. It consists of PMLA that harbors antisense morpholino oligonucleotides and several biochemically functional units which are chemically conjugated with the carboxyl groups. These functional units are tumor-specific antibodies that promote receptor-mediated endosomal uptake by the tumor cells, a unit that allows the vehicle to escape from the endosomal vesicles into the cytoplasm and another unit that, after cleavage, releases free oligonucleotides. This nanoconjugate can carry several antisense oligonucleotides or other drugs at the same time and also simultaneously several antibodies, which can bind specifically to cell surface antigens during drug delivery [5-9].
Both synthetic and biosynthetic polymalic acids are spontaneously or enzymatically degraded in aqueous environment (see review [2]). Regardless chirality, degradation is moderate at physiological pH and proceeds rapidly in acidic (pH < 5) and basic (pH > 9) solutions [2] by random scission of the main chain ester bonds to yield malic acid as the final degradation product [10]. By chemical blocking of the pendant carboxylic groups, the properties of PMLA could be changed to slow down its hydrolysis rate. Methylation with diazomethane has proven to be an efficient method that produced poly(α-methyl β,L-malic acid) (PMLA-Me) without significant cleavage of the polyester backbone [11, 12]. While low degrees of methylation resulted in water soluble products, 75% methylation of pendant carboxyl groups allowed formulation of stable, water insoluble nanoparticles that could be loaded with proteins for delivery applications [13].
The soluble PMLA methyl esters containing 25% and 50% of methylated carboxylic side groups (coPMLA-Me25H75 and coPMLA-Me50H50) are characterized here by investigating their light scattering properties, their degradation in human plasma and their cytotoxicity for several human cancer cell lines to be used as nanoplatforms for drug delivery.
2. Materials and Methods
2.1. PMLA Production
PMLA of microbial origin was used in this work. It was obtained by cultivation of Physarum polycephalum and subsequent purification as described in detail elsewhere [14]. The polyacid of NMR purity had a Mw = 34 300 Da and a polydispersity Mw/Mn = 1.1. All chemicals of highest purity, including human plasma, were bought from Merck (Germany) and Sigma-Aldrich (Germany). Organic solvents were of analytical grade and used without further purification. Water used for buffer preparation was double distilled and deionized in a “Milli-Q” system.
2.2. Esterification
Partial esterification of PMLA was performed as described recently by Portilla-Arias et al. [11]. In brief, a solution of diazomethane in ether (12.5meq) was added to a solution of PMLA in dry acetone (4.3 meq with regard to malic acid units) in different ratios according to the esterification degree to be obtained, and the mixture was left under stirring at room temperature for 1 hour. The reaction mixture was then evaporated under vacuum and the residue was dissolved in a small amount of N-methyl-2-pyrrolidone and precipitated with cold diethyl ether. The copolymer was recovered by filtration as a white powder. Yields for coPMLA-Me25H75 and coPMLA-Me50H50 were 97% and 92%, respectively. The 1H NMR analysis in deuterated water revealed that the conversion actually attained in the copolyesters was 20.2, 46.5, which are pretty close to the nominal values.
2.3. Absolute Molecular Weight, Hydrodynamic Diameter and Zeta Potential Measurements
The copolymers were characterized with respect to their absolute molecular weight (Mw), size and ζ potential using a Malvern Zetasizer Nano (Malvern Instruments, UK). Absolute weight averagemolecular weight was calculated with a modification of the Rayleigh equation that can be used to generate a Debye plot, which is a linear fit of KC/Rθ versus concentration according to the equation Kc/ Rθ = 1/Mw + 2A2cRθ is the Rayleigh ratio of scattered to incident light intensity, K is a constant defined by the solvent and analyte dependent refractive index increment (dn/dc = 0.169 mL/g for PMLA), Avogadro’s number and the solvent refractive index. c is the particle concentration and A2 is the second virial coefficient [15]. The intercept obtained from the Debye plot is equal to the inverse of the molecular weight and the slope is twice the second virial coefficient.
The size was calculated on the basis of noninvasive back-scattering (NIBS) measurements using the Stokes-Einstein equation, d(H) = kT/3πηD. d(H) is the hydrodynamic diameter, D the translational diffusion coefficient, k Boltzmann’s constant, T absolute temperature and η the viscosity. The diameter that ismeasured in DLS (Dynamic Light Scattering) refers to the particle diffusion within a fluid and is referred to as the hydrodynamic diameter corresponding to the diameter of a sphere that has the same translational diffusion coefficient as the particle [16]. The ζ potential was calculated from the electrophoretic mobility based on the Helmholtz-Smoluchowski formula, using electrophoresis M3-PALS. All calculations were carried out by the Zetasizer 6.0 software. For the molecular weight determination, 5 solutions of the copolymers in phosphate buffered saline (PBS, pH 7.4) were generated by serial dilution starting with 4mg/mL. For the measurement of the ζ potential, the concentration of the sample was 2 mg/mL dissolved in water containing 10mM NaCl, and the voltage applied was 150 V. For the particle size measurements, the solutions were prepared in PBS at a concentration of 2mg/mL, filtered through a 0.2 μm pore membrane. All the copolymer solutions were prepared immediately before analysis at 25°C. Data represent the mean ± standard deviation obtained for three measurements.
2.4. Copolyesters Stability in PBS and Human Plasma
The degradation essays in human plasma were carried out at 37°C with a polymer concentration of 1mg/mL. The sample vials were sealed to avoid evaporation and stored at 37°C in an incubator. For the isolation from the plasma, aliquots of 1 mL were extracted with 5 mL of chloroform/ethyl acetate (1 : 1 v/v). The copolyester contained in the organic phase was dried and redissolved in PBS and the Mw measured by sec-HPLC (Calibrated with polystyrene sulphonate-sodium salt standards). Sample preparation with the polymers of known Mw verified that the isolation had no effect on molecular weights. For comparison, the degradation study was performed in PBS (pH 7.4) at a concentration of 1mg/mL for each copolymer. Chromatography was performed on a Hitachi analytical Elite LaChrom HPLC-UV system and size exclusion column BioSep-SEC-S 3000 column (300 × 7.80mm) following the elution at 220nm wavelength. Molecular weights Mw(t) were plotted as a function of degradation time with reference to Mw(t = 0) at zero incubation time.
2.5. Cell Lines and Culture Conditions
Primary glioma cell lines—U-87 MG and T98G—and invasive breast carcinoma cell lines—MDA-MB-231 and MDA-MB-468—were obtained from American Type Culture Collection (ATCC) USA. U-87 MG and T98G cells were cultured in MEM supplemented with the following ingredients (final concentrations): 10% fetal bovine serum, 1% MEM NEAA, 1mM sodium pyruvate and 2mM L-glutamine. For MDA-MB-231 and MDA-MB-468, Leibovitz’s L-15 medium with 10% final concentration fetal bovine serumwas used. Cells were seeded at 10,000 cells per well (0.1 mL) in 96-well flat-bottomed plates and incubated overnight at 37°C in humid atmosphere with 5% CO2 (breast cancer cell lines MDA-MB-231 and MDA-MB-468 were incubated without CO2).
2.6. Cytotoxicity Test
The copolymers (1 mg/mL and serial dilutions) were dissolved in culture media and incubated with cells for 24 hours. Cell viability was measured using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega Corporation, Cat. No.PR-G3580). Yellow [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (MTS) is bioreduced by cells into formazan that is soluble in the tissue culture medium. The absorbance reading at 490 nm from the 96-well plates was directly proportional to the number of living cells [17]. The viability of the untreated cells was referenced as 100%. The results shown are the means and deviations standard of three independent measurements, calculated with the statistical software GraphPad PRISM 3.0.
2.7. Liposome Leakage Assay
The capability of the copolymers to escape from endosome was measured by the liposome leakage assay. This method generally assumed to represent main features of the endosome membrane and gives similar results as the red blood cell lysis method, because it is less biased by adverse effects of proteins contained in erythrocytes/membranes. Liposome suspensions were prepared by the extrusion method. Briefly, the mixture of egg phosphatidylcholine and cholesterol (molar ratio 2 : 1) dissolved in CHCl3/MeOH (v/v, 2 : 1) was dried under a stream of nitrogen. The lipid mixture was hydrated with HBS buffer (5mM HEPES, 150mM NaCl, pH 7.4) containing 90mM calcein, followed by 19 extrusions through 0.1 μm polycarbonate membrane using mini-extruder (Avanti Polar Lipids). Serial dilutions were carried out using 95 μL of two buffers of different pH, 137 mM HEPES buffer pH 7.4 and 137 mM citrate buffer pH 5.0. Liposome 5 μL (lipid concentration 200 μM) was added to each sample and the plate was incubated at room temperature for 1 hour. Complete leakage of calcein (100% reference) was achieved with the addition of 0.25% (v/v) Triton-X100 solution of respective buffers. Fluorescence of released calcein release was measured using excitation wavelength 488 nm and an emission wavelength 535 nm. The results shown are the means and deviations standard of three independent measurements, calculated with the statistical software GraphPad PRISM 3.0.
2.8. Method of Fluorescence Labeling
The copolymers were fluorescence labeled with rhodamine as follows: Pendant carboxyl groups of copolymer (1 mmol malyl residues) in 1 mL of dimethyl formamide (DMF) were activated with a mixture of N-hydroxysuccinimide (NHS, 1 mmol) and dicyclohexylcarbodiimide (DCC, 1 mmol) dissolved in 2 ml of DMF at room temperature for 3-4 hours under vigorous stirring. 2-Mercapto-1-ethylamine (0.1 mmol) and 0.1 mmol of dithiothreitol (DTT). were added and the reaction was completed in 30–40 minutes. After 30 minutes of stirring in 6 mL of water at room temperature, the mixture was centrifuged and the clear supernatant passed over Sephadex G10 columns in water. The product containing fractions were freeze dryed yielding a white powder. Rhodamine Red C2 maleimide (40 μL of 1 mg/mL solution in DMF) was added to 2 mg of activated copolymer dissolved in 2 mL of PBS of pH 5.5 and stirred for 3 hours at room temperature. The fluorescent polymers were purified over Sephadex PD-10 columns pre-equilibrated with PBS (pH 7.4).
2.9. Fluorescence Microscopy
MDA-MB-231 cells were seeded into the microscopic chamber slide. For fluorescence microscopy study, cells were incubated with rhodamine-labeled-polymer (1 mg/mL) in fetal bovine serum (FBS) free medium for 3 hours. The stained cells were washed with PBS and fixed in 4% paraformaldehyde (PFA) for 15 minutes. Then the cells were counterstaining with DAPI to visualize the nuclei and observed under a DM6000 Leica fluorescence microscope (Wetzlar, Germany). The amount of polymer uptake was calculated from the rhodamine fluorescence intensity, with subtracted background and divided by the area of the cell under study using the Image J1.43c software from NIH (values averaged over >50 cells). The experiment was done by triplicate, with three independent preparations of cells. Mean and deviation standard were calculated with the statistical software GraphPad PRISM 3.0.
3. Results and Discussion
3.1. Chemical Characterization
Partially methylated poly(β-l-malic acid) copolymers were synthesized in high yield and purity by the diazomethane in acetone method. The two products are denoted as coPMLA-Me25H75 and coPMLA-Me50H50, their chemical formulae are depicted in Figure 1 and their properties are shown in Table 1. The degree of methylation attained in the two cases (20.2 and 46.5%) in the reaction was close to the input ratio of diazomethane to total carboxylic acid (25 and 50%). Molecular weights of Mw (weight-average) = 32,600 Da and 33,100 Da with polydispersity indexes (Mw/Mn) of 1.3 and 1.1, respectively, were determined by sec-HPLC for the two copolyesters which are values comparable to those measured for the original polymalic acid (Mw = 34 300, Mw/Mn = 1.1). The slightly decrease in Mw and increase in polydispersity observed after methylation suggest partial cleavage during the chemical synthesis. The 13C-NMR analysis revealed that the distribution of methyl groups along the copolyesters chain is not random. The average number of contiguous methylated carboxylic groups indicated hydrophobic patches as opposed to regions with contiguous free carboxyl groups and interdispersed free/methylated carboxyl groups. As expected, the length of hydrophobic sequences was larger for the polymer with the higher degree of methylation.
Figure 1.
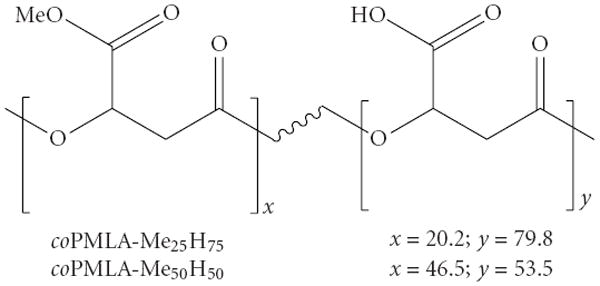
Chemical structure of copolyesters studied in the present work with indication of their contents in methylated and free-carboxyl malic units.
Table 1.
Physical properties of copolymers.
| coPMLA-Me25H75 | coPMLA-Me50H50 | |
|---|---|---|
| Methylation(a) (%) | 20.2 | 46.5 |
| Mw (b) | 32,600 | 33,100 |
| Mn (b) | 26,100 | 24,300 |
| Tm (c) (°C) | 174 | 172 |
| Td (d) (°C) | 186 | 198 |
| Microstructure(e) | ||
| Contiguous patches nf Me | 3.9 | 11.8 |
| Contiguous patches ng COOH | 11.5 | 14.5 |
| R | 0.3 | 0.2 |
Copolymer composition determined by 1H NMR;
Weight- and number-average molecular weight measured by sec-HPLC;
Melting temperature measured by differential scanning calorimetry;
Onset decomposition temperature measured at 5% of loss of initial weight;
nf and ng refer to averaged numbers of methylated and free carboxyl group within homogeneous sequences, respectively, and (R) refers the randomness determined by 13C NMR analysis [11].
3.2. Absolute Molecular Weight, Hydrodynamic Diameter and Zeta Potential
The values for absolute weight averaged molecular weight, particle size and zeta potential of the resulting copolyesters together with PMLA are shown in Table 2. The values of molecular weight and polydispersity found by DLS are similar to those obtained by GPC (see Table 1) and corroborate the notion that some cleavage occurred during synthesis. While the hydrodynamic diameter does not follow a clear trend upon methylation, the second virial coefficient A2 and the ζ-potential correlate well with the degree of methylation. The second virial coefficient is a parameter describing the interaction strength between the molecule and the solvent [16]. The relatively high values obtained for PMLA and coPMLA-Me25H75 indicate the ability of these polymers to stay in solution, whereas the much lower value obtained for coPMLA-Me50H50 indicates that this copolymer has some tendency for aggregation. The conclusion is corroborated by the progressive decrease of negative ζ-potential observed for increasing methylation, that is, less repulsion and thus higher tendency for aggregation.
Table 2.
Light scattering and zeta potential measurements.
| PMLA | coPMLA-Me25H75 | coPMLA-Me50H50 | |
|---|---|---|---|
| Molecular weight (Da) | 34,200 | 30,100 | 31,900 |
| Polydispersity index | 1.1 | 1.22 | 1.41 |
| 2nd virial coeffient A2 (mL · mol/g2) | 6.50E-05 | 2.85E-05 | 3.50E-08 |
| Hydrodynamic diameter (nm) | 3.4 (±0.1) | 3.0 (±0.1) | 5.2 (±0.1) |
| Zeta potential (mV) | −22.9 (±1.7) | −15 (±1.1) | −8.25 (±1.3) |
The effects of esterification on solubility are both an increase in hydrophobicity and a decrease in electrostatic mutual repulsion between polymers at higher degrees of methylation. The blocky microstructure of the copolymer harboring highly hydrophobic domains of contiguously methylated units could contribute to aggregation by favoring intermolecular hydrophobic contacts. The polyacid is highly soluble in water and acetone, coPMLA-Me50H50 is less soluble in water than coPMLA-Me25H75, and coPMLA-Me75H25 (not studied here) has been reported to be water insoluble [11].
3.3. Copolyesters Degradation in PBS and Human Plasma
Copolyester degradation was followed by measuring molecular weights Mw by sec-HPLC as a function of incubation time (Figure 2). During incubation in PBS (pH 7.4) at 37°C the polymers were slowly degraded as indicated by an increase in the retention time [11]. The measurements for degradation in serum at 37°C were complicated by the presence of proteins which co-eluted and interfered with the polymers in sec-HPLC, rendering polymer detection impossible. The degraded polyesters except PMLA could be separated from proteins by extraction of the plasma mixture with chloroform/ethyl acetate. As an example the chromatograms of coPMLA-Me50H50 incubated in serum at time zero (a) and after incubation (b) are compared in Figure 3. This case once more demonstrates the degradability of PMLA and its derivatives in physiological conditions. Specific PMLA hydrolases have been reported for microorganisms [18, 19]. As in the case of degradation in PBS (not shown), the elution profile indicated a single peak. This suggested cleavage from the ends and not fragmentation at internal cleavage sites.
Figure 2.
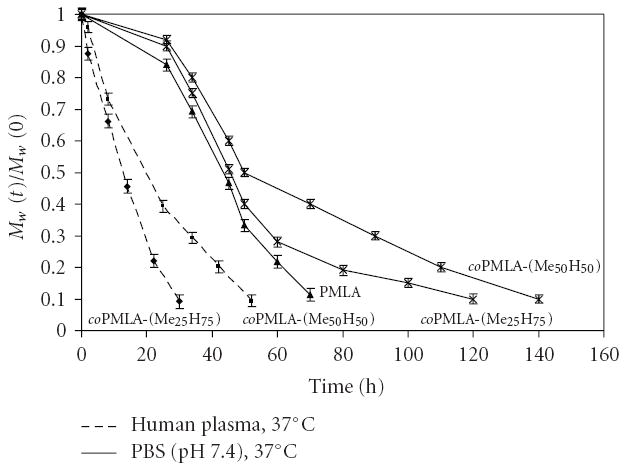
Degradation at 37°C in PBS (pH 7.4) and human plasma for PMLA and the copolyesters.
Figure 3.
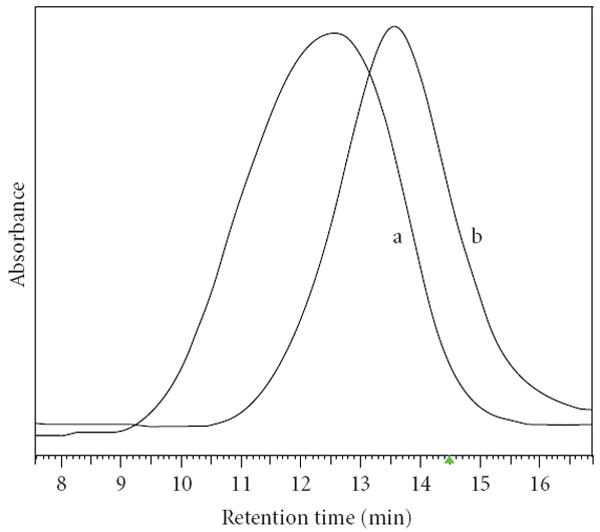
Elution profiles obtained by sec-HPLC for coPMLA-Me50H50 at time zero (a) and after 24 hours of incubation (b) in human plasma at 37°C. Samples were extracted with chloroform/ethylacetate before chromatography (see Section 2).
Comparison of kinetics in Figure 2 revealed that (i) degradation in human plasma was faster than in PBS with half lives of 13 h in plasma for coPMLA-Me25H75 and 20 hours for coPMLA-Me50H50 compared with 45 hours and 50 hours in PBS, respectively and (ii) Half life was lowest for PMLA and increased with higher levels of methylation in agreement with the notion that alkylation of the α-carboxyl group stabilized the main chain ester bond. Hydrolysis was significantly enhanced by constituents of human plasma, either by general catalysis or by hydrolytic enzymes, most likely esterases, such as plasma lipases or cholinesterases [20].
3.4. Membrane Disruption
The membrane disruption activity of coPMLA-Me25H75 and coPMLA-Me50H50 was measured by the phosphatidylcholine liposome leakage assay. The liposomes are filled with calcein, which leaks out if the liposome becomes destabilized or disrupted. Leakage was measured at pH 7.5, resembling physiological pH, and at pH 5.0, resembling pH of late endosomes/lysosomes. While coPMLA-Me25H75 did not show leakage activity, coPMLA-Me50H50 was active at concentrations above 0.1 mg/mL and the activity was pH independent (Figure 4). The finding suggested that membrane leakage was inferred by methylation of pendant carboxyl groups that increased hydrophobicity and neutralized negative charges (decrease in ζ-potential, Table 2). The absence of effect of pH change indicated that the carboxylic groups of the methylated PMLA did not protonate in this pH range, or if they did, they had no effect on the leakage activity.
Figure 4.
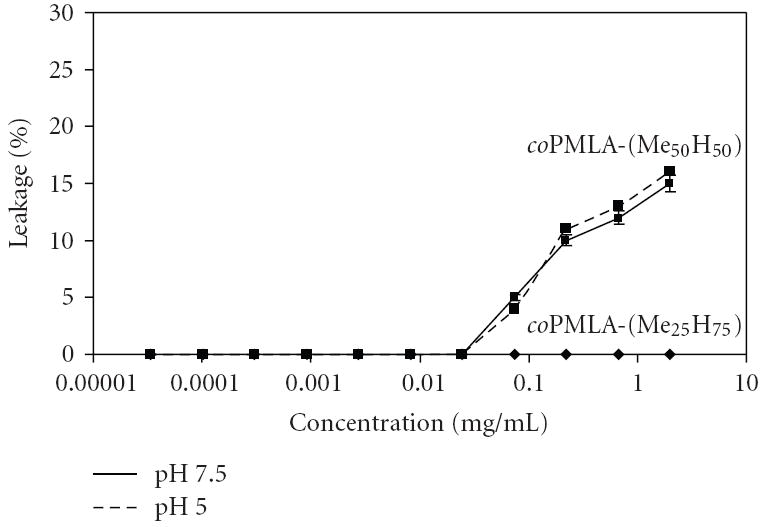
Liposome leakage at pH 5.0 and 7.5 and 37°C. 100% leakage is obtained after the addition of 0.25% Triton X100.
The role of hydrophobicity together with charge neutralization has been considered in drug delivery to be the mechanism for membrane disruption by a variety of molecular devices [21-25]. In the case of our methylated PMLA we think that the methylation dependent leakage refers mainly to the formation of the contiguous, electrically neutral hydrophobic patches (Table 2) which are prone to intrude into the lipid bilayer and cause the membrane damage. The membrane disruption activity especially coPMLA-Me50H50 is thought to be useful in the design of nanoconjugates that can deliver drugs to intracellular targets.
3.5. Cytotoxicity
Partially methylated PMLA contains carboxylic groups that can be conjugated to several prodrugs and in addition to a variety of biologically active units such as antibodies for targeting or PEG for protection against enzymatic degradation and resorption by the reticuloendothelial system (RES). The pro-drug is activated within the targeted cell compartment and only then unfolds its cytotoxic or any other activity.
It is desirable to know whether methylated PMLA as the platform is itself not toxic. To this end, the in vitro toxicity of the copolymers was tested. Figure 5 shows the viability of cultured brain and breast cancer cells: T98G, U-87 MG, MDA-MB-231 and MDA-MB-468 cells after 24 hours of incubation as a function of copolymer concentrations.
Figure 5.
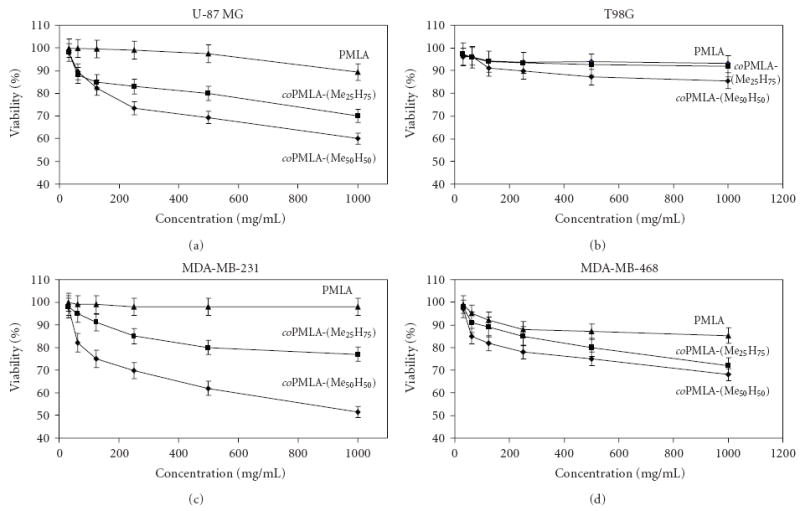
Cell viability of cultured cells after 24 hours incubation at 37°C for different cell lines in the presence of the indicated polyesters.
While PMLA only marginally decreased cell viability, the copolymers showed toxicity that increased with the degree of methylation, but the decrease in percentage cell viability was moderate and above 50% at concentrations ≤1 mg/mL. Effects on cell viability depended also on the type of cell line, glioma U-87 MG cells, and breast cancer MDA-MB-231 cells being more affected than glioma T98G cells and breast cancer MDA-MB-468 cells.
There are two main toxicity mechanisms commonly considered: (i) toxicity due to physical damage such as destabilization of membranes and (ii) toxicity resulting from the degradation products after intracellular uptake. The possibility of physical damage due to membrane disrupture of the kind as seen by the liposome leakage assay (Figure 5) may not be significant since the effect of copolymers on cell viability is not manifested in the time scale of minutes or a few hours (results not shown). It is highly likely that toxicity was the result of methanol formation during degradation. It is known that degradation in pure deuterated water generates as main products methanol and L-malic acid [11]. While l-malic acid is converted into water and carbon dioxide in the tricarboxylic acid cycle, methanol is known to be toxic for living cells. In the same line it has been reported that benzylesters of PMLA were toxic due to release of free benzylalcohol during degradation [26]. However, for consideration of the copolymer application in drug delivery, the short residence times of only a few hours before complete clearance through the renal system relatives their toxicity in vivo.
3.6. Cellular Uptake Study
Since we have found that the copolymers coPMLA-Me25H75 and coPMLA-Me50H50 contained hydrophobic patches that could be active in membrane disruption, it was important to test whether the polymers could enter cells in vitro. Rhodamine labeled copolymers were incubated with MDA-MB-231 cells for 3 hours. The cell’s uptake of the fluorescent polymers was seen under the fluorescence microscope (Figure 6). Rhodamine alone did not stain the cells (picture not shown). The distribution of copolymers-rhodamine in all cells, appeared to be homogeneous and probably involved nuclei. The fluorescence intensity was semi quantitatively measured by triplicate in >50 cells, and appeared to be 1.8 (Standard Deviation= ±0.3) folds more intensive for coPMLA-Me50H50 than for coPMLA-Me25H75 accordingly to the Image J1.43c software, thus correlating with the higher degree of hydrophobicity of the higher methylated polymer. The results are evidences that the copolymers are highly versatile materials and suitable for various applications.
Figure 6.
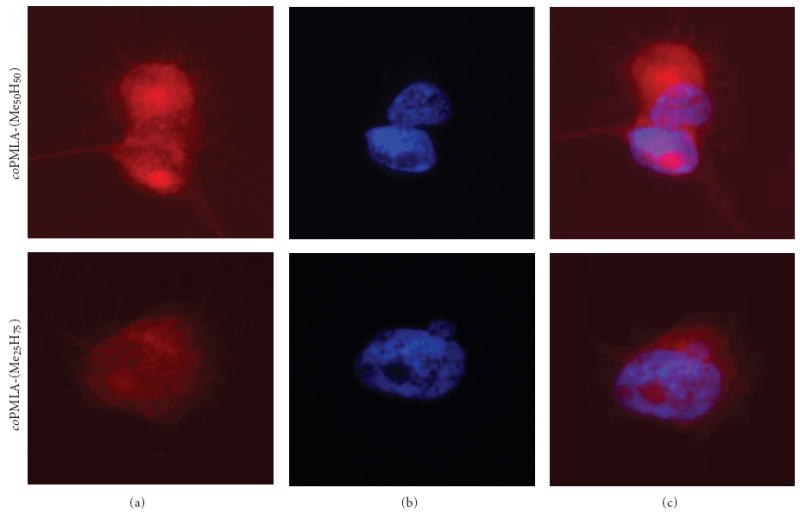
Localization of rhodamine tagged copolymers in MDA-MB-231 cells. Panels (a) and (b) separated copolymer-rhodamine uptake and DAPI staining. Panel (c) The copolymers are indicated by red fluorescence co-localizing with nuclei stained in blue with DAPI.
4. Conclusion
New designs of nanoconjugate drug delivery systems are introduced here. The systems involve a polymer platform containing pendant chemically reactive groups to be conjugated with molecular units that function in pro-drug attachment, cell recognition, membrane penetration and protection. The copolymers coPMLA-Me25H75 and coPMLA-Me50H50 are biodegradable and biocompatible and its half-life is limited, so they may minimize adverse host’s responses and development of liver storage diseases. The copolymers are endowed with membrane disrupting/penetrating activities which allow them to deliver drugs directly to intracellular targets by passing the plasma membrane. By raising the degree of methylation above 50%, the copolymers become insoluble and can be used as drug delivering nanoparticles. These results support the fact that poly (β,L-malic acid) is a highly versatile material and can be used for the design of a variety of nanoconjugate platforms for drug delivery systems.
Acknowledgments
The project was funded by grants from NIH/NCI R01 CA123495 to JYL; MAT2006-13209-C02-02 from CICYT (Comisión Interministerial de Ciencia y Tecnología) of Spain to SM and CONACYT-México (Concejo Nacional de Ciencia y Tecnología) for fellow granted to J. Portilla-Arias.
Footnotes
This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2(12):751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Lee B-S, Vert M, Holler E. Water-soluble aliphatic polyesters: poly(malic acid)s. In: Doi Y, Steinbüchel A, editors. Biopolymers. Wiley-VCH; Weinheim, Germany: 2002. pp. 75–103. [Google Scholar]
- 3.Gasslmaier B, Krell CM, Seebach D, Holler E. Synthetic substrates and inhibitors of β-poly(L-malate)-hydrolase (polymalatase) European Journal of Biochemistry. 2000;267(16):5101–5105. doi: 10.1046/j.1432-1327.2000.01573.x. [DOI] [PubMed] [Google Scholar]
- 4.Gasslmaier B, Holler E. Specificity and direction of depolymerization of β-poly(L-malate) catalysed by polymalatase from Physarum polycephalum—fluorescence labeling at the carboxy-terminus of β-poly(L-malate) European Journal of Biochemistry. 1997;250(2):308–314. doi: 10.1111/j.1432-1033.1997.0308a.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee B-S, Fujita M, Khazenzon NM, et al. Polycefin, a new prototype of a multifunctional nanoconjugate based on poly(β-L-malic acid) for drug delivery. Bioconjugate Chemistry. 2006;17(2):317–326. doi: 10.1021/bc0502457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita M, Khazenzon NM, Ljubimov AV, et al. Inhibition of laminin-8 in vivo using a novel poly(malic acid)-based carrier reduces glioma angiogenesis. Angiogenesis. 2006;9(4):183–191. doi: 10.1007/s10456-006-9046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita M, Lee B-S, Khazenzon NM, et al. Brain tumor tandem targeting using a combination of monoclonal antibodies attached to biopoly(β-L-malic acid) Journal of Controlled Release. 2007;122(3):356–363. doi: 10.1016/j.jconrel.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ljubimova JY, Fujita M, Khazenzon NM, et al. Nanoconjugate based on polymalic acid for tumor targeting. Chemico-Biological Interactions. 2008;171(2):195–203. doi: 10.1016/j.cbi.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ljubimova JY, Fujita M, Ljubimov AV, Torchilin VP, Black KL, Holler E. Poly(malic acid) nanoconjugates containing various antibodies and oligonucleotides for multi-targeting drug delivery. Nanomedicine. 2008;3(2):247–265. doi: 10.2217/17435889.3.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braud C, Vert M. Degradation of poly(β-malic acid)-monitoring of oligomers formation by aqueous SEC and HPCE. Polymer Bulletin. 1992;29(1-2):177–183. [Google Scholar]
- 11.Portilla-Arias JA, García-Álvarez M, de Ilarduya AM, Holler E, Galbis JA, Muñnoz-Guerra S. Synthesis, degradability, and drug releasing properties of methyl esters of fungal poly(β, L-malic acid) Macromolecular Bioscience. 2008;8(6):540–550. doi: 10.1002/mabi.200700248. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez CE, Mancera M, Holler E, Bou JJ, Galbis JA, Muñoz-Guerra S. Low-molecular-weight poly(α-methyl β,L-malate) of microbial origin: synthesis and crystallization. Macromolecular Bioscience. 2005;5(2):172–176. doi: 10.1002/mabi.200400166. [DOI] [PubMed] [Google Scholar]
- 13.Portilla-Arias JA, García-Álvarez M, Galbis JA, Muñoz-Guerra S. Biodegradable nanoparticles of partially methylated fungal poly(β-L-malic acid) as a novel protein delivery carrier. Macromolecular Bioscience. 2008;8(6):551–559. doi: 10.1002/mabi.200700249. [DOI] [PubMed] [Google Scholar]
- 14.Holler E. Handbook of Engineering Polymeric Materials. Vol. 997. Marcel Dekker; New York, NY, USA: 1997. [Google Scholar]
- 15.Hiemenz PC. Polymer Chemistry: The Basic Concepts. Vol. 659. Marcel Decker; New York, NY, USA: 1984. Light scattering by polymer solutions. [Google Scholar]
- 16.(ISO), I. O. f. S. International Standard ISO13321. 1996. Methods for Determination of Particle Size Distribution Part 8: Photon Correlation Spectroscopy. [Google Scholar]
- 17.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65(1-2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 18.Korherr C, Roth M, Holler E. Poly(β-L-malate) hydrolase from plasmodia of Physarum polycephalum. Canadian Journal of Microbiology. 1995;41(supplement 1):192–199. [Google Scholar]
- 19.Rathberger K, Reisner H, Willibald B, Molitoris H-P, Holler E. Comparative synthesis and hydrolytic degradation of poly (L-malate) by myxomycetes and fungi. Mycological Research. 1999;103(5):513–520. [Google Scholar]
- 20.Williams FM. Clinical significance of esterases in man. Clinical Pharmacokinetics. 1985;10(5):392–403. doi: 10.2165/00003088-198510050-00002. [DOI] [PubMed] [Google Scholar]
- 21.Tonge SR, Tighe BJ. Responsive hydrophobically associating polymers: a review of structure and properties. Advanced Drug Delivery Reviews. 2001;53(1):109–122. doi: 10.1016/s0169-409x(01)00223-x. [DOI] [PubMed] [Google Scholar]
- 22.Kusonwiriyawong C, van de Wetering P, Hubbell JA, Merkle HP, Walter E. Evaluation of pH-dependent membrane-disruptive properties of poly(acrylic acid) derived polymers. European Journal of Pharmaceutics and Biopharmaceutics. 2003;56(2):237–246. doi: 10.1016/s0939-6411(03)00093-6. [DOI] [PubMed] [Google Scholar]
- 23.Yessine M-A, Lafleur M, Meier C, Petereit H-U, Leroux J-C. Characterization of the membrane-destabilizing properties of different pH-sensitive methacrylic acid copolymers. Biochimica et Biophysica Acta. 2003;1613(1-2):28–38. doi: 10.1016/s0005-2736(03)00137-8. [DOI] [PubMed] [Google Scholar]
- 24.Yessine M-A, Leroux J-C. Membrane-destabilizing polyanions: interaction with lipid bilayers and endosomal escape of biomacromolecules. Advanced Drug Delivery Reviews. 2004;56(7):999–1021. doi: 10.1016/j.addr.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 25.Chen R, Khormaee S, Eccleston ME, Slater NKH. The role of hydrophobic amino acid grafts in the enhancement of membrane-disruptive activity of pH-responsive pseudo-peptides. Biomaterials. 2009;30(10):1954–1961. doi: 10.1016/j.biomaterials.2008.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez Barbosa ME, Cammas S, Appel M, Ponchel G. Investigation of the degradation mechanisms of poly(malic acid) esters in vitro and their related cytotoxicities on J774 macrophages. Biomacromolecules. 2004;5(1):137–143. doi: 10.1021/bm0300608. [DOI] [PubMed] [Google Scholar]


